Abstract
PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1α) is a master regulator of mitochondrial biogenesis and plays an important role in several other aspects of energy metabolism. To identify upstream regulators of PGC-1α gene transcription, 10,000 human full-length cDNAs were screened for induction of the PGC-1α promoter. A number of activators of PGC-1α transcription were found; the most potent activator was the transducer of regulated CREB (cAMP response element-binding protein) binding protein (TORC) 1, a coactivator of CREB. The other two members of the TORC family, TORC2 and TORC3, also strongly activated PGC-1α transcription. TORCs dramatically induced PGC-1α gene transcription through CREB. Forced expression of TORCs in primary muscle cells induced the endogenous mRNA of PGC-1α and its downstream target genes in the mitochondrial respiratory chain and TCA cycle. Importantly, these changes in gene expression resulted in increased mitochondrial oxidative capacity measured by cellular respiration and fatty acid oxidation. Finally, we demonstrated that the action of TORCs in promoting mitochondrial gene expression and function requires PGC-1α. Previous studies had indicated that TORCs function as a calcium- and cAMP-sensitive coincidence detector and mediate individual and synergistic effects of these two pathways. Our results, together with previous findings, strongly suggest that TORCs play a key role in linking these external signals to the transcriptional program of adaptive mitochondrial biogenesis by activating PGC-1α gene transcription.
Mitochondrial biogenesis is an adaptive biological process that occurs in response to developmental and physiologic cues. Over the past decade, significant insights have been gained into the molecular regulatory cascades controlling mitochondrial biogenesis and function (1). Peroxisome proliferator-activated receptor γ coactivator 1 α (PGC-1α) was originally identified as a coactivator of peroxisome proliferator-activated receptor (PPAR)γ and regulator of adaptive thermogenesis (2) and then subsequently found to coactivate many more transcription factors and play a central integrative role in mitochondrial biogenesis (3). In vitro and in vivo studies have demonstrated that PGC-1α also plays a multifaceted role in regulating energy metabolism in various tissues (4). Furthermore, the expression of PGC-1α and mitochondrial oxidative capacity were reduced in muscle from diabetic individuals (5, 6) and with aging (7). Whether the reduced expression of PGC-1α and mitochondrial biogenesis play a causal role in the initiation or development of diseases remains to be determined. Nevertheless, improvement or restoration of mitochondrial function may provide a therapeutic approach for the treatment of these diseases.
To identify upstream regulators controlling PGC-1α transcription, an unbiased functional genomic screen was performed by using a PGC-1α promoter–luciferase reporter gene assay. A number of activators were identified that induced PGC-1α promoter activity. Among the hits, including transcription factors/regulators, kinases, and ion channels, TORC1 was the most potent activator. The TORC family, including TORC1, TORC2, and TORC3, were originally identified as coactivators of CREB, which greatly enhance CREB-dependent gene transcription (8, 9). In this study, we investigated the effect of the TORC family on PGC-1α transcription and mitochondrial biogenesis in primary mouse muscle cells. Furthermore, we provided mechanistic insights by which TORCs modulate mitochondrial biogenesis.
Results
TORCs Strongly Activate PGC-1α Gene Transcription.
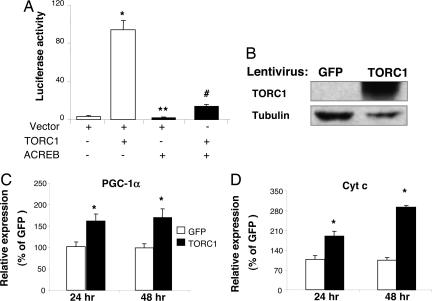
The Origene I clone collection, which contains 10,000 putative human full-length cDNA clones, was screened for the ability to induce the activity of the mouse 2 kb PGC-1α promoter (10) in HeLa cells. A TORC1 cDNA was identified as the most potent inducer of PGC-1α reporter from this screen. In addition to TORC1, several other genes were identified from the screen that strongly induced PGC-1α transcription (Table 1, which is published as supporting information on the PNAS web site). As shown in Fig. 1A, transfection of a TORC1 expression vector with the PGC-1α promoter luciferase construct (PGC1α-Luc) markedly induced the activity of PGC-1α promoter. To determine whether activation of PGC-1α transcription was CREB-dependent, a cDNA encoding a dominant negative CREB, ACREB, was cotransfected along with a TORC1 expression vector and PGC1α-Luc. The basal activity of the PGC-1α promoter was significantly suppressed by ACREB (Fig. 1A, lane 3 vs. lane 1). The activation of the PGC-1α promoter by TORC1 was markedly suppressed by ACREB (Fig. 1A, lane 4 vs. lane 2), suggesting that TORC1 activates PGC-1α transcription through CREB on the CRE site within the PGC-1α promoter.
Fig. 1.
TORC1 induces PGC-1α and Cyt c expression in HeLa cells. (A) HeLa cells were transfected with pGL3-basic-PGC1α-Luc (40 ng) and phRL-SV40 (3 ng, as control for transfection efficiency) along with the indicated cDNA constructs (TORC1, 60 ng; ACREB, 60 ng). The cells were lysed, and the luciferase activity was determined 48 h after transfection. The luciferase activity represents the ratio of firefly luminescence over renilla luminescence, an index of PGC-1α gene transcription; n = 6. ∗, P < 0.05 lane 2 vs. lane 1; ∗∗, P < 0.05 lane 3 vs. lane 1; #, P < 0.05 lane 4 vs. lane 2. (B–D) HeLa cells were transduced with a lentivirus expressing either GFP or TORC1. Total protein was extracted from cells 48 h after transduction and subjected to Western blot analysis to determine the expression of TORC1 protein (B). Total RNA was extracted from the cells at 24 and 48 h after transduction and subjected to Q-PCR analysis to determine the expression level of endogenous PGC-1α (C) and Cyt c mRNA (D). The average relative expression (mean ± SEM) is shown. ∗, P < 0.05 TORC1 vs. GFP; n = 3.
To determine whether activation of PGC-1α promoter is sufficient to induce expression of endogenous PGC-1α, HeLa cells were transduced with a lentivirus expressing either TORC1 or GFP. Total protein and RNA were extracted from the cells at 24 and 48 h after transduction. TORC1 protein was overexpressed in the cells transduced with the TORC1 lentivirus compared with those transduced with GFP (Fig. 1B). As shown in Fig. 1C, the endogenous level of PGC-1α mRNA was significantly elevated by overexpression of TORC1 in HeLa cells at both 24 and 48 h. We also examined the expression of cytochrome c (Cyt c), a PGC-1α target gene. Cyt c mRNA level increased by 2-fold in the cells transduced with TORC1 compared with the GFP controls (Fig. 1D). These data indicate that the activation of the PGC-1α promoter led to increased expression of PGC-1α and its target gene Cyt c, suggesting that mitochondrial gene expression and/or biogenesis might be increased upon TORC overexpression.
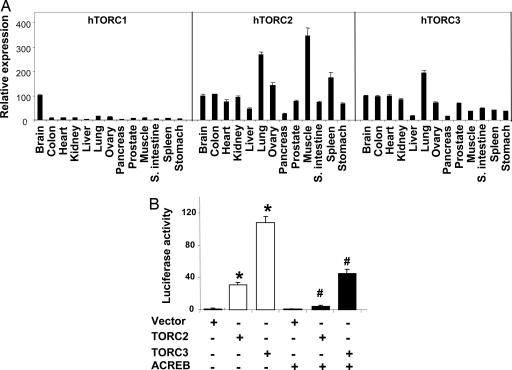
The TORC family has two other members, TORC2 and TORC3, which share 30–40% homology with the TORC1 protein. The distribution of the mRNAs from the three TORC genes in human tissues is shown in Fig. 2A. TORC1 mRNA expression was highest in brain and very low, by comparison, in all other tissues examined. Conversely, TORC2 and TORC3 are ubiquitously expressed in the tissues examined, and both are expressed at a significant level in the skeletal muscle. To determine whether TORC2 and TORC3 can also activate PGC-1α transcription, plasmids expressing hTORC2 or hTORC3 were cotransfected into the HeLa cells along with the PGC-1α promoter. As shown in Fig. 2B, both TORC2 and TORC3 strongly activated the PGC-1α promoter. In the presence of ACREB, the activation by TORC2 or TORC3 was markedly suppressed.
Fig. 2.
TORC2 and TORC3 strongly induce PGC-1α transcription. (A) Human tissue distribution of the TORC family. The mRNA expression levels of TORC1, 2, and 3 were measured by Q-PCR in a panel of human tissue cDNAs. The mRNA expression level of each TORC in brain is set at 100%. The mRNA expression levels in other tissues is relative to that of brain. (B) TORC2 and TORC3 induce PGC-1α promoter activity. HeLa cells were transfected with pGL3-basic-2kb-PGC1α-Luc (40 ng) and phRL-SV40 (3 ng) along with various cDNA constructs, as indicated (TORC2 or TORC3, 10 ng; ACREB, 110 ng). The cells were lysed, and the luciferase activity was determined at 48 h after transfection. The luciferase activity represents the ratio of firefly luminescence over renilla luminescence (mean ± SEM). ∗, P < 0.05, lanes 2 and 3 vs. lane 1; #, P < 0.05, lane 5 vs. lane 2 and lane 6 vs. lane 3; n = 6
TORCs Induce Mitochondrial Biogenesis in Muscle Cells.
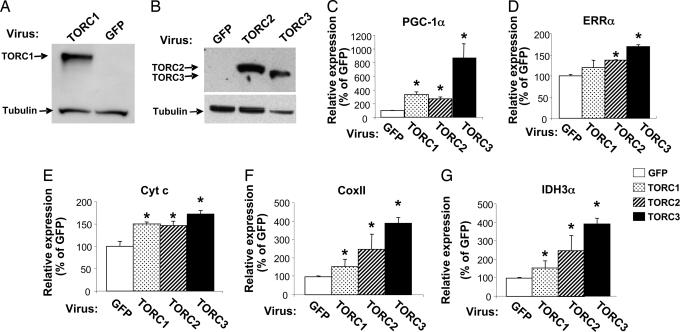
TORCs are regulated by nuclear import, which is induced by both cAMP- and calcium-signaling pathways (9, 11). Interestingly, both pathways are also activated during contraction and exercise as well as cold adaptation, and these processes lead to the induction of PGC-1α and mitochondrial biogenesis in skeletal muscle and brown adipose tissue (10, 12–15). The requirement of PGC-1α activation by TORCs in these processes has not been determined. Thus, we investigated the effect of TORCs on PGC-1α expression and mitochondrial biogenesis in mouse primary muscle cells. Mouse primary myotubes were transduced with an adenovirus expressing hTORC1, hTORC2, hTORC3, or GFP for 48 h. Total protein was extracted from the cells, and the human TORC1, TORC2, and TORC3 protein levels were determined by Western blot analysis. As shown in Fig. 3, the expression levels of human TORC1 (Fig. 3A), TORC2 and TORC3 (Fig. 3B) proteins were expressed in the cells transduced with the appropriate TORC adenovirus. To determine whether forced expression of TORCs induced endogenous PGC-1α expression, total RNA was extracted from these muscle cells and subjected to quantitative (Q)-PCR analysis. Fig. 3C shows that PGC-1α mRNA levels were increased approximately 3-fold with TORC1 or TORC2 and 8-fold with TORC3 compared with the GFP control. To determine whether induction of PGC-1α can activate the mitochondrial biogenic program in these cells, the expression levels of PGC-1α target genes were examined. Estrogen related receptor α (ERRα) an early target gene of PGC-1α during mitochondrial biogenesis, was significantly elevated by TORC2 and TORC3 (Fig. 3D). Genes that are involved in the mitochondrial respiratory chain and TCA cycle, such as Cyt c, Cox II (Cytochrome oxidase subunit II), and IDH3α (isocitrate dehydrogenase 3 α) were also significantly increased (Fig. 3 E–G). Both Cyt c and IDH3α are nuclear genome-encoded genes, whereas Cox II is a mitochondrial genome-encoded gene. The induction of Cox II suggests that mitochondrial DNA replication is increased. These results suggest that TORCs promote mitochondrial biogenesis by increasing the expression of both nuclear- and mitochondrial-encoded genes.
Fig. 3.
Adenoviral-mediated expression of TORCs increases mitochondrial gene expression. (A and B) Primary muscle cells were transduced with TORC1, TORC2, TORC3, or GFP adenovirus. Total protein or RNA was extracted from the cells at 48 h after transduction, and 50 μg of total protein extract from each sample was subjected to Western blot analysis to determine the expression levels of TORC1 (A), and ectopically expressed TORC2 and TORC3 protein (B). The primary antibodies used were anti-TORC1 in A and anti-V5 in B. The secondary antibody used was HRP-conjugated goat anti-rabbit IgG. Total RNA samples were subjected to Q-PCR analysis. (C–G) The relative expression levels of the endogenous PGC-1α (C) and ERRα (D), Cyt c (E), Cox II (F), and IDH3α (G) are shown. Mean ± SEM; n = 3; ∗, P < 0.05, TORC1, TORC2, or TORC3 vs. GFP.
TORCs Enhance Mitochondrial Oxidative Capacity in Muscle Cells.
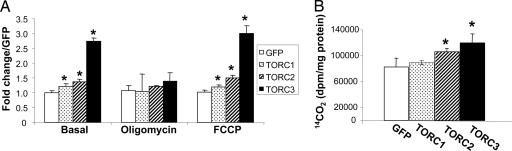
We further investigated whether induction of mitochondrial gene expression by TORCs has an impact on mitochondrial function, especially oxidative phosphorylation. Cellular respiration was measured from the mouse primary muscle cells transduced with either TORC or GFP adenovirus for 48 h (Fig. 4A). Coinciding with the induction of gene expression in the respiratory chain, the basal cellular respiration, without treatment, was elevated 22%, 37%, and 174% by overexpression of TORC1, TORC2, and TORC3, respectively. Oligomycin-insensitive respiration, the proton leak, was not affected by overexpression of TORCs. Consistent with this observation, we found that the expression of UCP2 and UCP3, two major UCPs expressed in these cells, was not altered (data not shown). Carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP)-stimulated cellular respiration, indicative of mitochondrial maximum capacity, was increased 20%, 49%, and 200% by TORC1, TORC2, and TORC3, respectively (Fig. 4A). These results demonstrate that TORCs not only induced mitochondrial gene expression, but also enhanced mitochondrial oxidative capacity in the muscle cells.
Fig. 4.
Adenoviral-mediated overexpression of TORCs increase mitochondrial oxidative capacity in muscle cells. Primary muscle cells were transduced with TORC1, TORC2, TORC3, or GFP adenovirus for 48 h. (A) Cellular respiration was measured without treatment (basal) or with treatment, as indicated. The average fold change over GFP (mean ± SEM) is shown. (B) Cells were labeled with [14C]palmitate for 2 h, and the complete fatty acid oxidation rate indicated by the amount of 14CO2 produced was determined. All experiments were performed in triplicate. ∗, P < 0.05 for TORC vs. GFP; n = 3.
Another important pathway in the mitochondria for energy metabolism is fatty acid β-oxidation. To determine whether TORCs affect this pathway, myotubes were transduced with TORC2, TORC3, or GFP adenovirus for 48 h. Cells were then labeled with [14C]palmitate for 2 h, and the complete oxidation product, 14CO2, was measured. As shown in Fig. 4B, the fatty acid oxidation rate was increased 28% and 45% in the cells overexpressing TORC2 and TORC3, respectively, compared with the GFP control. TORC1 overexpression, however, did not elevate the fatty acid oxidation significantly in these cells.
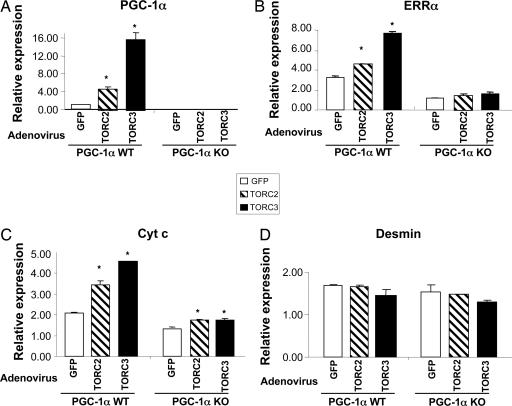
Induction of Mitochondrial Biogenesis by TORCs Requires PGC-1α.
CREB is known to regulate the transcription of a number of mitochondrial proteins directly (16–20). A crucial question is whether the induction of mitochondrial gene expression by TORCs depends on PGC-1α. To investigate this question, we used primary muscle cells from WT and PGC-1α KO mice. These cells were transduced with TORC2, TORC3, or GFP adenovirus for 48 h. Total RNA was extracted, and the expression of PGC-1α and its target genes, ERRα and Cyt c, was measured by Q-PCR. As expected, PGC-1α expression was significantly increased by overexpression of TORCs in the WT myotubes but not in the PGC-1α-null myotubes (Fig. 5A). ERRα mRNA levels were significantly elevated in the WT PGC-1α myotubes by TORCs, however, this increase was completely abolished in the null myotubes (Fig. 5B). Cyt c expression was elevated by TORCs in the WT myotubes, and this induction was markedly, but not completely, abolished, in the PGC-1α-null myotubes (Fig. 5C). In contrast to the PGC-1α target genes, the expression of desmin, a myogenic-specific marker, was not altered (Fig. 5D). These results indicate that the effect of TORCs on mitochondrial biogenesis largely depends on PGC-1α.
Fig. 5.
The effect of TORC2 and TORC3 on mitochondrial gene expression is primarily mediated through PGC-1α. WT and PGC-1α-null primary muscle cells were transduced with TORC2, TORC3, or GFP adenovirus for 48 h. Total RNA was extracted, and the gene expression levels (mean ± SEM) were measured by Q-PCR analysis. ∗, P < 0.05, TORC2 or TORC3 vs. GFP; n = 3.
Discussion
Based on an unbiased screen, we have identified TORCs as the most potent activators for PGC-1α gene transcription. All three members of the TORC family, TORC1, 2, and 3, markedly increased PGC-1α promoter activity. This transcription activation by TORCs was strongly suppressed by ACREB, indicating that TORCs increase PGC-1α transcription in a CREB-dependent fashion. The TORC family had been identified as coactivators of CREB and enhanced CREB-dependent gene transcription (8). TORCs function as downstream effectors of cAMP- and calcium-signaling pathways and appear to be an essential component of inducible, CREB-mediated gene expression. TORC2 was shown recently to promote hepatic gluconeogenesis in response to fasting by inducing the expression of PGC-1α (21). Interestingly, TORC2 also enhanced hepatic insulin sensitivity by increasing IRS-2 expression, which suppresses hepatic gluconeogenesis (22). By playing a dual role in controlling fasting hepatic gluconeogenesis, the CREB:TORC2 pathway thus triggers a feedback response that limits glucose output from the liver during fasting. However, the role of the TORC family in mitochondrial biogenesis and energy metabolism in muscle was not known.
The calcium-signaling pathways play an important role in triggering PGC-1α expression and mitochondrial biogenesis upon exercise (12, 13, 23–28). We hypothesized that TORCs might play a role in mediating PGC-1α expression and PGC-1α-dependent mitochondrial biogenesis in this process. Ectopic expression of TORCs induced the expression of the endogenous PGC-1α mRNA as well as its downstream target genes including ERRα, a transcription regulator of mitochondrial biogenesis (29, 30) and mitochondrial markers including Cyt c, COX II, and IDH3α. Importantly, coinciding with the changes of gene expression, the oxidative capacity of these cells was enhanced, indicated by increased cellular respiration as well as fatty acid oxidation. These results clearly demonstrate that TORCs can promote mitochondrial biogenesis and enhance oxidative capacity in muscle cells.
We further dissected the mechanism by which TORCs promote mitochondrial biogenesis in muscle. We found that ectopic expression of TORCs strongly induced the mRNA levels of PGC-1α, ERRα, and Cyt c in the PGC-1α WT myotubes. However, the induction of these genes by TORCs was drastically suppressed in the PGC-1α-null muscle cells. Conversely, the expression of desmin, a myogenic marker and non-PGC-1α target, was not altered by overexpression of TORCs in either the PGC-1α WT or the null muscle cells. These results clearly demonstrate that the action of TORCs in promoting mitochondrial biogenesis requires PGC-1α and further highlights the importance of PGC-1α in this process.
The three TORCs display differential tissue distribution patterns (Fig. 2A) and subcellular localization (11). The activity of TORC1 and TORC2 is regulated by calcineurin and salt-inducible kinases through dephosphorylation or phosphorylation, which promotes or blocks the translocation of TORC1 and 2, respectively, from the cytoplasm to the nucleus (9, 11). TORC3, on the other hand, was shown to be localized in the nucleus, mainly in the resting state in some cells, and had a higher potency in enhancing CREB transcription activity compared with TORC1 and 2 (11). Consistent with this finding, our results show that TORC3 exerts a stronger effect in activating the transcription of the PGC-1α gene as well as inducing mitochondrial biogenesis in muscle cells compared with TORC1 and TORC2. Interestingly, the effect of TORC1 and TORC2 in activating the PGC-1α promoter was diminished in the presence of ACREB, indicating that this activation is entirely mediated through CREB. The activation by TORC3, however, was suppressed to a lesser degree (≈60%) compared with TORC1 and TORC2 (>90%). It is conceivable that the remaining activity by TORC3 in the presence of ACREB is mediated through other transcription factor(s). The identity of these other transcription factors and their target sequences in the PGC-1α gene remains to be determined.
In skeletal muscle, a large body of evidence indicates that, in response to contractile activity, calcium-dependent signaling pathways trigger regulatory events leading to increased oxidative fiber content, mitochondrial biogenesis, and oxidative capacity (31–33). The molecular mechanism by which activation of the calcium signaling pathways, upon exercise in muscle cells, leads to the induction of PGC-1α-mediated mitochondrial biogenesis has been elucidated by Handschin et al. (10). CREB and MEF2 are the two key transcription factors activated by calcineurin and CaMKIV, which, in turn, activate PGC-1α transcription through the CRE and MEF2 sites, respectively, on the PGC-1α promoter. Akimoto et al. (13) showed that the activity of the PGC-1α promoter was activated in muscle in live mice upon electrical stimulation and that the CRE and MEF2 sites on the PGC-1α promoter were critical for this activation. We demonstrate here that TORCs strongly induced the PGC-1α-dependent mitochondrial biogenesis and oxidative capacity in muscle. Our results, together with the observation that TORC activation occurs through calcium in a calcineurin-dependent fashion (11), strongly suggest that TORCs play a key role in linking these external signals, such as exercise, to the transcriptional program of adaptive mitochondrial biogenesis by activating PGC-1α transcription.
Mitochondria are dynamic organelles and serve as the center stage for oxidative metabolism, nucleotide and lipid biosynthesis, apoptosis, and reactive oxygen species homeostasis. Dysregulation of mitochondrial function has been linked to various human diseases, such as neurodegeneration, cancer, cardiomyopathy, and diabetes (34). Although it remains to be determined whether mitochondrial dysfunction plays an etiologic role in the development of any of these diseases, restoration/improvement of mitochondrial function might delay the onset or have a better control of these diseases. Therefore, enhancing mitochondrial function through activation of PGC-1α by TORCs as well as other identified gene hits from this study could potentially be a therapeutic approach for the treatment of some these diseases.
Materials and Methods
cDNA Collection and Plasmids.
The OriGene I cDNA collection was purchased from OriGene Technologies (Rockville, MD). The collection contains ≈10,000 putative full-length human cDNA clones representing 8,980 gene loci, covering ≈35% of Refseq-annotated human genes. The mouse 2-kb PGC-1α promoter luciferase construct, pGL3-basic-PGC-1α-Luc (PGC1α-Luc), and ACREB were described in ref. 10.
High-Throughput Transfection and Luciferase Reporter Gene Assay.
High-throughput DNA transfection in 384-well plates was performed by using a Biomek FX liquid handling workstation (Beckman-Coulter, Fullerton, CA) as described (35). The other reporter gene assays were performed in 96-well plates.
Primary Mouse Myoblast Isolation and Differentiation.
Primary muscle cells were isolated from 2- to 3-week-old FVB mice as well as the wild-type and PGC-1α-null mice (36) as described (37). To induce differentiation, myoblasts were grown to 80% confluence and then switched to the differentiation medium, DMEM (GIBCO, Carlsbad, CA) containing 5% horse serum (HyClone, Logan, UT).
Lentiviral and Adenoviral Transduction.
Generation of TORC1 and GFP lentiviruses and transduction into HeLa cells were described (see supporting information). The adenoviruses expressing human TORC1 or TORC2 and TORC3 were generated by Welgen (Worcester, MA) and Invitrogen (Carlsbad, CA), respectively. Primary muscle cells were transduced with the adenovirus of TORC2, TORC3, or GFP at 4 × 108 particles per ml per well in 6-well plates. Medium was changed daily. Cells were harvested 48 h after transduction for analysis.
RNA Extraction and Real-Time PCR Analysis of Gene Expression.
Total RNA was extracted from cells by using TRIzol (Invitrogen) and subjected to TaqMan analysis. The relative mRNA expression levels were calculated by comparing the target genes/18S rRNA. The TaqMan probe sequence of cytochrome oxidase subunit II (Cox II) is 5′-TCAAGCAACAGTAACATCAAACCGACCA-3′. The TaqMan primer sequences of Cox II are 5′-CCATCCCAGGCCGACTAA-3′ (forward) and CAGAGCATTGGCCATAGAATAACC (reverse). The TaqMan primer/probe sets for all of the other genes examined were obtained from Applied Biosystems (Foster City, CA).
Protein Extraction and Western Blotting.
Cells were lysed by using cell extraction buffer (Cat no. FNN0011; Biosource International, Camarillo, CA) supplemented with protease inhibitors. Fifty micrograms of cell lysate was used for Western blot analysis. The rabbit anti-human TORC1 polyclonal antibody (9) was used at 1:2,000 dilution. Tubulin (Cat no. ab3194; Abcam, Cambridge, MA) and V5 antibodies (Cat no. 960-25; Invitrogen) were used at dilutions of 1:3,000 and 1:5,000, respectively. The secondary antibodies, HRP-conjugated goat anti-rabbit IgG (Cat no. 31460) and goat-anti-mouse IgG (Cat no. 31430) were purchased from Pierce (Rockford, IL) and used at 1:10,000 dilution. The immunoblots were developed by using enhanced chemiluminescence with Super Signal West Dura Chemiluminescence kit (Cat no. 34075; Pierce).
Cellular Respiration and Fatty Acid Oxidation Measurements.
Respiration was measured in myotubes according to the published method (38). Half a million mouse primary myotubes were used for each measurement. The concentration of oligomycin (MP Biomedicals, Solon, OH) and FCCP (Sigma, St. Louis, MO) were 2 μg/ml and 2–5 μM, respectively. Fatty acid oxidation assay was performed as described (39)
Statistical Analysis.
Statistical analysis was performed on all of the data by using a two-tailed, unpaired Student's t test. A P value <0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Brian Latario and Zhaohui Xiong (both of Novartis Institutes for BioMedical Research) for providing the lentiviruses; Maria Ceuto for assistance on the PGC-1α promoter analysis; and Shamina Rangwala, Shari Caplan, and Tom Hughes for helpful scientific discussions.
Abbreviations
- CREB
cAMP response element-binding protein 1
- Cyt c
cytochrome c
- ERRα
estrogen-related receptor α
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator 1α
- Q-PCR
quantitative PCR
- TORC
transducer of regulated cAMP response element-binding protein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kelly DP, Scarpulla RC. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 2.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 4.Lin J, Handschin C, Spiegelman BM. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 6.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling C, Poulsen P, Carlsson E, Ridderstrale M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. J Clin Invest. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates I, John R. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 13.Akimoto T, Sorg BS, Yan Z. Am J Physiol Cell Physiol. 2004;287:C790–C796. doi: 10.1152/ajpcell.00425.2003. [DOI] [PubMed] [Google Scholar]
- 14.Berchtold MW, Brinkmeier H, Muntener M. Physiol Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 15.Boss O, Bachman E, Vidal-Puig A, Zhang C-Y, Peroni O, Lowell BB. Biochem Biophys Res Commun. 1999;261:870–876. doi: 10.1006/bbrc.1999.1145. [DOI] [PubMed] [Google Scholar]
- 16.Scarpulla RC. Gene. 2002;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 17.Herzig RP, Scacco S, Scarpulla RC. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 18.Eggers A, Caudevilla C, Asins G, Hegardt FG, Serra D. Biochem J. 2000;345(Pt 2):201–206. [PMC free article] [PubMed] [Google Scholar]
- 19.Varone CL, Giono LE, Ochoa A, Zakin MM, Canepa ET. Arch Biochem Biophys. 1999;372:261–270. doi: 10.1006/abbi.1999.1470. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Kim C-H, Simon DK, Aminova L, Andreyev A, Kushnareva Y, Murphy AA, Lonze BE, Kim K-S, Ginty DD, et al. J Biol Chem. 2005;280:40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo S-H, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 22.Canettieri G, Koo S-H, Berdeaux R, Heredia J, Hedrick S, Zhang X, Montminy M. Cell Metab. 2005;2:331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 24.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 25.Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 26.Pilegaard H, Saltin B, Neufer PD. J Physiol (London) 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Biochem Biophys Res Commun. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 28.Terada S, Tabata I. Am J Physiol Endocrinol Metab. 2004;286::E208–E216. doi: 10.1152/ajpendo.00051.2003. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Handschin C, Arlow D, Xie X, St. Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holloszy JO, Coyle EF. J Appl Physiol. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 32.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Wu H, Tarr PT, Zhang C-Y, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 34.Wallace DC. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, et al. Proc Natl Acad Sci USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J, Wu P-H, Tarr PT, Lindenberg KS, St. Pierre J, Zhang C-Y, Mootha VK, Jager S, Vianna CR, Reznick RM. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. J Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 39.Thupari JN, Landree LE, Ronnett GV, Kuhajda FP. Proc Natl Acad Sci USA. 2002;99:9498–9502. doi: 10.1073/pnas.132128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.