Abstract
Observations from different taxa, including plants, protozoa, insects and mammals, indicate that proteins involved in reproduction evolve rapidly. Several models of adaptive evolution have been proposed to explain this phenomenon, such as sexual conflict, sexual selection, self versus non-self recognition and pathogen resistance. Here we discuss the potential role of sexual conflict in the rapid evolution of reproductive genes in two different animal systems, abalone (Haliotis) and Drosophila. In abalone, we reveal how specific interacting sperm–egg proteins were identified and discuss this identification in the light of models for rapid protein evolution and speciation. For Drosophila, we describe the genomic approaches taken to identify male accessory gland proteins and female reproductive tract proteins. Patterns of protein evolution from both abalone and Drosophila support the predicted patterns of rapid protein evolution driven by sexual conflict. We stress however that other selective pressures may contribute to the rapid evolution that is observed. We conclude that the key to distinguishing between sexual conflict and other mechanisms of protein evolution will be an integration of genetic, experimental and theoretical data.
Keywords: speciation, fertilization, adaptive evolution, sexual conflict, sexual selection
1. Introduction
Sequence data from diverse taxonomic groups reveal that reproductive genes are rapidly changing by adaptive evolution (Swanson & Vacquier 2002a,b). For example, comparisons of sequence data between Drosophila species show that male reproductive genes are evolving faster than non-reproductive proteins (Civetta & Singh 1995; Swanson et al. 2001b). Mammalian reproductive proteins involved in sperm–egg interactions are also evolving rapidly by adaptive evolution, as seen in a high per cent of divergence at the amino acid level between human and rodent (Makalowski & Boguski 1998; Swanson et al. 2001c, 2003). Other taxonomic groups that show this pattern are marine invertebrates. In abalone (Haliotis), turban snails (Tegula) and sea urchins (Echinometra), sperm proteins are extremely divergent between species due to adaptive evolution (Lee & Vacquier 1992; Metz et al. 1998a; Hellberg & Vacquier 1999; Hellberg et al. 2000; Yang et al. 2000).
The adaptive mechanism for rapid evolution of reproductive proteins has not been directly identified, but potential mechanisms include sexual selection, sexual conflict, immune defence and self versus non-self recognition (Swanson & Vacquier 2002b). Given that many reproductive proteins are involved in interactions (either directly or indirectly) with the opposite sex, insight into the evolutionary mechanisms can be gained by studying DNA sequence evolution of both male and female reproductive proteins. Here we will discuss the potential role of sexual conflict in the rapid evolution of reproductive genes in two systems, abalone and Drosophila, and discuss new data on female reproductive proteins in Drosophila (Swanson et al. 2004).
Sexual conflict is involved in any aspect of reproduction where evolutionary interests between males and females diverge (Parker 1979), such as the rate of mating and fertilization, the number of offspring produced from a given mating and the amount of parental investment in offspring (Haygood 2004). One way this occurs is when traits that evolve to benefit male fitness have a pleiotropic effect of causing female harm (Linder & Rice 2005). Females in turn will evolve traits to resist male harm (Rice 1998; Linder & Rice 2005). A potential evolutionary outcome of sexual conflict is a continual coevolutionary chase between the sexes where adaptations in one sex lead to counter-adaptations in the other sex, followed by counter–counter adaptations (Trivers 1972; Dawkins 1976; Parker 1979; Gowaty 1997; Rice & Holland 1997; Rice 1998; Arnqvist & Rowe et al. 2003; Chapman et al. 2003; Linder & Rice 2005). However, additional regimes distinct from continual coevolution are also predicted from models and simulations of sexual conflict (Gavrilets 2000a; Gavrilets & Waxman 2002). A prediction of sexually antagonistic coevolution is that it will facilitate rapid evolutionary change in the underlying male–female genes involved in the conflict, such as genes for sperm–egg proteins, reproductive tract proteins, mating behaviours and morphological traits (Rice 1998). This rapid change could generate divergence between isolated populations and possibly subsequent speciation (Parker & Partridge 1998; Rice 1998; Gavrilets 2000b; Gavrilets & Waxman 2002; Rowe et al. 2003).
In the two systems presented here, abalone and Drosophila, there has been considerable focus on the role of sexual conflict in the evolution of their reproductive proteins (Swanson & Vacquier 2002a). In Drosophila, the main male reproductive proteins that have been studied are accessory gland proteins (Acps). Acps are predicted to mediate sexual conflict (Chapman et al. 1995, 2003). When Acps are ejaculated into females upon mating they have significant effects on female behaviour and physiology, including oogenesis, ovulation, remating rate and lifespan (Wolfner 1997). Several of these Acp effects are detrimental to female fitness, but beneficial to male fertilization success, potentially resulting in a sexually antagonistic coevolutionary race (Linder & Rice 2005). Consistent with this arms race are DNA analyses that reveal a twofold increase in Acp divergence relative to non-reproductive proteins (Swanson et al. 2001b), and the observation that several Acps studied in detail are rapidly evolving by positive selection (Tsaur & Wu 1997; Tsaur et al. 1998; Aguade 1999; Begun et al. 2000). What has been missing, until recently, is an analysis of the evolution of female reproductive proteins. To fully understand a process that facilitates coevolution between interacting proteins, we need information on both male and female protein evolution (Chapman et al. 2003). Here we will discuss exciting new research on female reproductive proteins (Swanson et al. 2004) and comment on what it can tell us about the role of sexual conflict in reproductive protein evolution.
In abalone (Haliotis), the potential for sexual conflict to promote rapid evolutionary change in reproductive proteins also exists. This gastropod mollusc is a free-spawning external fertilizer. The dynamics of free spawning sets up an environment for polyspermy (multiple sperm fertilizing a single egg) and sperm competition. The opportunity for polyspermy and sperm competition may be responsible for creating a conflict in fertilization rate between the sexes and can affect the evolution of sperm–egg proteins (Frank 2000). For instance, an increased fertilization rate may be beneficial to sperm if it increases the success in sperm competition. In females, however, where polyspermy can cause death to an egg, changes in the egg proteins that lower the fertilization rate may evolve (Rice & Holland 1997; Haygood 2004). This conflict may result in rapid coevolutionary changes in the sperm and egg proteins of abalone species. DNA analysis has revealed rapid divergence of the sperm acrosomal protein, lysin, which is necessary for egg penetration (Lewis et al. 1982; Lee & Vacquier 1992). The female receptor to the male protein has been identified and the evolution in this protein has been able to provide an explanation for the rapid adaptive evolution in lysin, which we discuss in detail below (Swanson & Vacquier 1997, 1998; Galindo et al. 2002, 2003).
2. Coevolution of abalone fertilization proteins
Abalone are large marine molluscs of the genus Haliotis. Seven species co-exist in the Eastern Pacific Ocean, with overlapping breeding seasons and habitats (Vacquier & Lee 1993). For example, it is possible to collect Haliotis rufescens and Haliotis corrugata in the same location and both will have gravid gonads when examined in the laboratory. Since fertilization occurs externally in abalone, there may be an increased potential for hybridization. Investigations into the mechanism that maintains these groups as distinct species may provide clues into the process of speciation.
Although hybrids can be generated in laboratory crosses and are found in the wild at low frequency, the species are distinct in nature and exhibit species-specific fertilization. Sperm from H. rufescens fertilize H. rufescens eggs much more efficiently than sperm from H. corrugata. Species-specific fertilization could result from a variety of steps in the fertilization cascade (Vacquier 1998). First, there is chemotaxis of the sperm to the egg. In abalone, chemotaxis is species-specific with H. rufescens attracted to l-tryptophan released from eggs while H. corrugata sperm are not (Riffell et al. 2002). Once the sperm reach the egg, they encounter the egg vitelline envelope (VE). The VE is a tough, elevated, glycoproteineous envelope that surrounds the egg. Once the sperm contacts the VE, a signal transduction event causes the contents of the acrosome to be released. One of the proteins in the acrosome, lysin, dissolves a hole in the egg by a species-specific non-enzymatic mechanism. The sperm is able to then pass through the VE and fuse with the egg plasma membrane, an event hypothesized to be regulated by the sperm protein sp18 (Swanson & Vacquier 1995). Any or all of these steps could demonstrate species-specificity of sperm–egg interaction. We focus on the dissolution of egg VE by the sperm protein lysin, since it is such a well-characterized event.
Both lysin and egg VEs can be purified in large quantities, which allows for detailed biochemical assays. When lysin is mixed with egg VEs, the VEs are dissolved in a species-specific manner. For example, it takes approximately 7 μg of H. rufescens lysin to get 50% dissolution of a mixture of H. rufescens egg VEs. To dissolve the H. rufescens VEs with lysin from H. corrugata, it takes approximately 21 μg of lysin to achieve the same degree of dissolution (Vacquier & Lee 1993). It is important to note that in the heterospecific mixture of lysin and VE, there is complete dissolution, indicating species-specificity is not an all or none phenomenon. Through detailed biochemical analyses (Kresge et al. 2001), it was determined that lysin dissolves a hole in VE by competing for hydrogen bonds of tightly intertwined fibrous molecules in the VE. The fibrous molecules have the same dimensions as the purified vitelline egg receptor for lysin (VERL), which was identified through affinity chromatography and density gradient sedimentation assays (Swanson & Vacquier 1997). VERL is a giant glycoprotein of 1.5 million Daltons, with 50% of the mass being carbohydrate. Purified VERL and lysin interact with the same species-specificity as lysin mediated VE dissolution. Having now identified interacting sperm–egg proteins, we can study their coevolution to gain insights into the molecular basis for the evolution of species-specific fertilization and potentially reproductive isolation (speciation).
The sperm protein lysin is extraordinarily divergent between closely related species (Lee & Vacquier 1992). A striking example of this rapid divergence is the observation that exons evolve up to 15 times faster than introns (Metz et al. 1998b). In order to determine if the rapid divergence is due to adaptive evolution or relaxed functional constraint, the rate of non-synonymous (amino acid altering; dN) to synonymous (silent; dS) substitutions per site was calculated. The neutral theory predicts that the dN/dS ratio should equal one (Yang & Bielawski 2000). For lysin, it was determined that the dN/dS ratio significantly exceeds one indicating a potential adaptive change in the amino acid sequence. Using methods to identify particular sites subjected to adaptive evolution, the N-terminal and C-terminal were predicted to contain a large number of sites subjected to positive Darwinian selection (Yang et al. 2000). By using site-directed mutagenesis to make chimeric lysins between H. rufescens and H. corrugata, it was demonstrated that these regions regulate species-specificity (Lyon & Vacquier 1999). This demonstrates that comparative analyses of adaptive evolution can provide insight into the functionally important regions of the proteins (figure 1), and further suggests that the proteins' adaptive evolution results in species-specific fertilization. The rapid adaptive evolution of the sperm protein begged the question of whether such extraordinary rates of evolution would be observed in the egg receptor, VERL.
Figure 1.
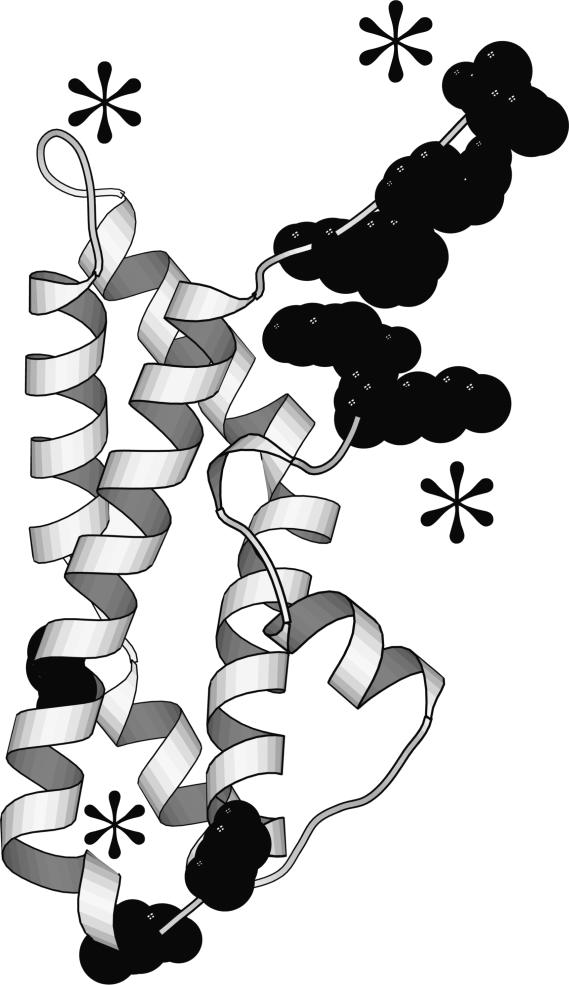
Three-dimensional structure of abalone sperm lysin showing sites predicted to be under adaptive evolution from an analysis of seven species. These sites correspond to regions experimentally demonstrated using site directed mutagenesis to regulate species-specificity (indicated by asterisks).
When cloned, egg VERL was discovered to be an enormous, repetitive molecule containing 22 repeats of approximately 153 amino acids (Galindo et al. 2002). The repetitive nature was consistent with the previously determined stoichiometry of approximately 60 lysin molecules binding each VERL, indicating each VERL repeat may bind two lysin molecules. Phylogenetic analysis showed that repeats 3–22 were homogenized within the genome, with each repeat being more than 95% identical to other repeats 3–22 in the array (Swanson & Vacquier 1998). The homogenization is due to the random process of unequal crossing over and/or gene conversion acting upon repeats in the genome, and is often referred to as concerted evolution (Elder & Turner 1995). Statistical tests of neutrality showed that this region of the repeat array appears to be under slight purifying selection, with a dN/dS ratio not significantly different from one (Swanson & Vacquier 1998). Additional tests utilizing polymorphism data from the C-terminal repeats did not show any departure from equilibrium neutral expectations (Swanson et al. 2001a). The repetitive nature of the array suggested that the repeats might be redundant and subjected to relaxed purifying selection (Swanson & Vacquier 2002a). For example, if a mutation that was suboptimal for lysin–VERL binding arose in one VERL repeat, it may not be selected against since lysin could bind the remaining 21 repeats. By this mechanism, repeats that are suboptimal for lysin–VERL interaction could arise in the population. By chance, through the process of concerted evolution the repeat could be corrected back to wild-type or begin to spread through the repeat array. As the repeat becomes more prevalent, there would be selective pressure on lysin to adapt to the repeat type in order to efficiently compete with other sperm. This hypothesis suggests that lysin is adapting to a neutrally drifting egg receptor. It is reliant upon the idea that there are excess sperm in successful free-spawning events, so that eggs will eventually be fertilized. The adaptive pressure on the sperm is the competition with other sperm to be the first to fertilize the egg, thus the need to interact as efficiently as possible with the egg coat. This model's predictions are similar to predictions of other models of rapid reproductive protein evolution (e.g. sexual conflict and sexual selection), but differ in respect to the lack of adaptive evolution on the female locus.
When the complete VERL gene was sequenced from H. rufescens, it became clear that the two N-terminal repeats were distinct from the main array of repeats 3–22. These repeats do not cluster indicating they are not subjected to homogenization through the process of concerted evolution (Galindo et al. 2002). To examine the evolutionary rates for this region of VERL, repeats one to three were PCR amplified and sequenced from eight abalone species. While the dN/dS ratio averaged across all sites for this region are less than one, analysis for variation in the dN/dS ratio between sites revealed a subset of sites subjected to adaptive evolution with dN/dS ratios of 3.3 (Galindo et al. 2003). This is the first, and to date only, example of interacting sperm and egg recognition molecules where both components have been shown to be subjected to adaptive evolution as predicted by the sexual conflict theory.
The combined biochemical and evolutionary analyses have generated a hypothesis about the origin of species-specific fertilization in abalone, which could lead to reproductive isolation and speciation. There could be an initial interbreeding population that becomes geographically split, although theory also suggests the following process could occur in sympatry (Gavrilets & Waxman 2002). The egg VERL molecule is subjected to two forces: adaptive evolution of repeats one and two and drift due to the repetitive nature of repeats 3–22. In the different populations, the evolution of VERL could lead to different targets to which lysin must adapt. As the sperm and egg molecule co-evolve over time, species-specific fertilization may arise, and when the two populations come in contact there may be sufficient divergence to generate reproductive isolation.
In this scenario, speciation is the by-product of the coevolution of sperm–egg recognition molecules. The selective pressure driving the divergence of abalone reproductive genes remains unknown. One possibility is a sexual conflict hypothesis involving polyspermy. Polyspermy occurs if more than one sperm fuses with the egg. Eggs have evolved elaborate blocks to polyspermy, including rapid reversal of the egg membrane potential and slower blocks involving destroying the receptors for sperm on the egg surface. It is also possible that eggs are selected to slow down the fertilization process to regulate sperm entry in order to allow time to raise blocks to polyspermy. One method to regulate sperm entry may be to change the receptors for sperm on the egg surface, effectively slowing down the process. However, from the sperm perspective, they would be selected to get through the egg investments as quickly as possible in order to be the fertilizing sperm. This may particularly be the case in free-spawning invertebrates where there are sperm from multiple males competing to fertilize eggs.
If the rapid divergence between species of these reproductive molecules drove speciation, it should be possible to capture incipient speciation events by analyses of polymorphism levels within species at these loci. In an initial survey of 11 H. corrugata individuals, a dramatic level of polymorphism at the egg VERL locus was identified (Swanson et al. 2001a). The sequence of the last VERL repeat and portion of the non-repetitive region showed two very distinct clades, with six amino acid differences and an 11 amino acid indel separating the two clades. It was also striking to observe that all the amino acid differences occurred in the VERL repeat region (which is the presumptive functional unit binding lysin) and none occurred in the non-repetitive region. This observation suggests that we may be observing an incipient speciation event. Current work is aimed at increasing the sample size and genotyping sperm lysin and other non-reproductive loci, with the long-term goal of correlating levels of genetic diversity with different regimes predicted by simulations of sexual conflict (Gavrilets 2000a; Gavrilets & Waxman 2002).
The abalone system is a well-characterized system for studying the function and coevolution of reproductive genes. Although the selective pressure driving the divergence of sperm lysin and egg VERL remains a mystery, there are several striking observations that are consistent with mathematical and verbal models of sexual conflict. First, we observe rapid, adaptive evolution of both the male and female reproductive loci (Lee & Vacquier 1992; Galindo et al. 2003). Second, there is variation in the rate of lysin evolution between lineages. This observation is consistent with mathematical models suggesting rapid evolution can occur sporadically, with periods of absent or reduced positive selection (Gavrilets 2000a). Third, the polymorphism data show a split in the female genotype with two distinct alleles. This is highly reminiscent of the Buridan's ass regime suggested by Gavrilets (Gavrilets & Waxman 2002). In this regime of sexual conflict, females diverge into two types and effectively trap males, who are unable to adapt to both types simultaneously. Over time, the males also split resulting in a sympatric speciation event. Initial analyses of sperm lysin in this population only discovered one allele, but not all exons have been sequenced (Swanson et al. 2001a). Overall, there is surprisingly good fit between the experimental observations of adaptive evolution in abalone and mathematical models of sexual conflict.
3. Identification and evolution of Drosophila reproductive proteins
While the abalone is a fantastic system for biochemistry and comparative analyses, it lacks the power of genetics and experimental manipulation needed for robust tests of the sexual conflict theory. Therefore, we have also pursued studies of reproductive genes in Drosophila in order to rigorously test sexual conflict theory. The primary focus has been on the identification of reproductive genes in Drosophila and studies of their evolutionary rates. In particular, the focus is on identifying male (Swanson et al. 2001b) and female reproductive proteins (Swanson et al. 2004), with the long-term goal of isolating interacting partners. We briefly describe the approaches for identification of male Acps (Swanson et al. 2001b) and receptors in the female reproductive tract (Swanson et al. 2004).
The male accessory gland produces a majority of the Drosophila seminal fluid components, which are transferred along with sperm during mating (Wolfner 1997). The seminal fluid contains many Acps that have several effects on the mated female's physiology, sperm storage and sperm competition (Wolfner 1997). For example, the Acp sex peptide is known to reduce the female's propensity to remate (Soller et al. 1997); Acp26Aa is involved in induction of ovulation (Herndon & Wolfner 1995); and Acp36DE has been implicated in sperm storage (Neubaum & Wolfner 1999) and potentially sperm competition (Chapman et al. 2000). Additionally, the receipt of seminal fluid appears to reduce female lifespan (Chapman et al. 1995), potentially due to the transfer of the protease inhibitor Acp62F (Lung et al. 2002). These observations suggest that studies of Acps will be a particularly fertile area of research for studies of sexual conflict.
In order to obtain a genomic view of Acp function and evolution, a study was designed to identify the entire suite of Acp genes through analyses of expressed sequence tags (ESTs) (Swanson et al. 2001b). ESTs are small portions (approx. 200–500 bp) of an entire expressed gene that are useful for identifying unknown genes. Previously, 18 Acps had been identified through differential hybridization studies (DiBenedetto et al. 1987; Wolfner et al. 1997). Acp numbers were estimated to be approximately 100 through differential cDNA hybridization and two-dimensional protein electrophoresis. To identify the entire suite of Acps, a cDNA library was made from dissected accessory glands and subjected to differential hybridization (Swanson et al. 2001b). In this differential hybridization step, clones were hybridized to cDNA from both male and female flies. Clones that appeared to be enriched for male-biased expression were selected for sequencing and further study. An additional differential hybridization step was performed comparing hybridization of males with accessory gland to males with accessory glands ablated by expression of diphtheria toxin (DTA males; Kalb et al. 1993), this allowed a strict test of accessory gland specific expression as those ESTs that hybridize with wild-type males and not DTA males. Clones that hybridized to cDNA from wild-type males but not DTA males are by definition accessory gland specific. Acps were then identified as those ESTs that encoded proteins either containing signal sequences indicating secreted molecules or lost hybridization to cDNA from DTA male flies. By these criteria, 75 potential Acps were identified. By analyses of multiple hit rates, these Acps are estimated to represent 90% of the expressed Acps. Based upon the assumption that the genes will have equal expression, a Poisson distribution is expected for the number of times each gene is recovered in the clones (multiple hit rate). This expectation allows for an estimate of the total number of Acp genes expected (see Swanson et al. 2001b for details). The genes identified encode a variety of molecules with predicted functions consistent with activities known to exist in seminal fluid, such as proteases, protease inhibitors and lipases (Wolfner 1997). Although further refinement of this catalogue of Acps will be important, we are beginning to have a genomic level description of Drosophila male Acps.
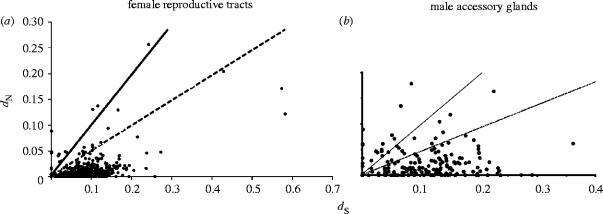
While the EST screen was instrumental in the identification of male Acps, the rates of evolution for these genes is also of interest. Therefore, ESTs were generated from a cDNA library constructed from Drosophila simulans (Swanson et al. 2001b). Drosophila simulans is a close relative to Drosophila melanogaster, with approximately 11% divergence at synonymous sites (Bauer & Aquadro 1997). This allowed direct comparison of the cDNA coding sequences between the two species in order to calculate dN/dS ratios (figure 2). Overall, the Acps showed a twofold increase in dN compared to non-Acps while dS was not statistically different between the two categories. The twofold increase in dN was consistent with previous protein electrophoresis studies (Civetta & Singh 1995); however, this observation by itself does not indicate adaptive evolution (Swanson et al. 2001b). Several candidates for recent targets of adaptive evolution were identified by virtue of having dN/dS ratios>1 (11%), but these need further investigation before it can be concluded that adaptive evolution drives their divergence.
Figure 2.
Evolutionary EST analyses from Drosophila (a) female reproductive tracts (Swanson et al. 2004) and (b) male accessory glands (Swanson et al. 2001b). For each coding region identified in the EST, dN was plotted against dS. Genes with a dN/dS ratio>1 (solid line) or 0.5 (dashed line) are prime candidates to have been recent targets of adaptive evolution. In both male accessory glands and female reproductive tracts, there are several genes that potentially have been subjected to adaptive evolution.
As with abalone sperm lysin, the identification of substantial numbers of male Acps potentially being subjected to positive Darwinian selection begs the question of whether female receptors for the Acps will show similar rates of evolution. In order to initiate studies of the function and evolution of female receptors, an EST approach analogous to the male accessory gland EST project described previously was performed. A cDNA library was made from dissected female reproductive tracts (minus ovaries) and subjected to differential hybridization to enrich for genes with female-biased expression (Swanson et al. 2004). Candidate female reproductive genes were identified as those containing either a signal sequence or transmembrane domains, indicating secreted proteins. By this approach, 169 putative female reproductive genes were identified. These putative receptors are now prime targets for future functional assays to determine if they interact with male Acps. It will be important to generate comparisons with other approaches, such as microarray experiments, to analyse for genes upregulated following mating (Lawniczak & Begun 2004; McGraw et al. 2004). Microarray methods have already provided insights into the identification of potential female receptors for Acps.
Like the male accessory gland, the female reproductive tract was an evolutionary EST screen with the ESTs derived from D. simulans and compared to the completed D. melanogaster genome (Swanson et al. 2004). From aligned coding sequences, the dN/dS ratio was calculated as a measure of selective pressure (figure 2). From comparison of published examples of adaptive evolution, it was proposed that genes with a dN/dS ratio>0.5 were good candidates to have been the target of recent adaptive evolution. When genes from the literature with an overall dN/dS ratio>0.5 were analysed by more powerful maximum-likelihood analyses incorporating variation in the dN/dS ratio between sites, more than 90% showed robust signatures of adaptive evolution (Swanson et al. 2004). This indicates a dN/dS ratio>0.5 is a good indicator for targets of recent adaptive evolution, but that additional studies are necessary prior to invoking adaptive evolution. From the female EST screen, 27 genes had a dN/dS ratio>0.5, indicating that they may have been the target of recent adaptive evolution. Eight genes were selected for in depth polymorphism and divergence analyses for signatures of adaptive evolution, and six were confirmed to have been the target of recent adaptive evolution (Swanson et al. 2004). Thus, as predicted by the sexual conflict theory, genes expressed in both the male and female reproductive tracts have been subjected to adaptive evolution (figure 2). Important future studies will be to identify interacting male–female reproductive molecules.
4. Conclusions and future directions
We have discussed the recent molecular work on the evolution of male and female reproductive proteins in two different animal systems, abalone and Drosophila. The abalone system is the first to show that female–male interacting gametic proteins are rapidly evolving. The mechanism behind this rapid evolution remains unknown; however, we know that both proteins show signs of change by adaptive evolution (in partial regions of VERL). In Drosophila, we are making important strides toward identifying female reproductive proteins. Similar to male reproductive proteins, female proteins also are rapidly evolving by Darwinian selection. This molecular work is an important first step for investigating the coevolution of interacting male–female reproductive proteins. Patterns of protein evolution from both systems support the predicted patterns of rapid protein evolution driven by sexual conflict. We stress, however, that other selective pressures may contribute to the rapid evolution we observe in the reproductive proteins of abalone and Drosophila. For instance, models of sexual selection, pathogen resistance and self versus non-self recognition make similar predictions as sexual conflict (Chapman et al. 2003). The key to distinguishing between sexual conflict and other mechanisms will be an integration of genetic, experimental and theoretical data. An integrative approach will help provide a link between genotype and phenotype of male and female reproductive molecules. Correlations of genotype frequencies and predictions from theoretical models will also provide support for models of sexual conflict.
For genetic data we need to identify interacting male and female reproductive proteins (as has been done for abalone egg VERL and sperm lysin) from several taxonomic groups. This data will allow us to determine if female–male coevolution supports or contradicts models of sexual conflict. For example, in Drosophila, under predictions of sexual conflict, female receptors to male Acps involved in mediating costs to females should be rapidly evolving. Functional studies have identified that the male reproductive protein Acp62F is a candidate for influencing the cost of mating in females (Lung et al. 2002). When the female receptor to Acp62F is identified we will be able to determine if it shows signs of rapid evolution as would be predicted by sexual conflict.
Theoretical predictions about the rates and patterns of variability in reproductive genes can provide clues into the identification of new genes involved in reproduction. An example where prediction of variability was useful in the experimental identification of new reproductive genes is in plant species with self-incompatibility. Self-incompatibility in plants probably evolved to prevent the costs associated with inbreeding depression that can arise from self-fertilization. Self-incompatible genes are predicted to be subjected to adaptive evolution, have high levels of polymorphism and the male and female loci should be tightly linked (Nasrallah 2002). This pattern of molecular evolution has been characterized for self-incompatible loci in several plant genera, including Brassicaceae and Solanaceae (Schopfer et al. 1999; Richman & Kohn 2000). In Brassicaceae, combining the predicted molecular properties of these loci with experimental work has proven useful for identifying and characterizing the self-incompatible loci for both the male pollen and female stigma proteins (Kachroo et al. 2001).
For experimental tests of sexual conflict, it will be informative to include genetic analyses of reproductive genes. In many experimental tests, it is often indirectly assumed that genes, such as those encoding Acps in Drosophila, have responded during the experimental procedure. For example, in one of the original laboratory experiments demonstrating sexual conflict, Drosophila Acps became more toxic upon adaptation to a non-responding female genotype (Rice 1996). Now that several male Acps and potential female reproductive molecules have been identified, it will be relatively easy to genotype these loci during the course of experimental evolution studies and shed light on the role of sexual conflict in protein evolution.
With the emergence of data on the rapid evolution of reproductive genes from many different taxa, including ciliate protozoa, green algae, fungi, plants, insects, marine invertebrates and mammals (see review in Swanson & Vacquier 2002a), we now have a good starting place for experimental and functional tests of reproductive genes which will be useful for identifying the adaptive mechanisms involved in rapid reproductive protein evolution. Continued integration of genetic, experimental and theoretical studies of sexual conflict will lead to a robust understanding of sexual conflict and the evolutionary implications.
Acknowledgments
We thank Jennifer D. Calkins for helpful discussions. W.J.S. is supported by NSF grant DEB-0410112 and NIH grant HD42563. N.L.C. and T.M.P. were supported by training grants from the NIH.
Footnotes
One contribution of 13 to a Discussion Meeting Issue ‘Sexual conflict: a new paradigm?’.
References
- Aguade M. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics. 1999;152:543–551. doi: 10.1093/genetics/152.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G, Rowe L. Sexual conflict and arms races between the sexes: a morphological adaptation for control in mating in a female insect. Proc. R. Soc. B. 1995;261:123–127. [Google Scholar]
- Bauer V.L, Aquadro C.F. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- Begun D.J, Whitley P, Todd B.L, Waldrip-Dail H.M, Clark A.G. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Liddle L.F, Kalb J.M, Wolfner M.F, Partridge L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature. 1995;373:241–244. doi: 10.1038/373241a0. 10.1038/373241a0 [DOI] [PubMed] [Google Scholar]
- Chapman T, Neubaum D.M, Wolfner M.F, Partridge L. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. R. Soc. B. 2000;267:1097–1105. doi: 10.1098/rspb.2000.1114. 10.1098/rspb.2000.1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol. Evol. 2003;18:41–47. 10.1016/S0169-5347(02)00004-6 [Google Scholar]
- Civetta A, Singh R.S. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J. Mol. Evol. 1995;41:1085–1095. doi: 10.1007/BF00173190. 10.1007/BF00173190 [DOI] [PubMed] [Google Scholar]
- Dawkins R. The selfish gene. Oxford University Press; Oxford: 1976. [Google Scholar]
- DiBenedetto A.J, Lakich D.M, Kruger W.D, Belote J.M, Baker B.S, Wolfner M.F. Sequences expressed sex-specifically in Drosophila melanogaster adults. Dev. Biol. 1987;119:242–251. doi: 10.1016/0012-1606(87)90225-9. 10.1016/0012-1606(87)90225-9 [DOI] [PubMed] [Google Scholar]
- Elder J.F, Jr, Turner B.J. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 1995;70:297–320. doi: 10.1086/419073. 10.1086/419073 [DOI] [PubMed] [Google Scholar]
- Frank S.A. Sperm competition and female avoidance of polyspermy mediated by sperm–egg biochemistry. Evol. Ecol. Res. 2000;2:613–625. [Google Scholar]
- Galindo B.E, Moy G.W, Swanson W.J, Vacquier V.D. Full length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. 10.1016/S0378-1119(02)00459-6 [DOI] [PubMed] [Google Scholar]
- Galindo B.E, Vacquier V.D, Swanson W.J. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl Acad. Sci. USA. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. 10.1073/pnas.0830022100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. Rapid evolution of reproductive barriers driven by sexual conflict. Nature. 2000a;403:886–889. doi: 10.1038/35002564. 10.1038/35002564 [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Waiting time to parapatric speciation. Proc. R. Soc. B. 2000b;267:2483–2492. doi: 10.1098/rspb.2000.1309. 10.1098/rspb.2000.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S, Waxman D. Sympatric speciation by sexual conflict. Proc. Natl Acad. Sci. USA. 2002;99:10 533–10 538. doi: 10.1073/pnas.152011499. 10.1073/pnas.152011499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowaty P.A. Sexul dialects, sexual selection, and variation in reproductive behavior: feminism and evolutionary biology. Chapman & Hall; New York: 1997. [Google Scholar]
- Haygood R. Sexual conflict and protein polymorphism. Evol. Int. J. Org. Evol. 2004;58:1414–1423. doi: 10.1111/j.0014-3820.2004.tb01723.x. [DOI] [PubMed] [Google Scholar]
- Hellberg M.E, Vacquier V.D. Rapid evolution of fertilization selectivity and lysin cDNA sequences in teguline gastropods. Mol. Biol. Evol. 1999;16:839–848. doi: 10.1093/oxfordjournals.molbev.a026168. [DOI] [PubMed] [Google Scholar]
- Hellberg M.E, Moy G.W, Vacquier V.D. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol. Biol. Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- Herndon L.A, Wolfner M.F. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc. Natl Acad. Sci. USA. 1995;92:10 114–10 118. doi: 10.1073/pnas.92.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Schopfer C.R, Nasrallah M.E, Nasrallah J.B. Allele-specific receptor–ligand interactions in Brassica self-incompatibility. Science. 2001;293:1824–1826. doi: 10.1126/science.1062509. 10.1126/science.1062509 [DOI] [PubMed] [Google Scholar]
- Kalb J.M, DiBenedetto A.J, Wolfner M.F. Probing the function of Drosophila melanogaster accessory glands by directed cell ablation. Proc. Natl Acad. Sci. USA. 1993;90:8093–8097. doi: 10.1073/pnas.90.17.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N, Vacquier V.D, Stout C.D. Abalone lysin: the dissolving and evolving sperm protein. Bioessays. 2001;23:95–103. doi: 10.1002/1521-1878(200101)23:1<95::AID-BIES1012>3.0.CO;2-C. 10.1002/1521-1878(200101)23:1%3C95::AID-BIES1012%3E3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- Lawniczak M.K, Begun D.J. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. 10.1139/g04-050 [DOI] [PubMed] [Google Scholar]
- Lee Y.H, Vacquier V.D. The divergence of species-specific abalone sperm lysins is promoted by positive Darwinian selection. Biol. Bull. (Woods Hole) 1992;182:97–104. doi: 10.2307/1542183. [DOI] [PubMed] [Google Scholar]
- Lewis C.A, Talbot C.F, Vacquier V.D. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev. Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. 10.1016/0012-1606(82)90167-1 [DOI] [PubMed] [Google Scholar]
- Linder J.E, Rice W.R. Natural selection and genetic variation for female resistance to harm from males. J. Evol. Biol. 2005;18:568–575. doi: 10.1111/j.1420-9101.2004.00872.x. 10.1111/j.1420-9101.2004.00872.x [DOI] [PubMed] [Google Scholar]
- Lung O, Tram U, Finnerty C.M, Eipper-Mains M.A, Kalb J.M, Wolfner M.F. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics. 2002;160:211–224. doi: 10.1093/genetics/160.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon J.D, Vacquier V.D. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev. Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. 10.1006/dbio.1999.9411 [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski M.S. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. 10.1073/pnas.95.16.9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw L.A, Gibson G, Clark A.G, Wolfner M.F. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. 10.1016/j.cub.2004.08.028 [DOI] [PubMed] [Google Scholar]
- Metz E.C, Gomez-Gutierrez G, Vacquier V.D. Mitochondrial DNA and bindin gene sequence evolution among allopatric species of the sea urchin genus Arbacia. Mol. Biol. Evol. 1998a;15:185–195. doi: 10.1093/oxfordjournals.molbev.a025914. [DOI] [PubMed] [Google Scholar]
- Metz E.C, Robles-Sikisaka R, Vacquier V.D. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1998b;95:10 676–10 681. doi: 10.1073/pnas.95.18.10676. 10.1073/pnas.95.18.10676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah J.B. Recognition and rejection of self in plant reproduction. Science. 2002;296:305–308. doi: 10.1126/science.296.5566.305. 10.1126/science.296.5566.305 [DOI] [PubMed] [Google Scholar]
- Neubaum D.M, Wolfner M.F. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics. 1999;153:845–857. doi: 10.1093/genetics/153.2.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G.A. M.S. Blum & N.A. Blum; London: 1979. Sexual selection and reproduction competition in insects. [Google Scholar]
- Parker G.A, Partridge L. Sexual conflict and speciation. Phil. Trans. R. Soc. B. 1998;353:261–274. doi: 10.1098/rstb.1998.0208. 10.1098/rstb.1998.0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W.R. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature. 1996;381:232–234. doi: 10.1038/381232a0. 10.1038/381232a0 [DOI] [PubMed] [Google Scholar]
- Rice W.R. Intergenomic conflict, interlocus antagonistic coevolution, and the evolution of reproductive isolation. In: Howard D.J, Berlocher S.H, editors. Endless forms: species and speciation. Oxford University Press; Oxford: 1998. pp. 161–270. [Google Scholar]
- Rice W.R, Holland B. The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav. Ecol. Sociobiol. 1997;41:1–10. 10.1007/s002650050357 [Google Scholar]
- Richman A.D, Kohn J.R. Evolutionary genetics of self-incompatibility in the Solanaceae. Plant Mol. Biol. 2000;42:169–179. 10.1023/A:1006336206637 [PubMed] [Google Scholar]
- Riffell J.A, Krug P.J, Zimmer R.K. Fertilization in the sea: the chemical identity of an abalone sperm attractant. J. Exp. Biol. 2002;205:1439–1450. doi: 10.1242/jeb.205.10.1439. [DOI] [PubMed] [Google Scholar]
- Rowe L, Cameron E, Day T. Detecting sexually antagonistic coevolution with population crosses. Proc. R. Soc. B. 2003;270:2009–2016. doi: 10.1098/rspb.2003.2453. 10.1098/rspb.2003.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer C.R, Nasrallah M.E, Nasrallah J.B. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. 10.1126/science.286.5445.1697 [DOI] [PubMed] [Google Scholar]
- Soller M, Bownes M, Kubli E. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur. J. Biochem. 1997;243:732–738. doi: 10.1111/j.1432-1033.1997.00732.x. 10.1111/j.1432-1033.1997.00732.x [DOI] [PubMed] [Google Scholar]
- Swanson W.J, Vacquier V.D. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating the acrosomal process of abalone spermatozoa. Proc. Natl Acad. Sci. USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J, Vacquier V.D. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc. Natl Acad. Sci. USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. 10.1073/pnas.94.13.6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J, Vacquier V.D. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. 10.1126/science.281.5377.710 [DOI] [PubMed] [Google Scholar]
- Swanson W.J, Vacquier V.D. Rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002a;3:137–144. doi: 10.1038/nrg733. 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- Swanson W.J, Vacquier V.D. Reproductive protein evolution. Annu. Rev. Ecol. Syst. 2002b;33:161–179. 10.1146/annurev.ecolsys.33.010802.150439 [Google Scholar]
- Swanson W.J, Aquadro C.F, Vacquier V.D. Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol. Biol. Evol. 2001a;18:376–383. doi: 10.1093/oxfordjournals.molbev.a003813. [DOI] [PubMed] [Google Scholar]
- Swanson W.J, Clark A.G, Waldrip-Dail H.M, Wolfner M.F, Aquadro C.F. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl Acad. Sci. USA. 2001b;98:7375–7379. doi: 10.1073/pnas.131568198. 10.1073/pnas.131568198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J, Yang Z, Wolfner M.F, Aquadro C.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl Acad. Sci. USA. 2001c;98:2509–2514. doi: 10.1073/pnas.051605998. 10.1073/pnas.051605998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swanson W.J, Wong A, Wolfner M.F, Aquadro C.F. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. 10.1534/genetics.104.030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers R.L. Cambell; Chicago: 1972. Sexual selection and the decent of man B. [Google Scholar]
- Tsaur S.C, Wu C.I. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 1997;14:544–549. doi: 10.1093/oxfordjournals.molbev.a025791. [DOI] [PubMed] [Google Scholar]
- Tsaur S.C, Ting C.T, Wu C.I. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 1998;15:1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- Vacquier V.D. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. 10.1126/science.281.5385.1995 [DOI] [PubMed] [Google Scholar]
- Vacquier V.D, Lee Y.H. Abalone sperm lysin: unusual mode of evolution of a gamete recognition protein. Zygote. 1993;1:181–196. doi: 10.1017/s0967199400001465. [DOI] [PubMed] [Google Scholar]
- Wolfner M.F. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem. Mol. Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. 10.1016/S0965-1748(96)00084-7 [DOI] [PubMed] [Google Scholar]
- Wolfner M.F, et al. New genes for male accessory gland proteins in Drosophila melanogaster. Insect Biochem. Mol. Biol. 1997;27:825–834. doi: 10.1016/s0965-1748(97)00056-8. 10.1016/S0965-1748(97)00056-8 [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. 10.1016/S0169-5347(00)01994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Swanson W.J, Vacquier V.D. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol. Biol. Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]