Abstract
Nonself recognition is exemplified in the fungal kingdom by the regulation of cell fusion events between genetically different individuals (heterokaryosis). The het-6 locus is one of ∼10 loci that control heterokaryon incompatibility during vegetative growth of N. crassa. Previously, it was found that het-6-associated incompatibility in Oak Ridge (OR) strains involves two contiguous genes, het-6 and un-24. The OR allele of either gene causes “strong” incompatibility (cell death) when transformed into Panama (PA)-background strains. Several remarkable features of the locus include the nature of these incompatibility genes (het-6 is a member of a repetitive gene family and un-24 also encodes the large subunit of ribonucleotide reductase) and the observation that un-24 and het-6 are in severe linkage disequilibrium. Here, we identify “weak” (slow, aberrant growth) incompatibility activities by un-24PA and het-6PA when transformed separately into OR strains, whereas together they exhibit an additive, strong effect. We synthesized strains with the new allelic combinations un-24PA het-6OR and un-24OR het-6PA, which are not found in nature. These strains grow normally and have distinct nonself recognition capabilities but may have reduced fitness. Comparing the Oak Ridge and Panama het-6 regions revealed a paracentric inversion, the architecture of which provides insights into the evolution of the un-24–het-6 gene complex.
A process analogous to somatic nonself recognition is fundamental to all living organisms and has been well characterized at the molecular level in fungi (heterokaryon incompatibility: Glass and Kaneko 2003; Micali and Smith 2005), in invertebrates (histocompatibility system controlling colony fusion: Grosberg and Quinn 1986), and in mammals (major histocompatibility complex: Hughes and Yeager 1998). The diversity of recognition mechanisms and the pervasiveness of recognition phenomena across taxa suggest that nonself recognition has emerged independently several times in evolutionary history and is therefore of biological significance. In filamentous fungi, as elsewhere, distinct nonself recognition mechanisms operate during the sexual and asexual phases of the life cycle. During the sexual phase, cell fusion between genetically different individuals is promoted by the mating-type system (Glass and Kaneko 2003). In contrast, during asexual (vegetative) growth, cell fusion and the establishment of cells containing genetically different nuclei (heterokaryons) is limited to interactions among individuals that are genetically very similar or identical. Controls on the formation and proliferation of heterokaryotic cells are thought to be modulated by the interaction of protein products of the same (allelic) or different (nonallelic) het (heterokaryon incompatibility) or vic (vegetative incompatibility) loci (Saupe 2000). One of the best-characterized fungal systems is represented by Neurospora crassa where at least 10 het loci and mat (mating type) control heterokaryon formation during vegetative growth (Beadle and Coonradt 1944; Perkins 1988). Strains must bear compatible alleles at all incompatibility loci to fuse and form stable heterokaryons. Genetic differences at any one of these het-associated loci lead to incompatibility reactions expressed phenotypically as inhibited, abnormal growth, breakdown of the heterokaryotic products, and in certain cases cell death (Glass et al. 2000).
The function of vegetative nonself recognition in fungi has been debated for some time (Caten 1972; Hartl et al. 1975; Wu et al. 1998; Saupe 2000). A favored explanation is that nonself recognition reduces the transmission of potentially deleterious genetic elements (organelles, viruses, plasmids) from one individual to another. Evidence in support of this hypothesis includes direct observation of reduced transmission of infectious agents between incompatible fungal strains (Debets and Griffiths 1998; Cortesi et al. 2001). In Ophiostoma novo-ulmi, the causative agent of Dutch elm disease, a shift to greater diversity in vegetative incompatibility groups is associated with increased virus abundance in populations (Paoletti et al. 2006). In another study, the frequency of programmed cell death (PCD) associated with vegetative incompatibility was found to be negatively correlated to virus transmission in Cryphonectria parasitica (Biella et al. 2002). Such observations provide a potential adaptive explanation for the maintenance of heterokaryon incompatibility in fungi.
Among the incompatibility factors in N. crassa, differences at het-6 cause one of the most intense heterokaryon incompatibility reactions. Initially identified as a single locus (Mylyk 1975), the het-6 region comprises two tightly linked genes, un-24 and het-6, each having two allelic variants, Oak Ridge (OR) and Panama (PA). The two loci are in severe linkage disequilibrium such that they have been found in only two of four possible allelic combinations, un-24ORhet-6OR and un-24PAhet-6PA (Mir-Rashed et al. 2000; Smith et al. 2000b), referred to as OR and PA haplotypes, respectively. In addition to its function in heterokaryon incompatibility, un-24 encodes the large subunit of ribonucleotide reductase, an essential enzyme that converts ribonucleotides to the deoxyribonucleotides required for DNA synthesis and repair (Smith et al. 2000a). In contrast, het-6 appears to be a member of a repetitive gene family and to encode a protein with no known function aside from nonself recognition. OR alleles at both het-6 and un-24 were previously characterized and shown to elicit incompatibility reactions when introduced separately or together into PA recipient cells; however, failure to detect reciprocal incompatibility reactions from the PA allelic counterparts suggested that un-24 and het-6 incompatibility reactions may involve nonallelic interactions with additional factors in the het-6 region (Smith et al. 2000b).
This article reports on the isolation and characterization of functional PA alleles of un-24 and het-6 and describes a paracentric inversion polymorphism that provides a proximate reason for decreased recombination within the region and linkage disequilibrium of these two genes. The inversion breakpoints are immediately adjacent to regions in both un-24 and het-6 that appear to be under diversifying selection. We constructed strains that express the novel allelic combinations un-24PA het-6OR or un-24OR het-6PA and observed that, unlike their naturally occurring counterparts, these strains have lower fitness and compromised incompatibility function. Our observations suggest that evolution within the un-24–het-6 gene complex has had an adaptive role in heterokaryon incompatibility function in the species.
MATERIALS AND METHODS
Strains and growth conditions:
Laboratory-derived (Table 1) and wild-collected (listed in Figure 6) N. crassa strains were cultured with Vogel's sucrose medium with supplements as required (Davis and de Serres 1970). Heterokaryon incompatibility tests and DNA transformations into N. crassa were done as described previously (Smith et al. 2000b). Crosses were performed on synthetic crossing medium with 0.1× supplements at 25° (Davis and de Serres 1970).
TABLE 1.
Laboratory strains used in this study
| Straina | Genotypeb | Origin |
|---|---|---|
| FGSC 1131 | het-6PAhet-cPAA | Mir-Rashed (1998) |
| FGSC 2489 | 74-OR23-IV A | Oak Ridge wild type, 74ORs |
| RLM 58-18 | het-6PAhet-cPAa | Smith et al. (1996) |
| C2(2)-1 | het-6PAhet-cORthr-2 a | Smith et al. (1996) |
| C2(2)-5 | het-6PAhet-cPAthr-2 a | Smith et al. (1996) |
| C9-2 | het-6ORhet-cPAthr-2 a | Smith et al. (2000b) |
| C2(2)-9 | het-6PAhet-cORthr-2 A | Smith et al. (1996) |
| C2(3)-25 | het-6PAhet-cPAthr-2 A | Saupe et al. (1996) |
| C9-15 | het-6ORhet-cPAthr-2 A | M. L. Smith |
| 6-13 | arg-1 ad-3B; het-6PAhet-cPApyr-4 A | Smith et al. (1996) |
| C8c-32 | ro-7 un-24 het-6ORhet-cPApyr-4 a | Smith et al. (2000b) |
| C01(4)-2 | un-24 het-6ORhet-cPApyr-4 a | This study |
FGSC, Fungal Genetics Stock Center, University of Kansas Medical Center.
The un-24 allelic designation is the same as that given for het-6; alleles at other undesignated het loci are OR for all strains except FGSC1131, where they are unknown. “un-24,” with no superscript, designates the temperature-sensitive allele that has OR incompatibility activity.
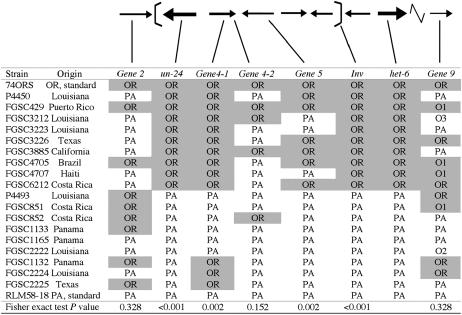
Figure 6.—
Analysis of polymorphisms in the un-24–het-6 region in 20 N. crassa strains. Eight different markers in and around In(het-6) were assayed for OR- and PA-specific PCR–RFLP patterns using primer pairs and restriction enzymes listed in Table 4. Inversion type was determined by PCR with primers Gene7b-P1/cys3-P4 (OR) and Gene7b-P1/6J-P11 (PA), corresponding to brk-1, or with Gene2-P3/6J-P11 (OR) or Gene2-P3/Gene 7a-P5 (PA), corresponding to brk-2 (see Figure 4 for primer locations). Strains RLM58-18 and 74OR23-1V were used as reference PA and OR strains, respectively. Correlation between each marker and the genotype at het-6 was calculated with the Fisher exact test.
Molecular biology methods:
Neurospora genomic DNA was isolated by the method of Oakley et al. (1987). Plasmid and cosmid DNAs were obtained by Wizard miniprep (Promega, Madison, WI) and λDNA was obtained by the liquid lysis method (Sambrook et al. 1989). Oligonucleotides used for PCR amplification (Table 2) were synthesized by Invitrogen (Carlsbad, CA) and PCR amplifications were done with Taq DNA polymerase (Invitrogen) or an Expand High Fidelity PCR system (Roche, Laval, PQ, Canada). When required, PCR products were cloned into pCRII or TOPO pCRII (Invitrogen) and subclones were prepared in pUC118 and pCB1004, which carries hygromycin resistance (Carrol et al. 1994). Southern hybridizations with [α-32P]dCTP-labeled probes were by standard methods (Sambrook et al. 1989). For a λ-library of FGSC1131, genomic DNA was partially digested with MboI and separated on a 10–40% sucrose gradient by ultracentrifugation at 26,000 rpm for 42 hr in a Beckman swinging bucket SW27 rotor. Fragments between 10 and 20 kbp in length were ligated into Lambda FixII arms and packaged into a λ-phage according to the manufacturer's recommendations (Stratagene, La Jolla, CA).
TABLE 2.
PCR primers used in this study
| Primer name | Primer sequence 5′ → 3′ |
|---|---|
| Gene2-P1 | TGCCTGTGCTGAACATGTG |
| Gene2-P2 | GCAGCCAGTTACTGTCCA |
| Gene2-P3 | TCACGTCGGATGGGCAGACTC |
| 6J-P11 | CTCCGGATGAGGTTGCCG |
| 6J-P6 | GTGCGGGCTTAACCGCTG |
| Gene4-P1 | ATCGATGTAAGTCACTCG |
| Gene4-P2 | ATCTCTTCATGTCGTCTC |
| Gene5-P1 | GGTGAAGTATCATGCGAGG |
| Gene5-P2 | ATCGACGATTGCTGTGGA |
| cys3-P4 | GCTGCCGTGCTTCTCCG |
| Gene7b-P1 | CTGATGCTGAACGCGCCA |
| Gene7a-P5 | CTGCGACGGACTTGTTCGTGA |
| 6V-P3 | CGGTAACCTGTTCAGCT |
| 6V-P5 | CCCGCTAAGCCAAGGAGTCC |
| Gene9-P5 | GGATATCGCGTGTAAGAGG |
| Gene9-P6 | TGCGTGGAGCTGATTAGG |
| 6J-inv2 | TCAGCCCATCCAGTTCACTGTA |
| 6V-PA3 | CGGCGCACAGACCACCAG |
| 6V-PA4 | CCCTAAACATGCAACTTCC |
Data analysis:
DNA sequences in the het-6OR region were obtained from the Munich Information Center for Protein Sequences (MIPS) (Schulte et al. 2002) and the Neurospora Genome database at the Broad Institute (Cambridge, MA) (Galagan et al. 2003). Gene/open reading frame designations used in this article are cross-referenced to contig and gene number at the MIPS site as: Gene 2, b2a19 020; un-24, b2a19 030; Gene 4, b2a19 040; Gene 5, b2a19 050; cys-3, b2a19 070; Gene 7a, b2a19 080; Gene 7b, b2a19 090; het-6, b2a19 100; and Gene 9, b2a19 160. BLAST searches were performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/). DNA sequencing of PA-derived clones was carried out at Canadian Molecular Research Services (Ottawa). DNA sequence alignments were done using the software ClustalX and ClustalW (http://clustalw.genome.ad.jp/; Thompson et al. 1997) with manual adjustments. The distribution of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) was calculated according to the method of Nei and Gojobori (Nei and Gojobori 1986; Korber 2000). Aligned sequences were submitted to the Synonymous/Non-synonymous Analysis Program (http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html). dS and dN were averaged for 10 codons and were plotted against codon number, and statistics were done with JMP (v5.1, SAS Institute, Cary, NC).
RESULTS
Isolation of functional PA incompatibility alleles from the het-6 region:
The localization of the het-6 locus within the T(IIL → IIIR)AR18 translocated region was previously determined by partial diploid analyses (Mylyk 1975; Smith et al. 1996). Colonies with heterozygous duplications of the region [Dp(AR18)] display inhibited growth due to het-6 self-incompatibility. Escape from self-incompatibility by Dp(AR18) colonies was associated with deletions within either the OR or the PA haplotype DNA of at least an ∼35-kbp region that includes both un-24 and het-6 (Smith et al. 2000b). However, PCR products obtained from PA strains that were equivalent to active un-24OR and het-6OR PCR products did not have incompatibility activity when transformed into OR spheroplasts, suggesting that un-24PA and het-6PA do not have incompatibility activity.
Using a different approach to isolate the PA-specific incompatibility factor(s) in the ∼35-kbp region, a λ-library of strain FGSC1131 was constructed. λ-Clones from the het-6PA region were identified by hybridization to probes corresponding to het-6OR, het-6PA, and un-24OR and a chromosome walk was done from het-6PA both toward and away from the centromere (Figure 1). Within this walk, clone λB was found to hybridize to both un-24 and het-6 and was chosen for further study. The λB insert was subcloned using NotI into pCB1004 as pλB and transformed into C9-2 (het-6OR) and C2(2)-1 (het-6PA) spheroplasts (Figure 2). pλB caused an ∼83% decrease (±16% in four independent transformation experiments) in the number of C9-2 (OR haplotype) transformants recovered as compared to the pCB1004 vector control, but no decrease in the number of transformants when introduced into PA-haplotype C2(2)-1 spheroplasts. This indicated that pλB has PA-specific incompatibility activity. Subclones of pλB were constructed and tested in transformation assays to further define the incompatibility region, as summarized in Figure 2. The incompatibility activity associated with un-24PA is notably weaker than that of un-24OR, which yields no, or few, viable colonies when transformed into un-24PA strains (Smith et al. 2000b). Construct pλBP/C contains the entire un-24PA coding region along with 550-bp upstream and 380-bp downstream regions. When transformed into OR spheroplasts, on average ∼52% (± 20% in three independent trials) fewer colonies were observed as compared to the pCB1004 vector control. Of these pλBP/C transformants, ∼63% appeared inhibited in their growth with a star-like appearance on the original transformation plates (Figure 2C). The remaining colonies had a cloud-like morphology characteristic of compatible transformants. When pλBP/C transformants were transferred to 1× Vogel's medium containing no sorbose, the star-like colonies grew significantly more slowly, did not conidiate, and displayed a “spidery” morphology whereas cloud colonies grew at wild-type rates and conidiated normally. A similar spidery morphology is typical of incompatibility due to heteroallelism at mat and het-c in N. crassa (Beadle and Coonradt 1944; Saupe et al. 1996). Early escape events or disruption of the un-24PA gene during integration of the transforming DNA may account for the cloud colonies obtained when un-24PA is transformed into OR cells. Within ∼3–6 days after transfer to Vogel's medium, the self-incompatible transformants were observed to give rise to fast-growing sectors, indicative of escape (Figure 2D). Thus, incompatibility activity is evident when un-24PA is transformed into un-24OR spheroplasts by a modest reduction in transformant numbers but a high frequency of transformants that are self-incompatible and consequently undergo escape.
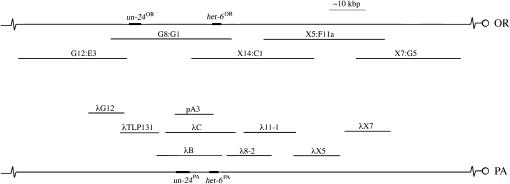
Figure 1.—
Chromosomal walks across the un-24–het-6 region for OR (top) and PA haplotypes (bottom).
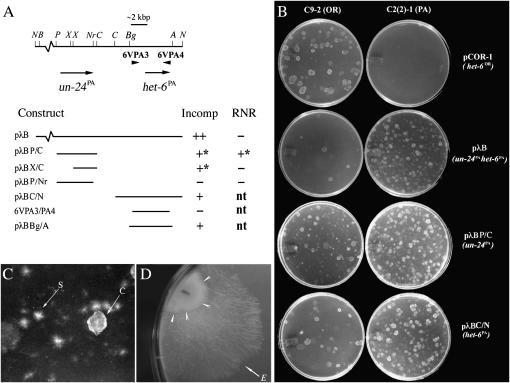
Figure 2.—
Incompatibility activity of constructs from the het-6 region. (A) Map at top gives selected restriction sites and locations of PCR primers used for making PA-haplotype constructs. Orientations of un-24PA and het-6PA are indicated by arrows. Restriction sites are A, ApaI; B, BamHI; Bg, BglII; C, ClaI; N, NotI; Nr, NarI; P, PstI; and X, XhoI. Incompatibility activity (Incomp) and ribonucleotide reductase catalytic function (RNR) for constructs are given below the map. Incompatibility activity was assayed by DNA transformations into OR [C9-2] and PA [C2(2)-1] spheroplasts and defined as strong (++), weak (+), weak with transformants having a star phenotype (+*; see text for details), or absent (−). Ribonucleotide reductase catalytic function is designated as “+” or “−” on the basis of whether or not the construct complements the un-24 temperature-sensitive mutation in C8c-164 spheroplasts. (B) Examples of DNA transformations that demonstrate strong (pCOR-1 and pλB) or weak (pλBP/C and pλBC/N) incompatibility activity. (C) Close up of star-like (“S,” self-incompatible) and cloud-like (“C,” wild type) morphologies observed when C9-2 (un-24OR) is transformed with un-24PA. (D) Fast-growing escape sector “E” evident after ∼7 days growth by a self-incompatible un-24PA transformant of C9-2 (arrowheads) following transfer from transformation plate to Vogel's medium.
Whether un-24PA complements a temperature-sensitive (ts) mutation in ribonucleotide reductase was also assayed (Figure 2A). Strain C8c-164 grows at wild-type rates at 30° but is unable to grow at 37° or above, due to a mutation that likely disrupts the allosteric activity site encoded in the 5′-end of un-24OR (Smith et al. 2000a). This ts mutant, designated un-24, was derived from un-24OR and maintains OR incompatibility activity. Transformation with wild-type un-24OR DNA enables un-24 strains to grow at 37°. When un-24PA was transformed into C8c-164 spheroplasts, growth of colonies with a star-like morphology was observed after 4–5 days of incubation at 37°, indicating that un-24PA is capable of complementing the un-24 ts mutation in spite of the PA-specific incompatibility activity. Therefore, un-24PA carries both incompatibility and catalytic functions, as does un-24OR.
Overall, the un-24PA (accession DQ525966) and un-24OR DNA sequences are 95% identical; however, they differ significantly at the 3′-end wherein the specificity region for nonself recognition is likely encoded (Smith et al. 2000b). Deletion analysis and transformation assays were used to delimit the incompatibility function of un-24PA to an ∼1.6-kbp region within the 3′-coding region (e.g., pλBX/C, Figure 2A). Loss of both incompatibility and catalytic functions are observed for the pλBP/Nr construct, which has a small 34-nucleotide region of 3′-coding sequence deleted. Similar to deletions within un-24OR (Smith et al. 2000b), the ∼1.6-kbp 3′-coding sequence with incompatibility activity does not have ribonucleotide reductase catalytic function. This provides further evidence that incompatibility function involves a mechanism that is independent of ribonucleotide reductase activity for the UN-24 protein.
Construct pλBC/N was derived from pλB and used to further test whether het-6PA has incompatibility activity (Figure 2A). This construct carries a full-length copy of het-6PA flanked by 3030 bp of 5′ upstream and 880 bp of 3′ downstream noncoding sequences. Transformation of pλBC/N into OR cells resulted in an average ∼69% (±10% in three independent trials) decrease in the number of OR transformants compared to the pCB1004 vector control but did not cause a significant decrease in the number of transformants when PA cells were used as recipients (Figure 2B). Therefore, pλBC/N was considered to carry het-6PA-associated incompatibility activity that is notably weaker than that of het-6OR, which, when introduced into PA spheroplasts, results in a nearly complete absence of transformants. Construct 6VPA3/6VPA4 spans the entire coding region of het-6PA as well as 340 bp upstream of the translation initiation site, but lacks heterokaryon incompatibility activity. In contrast, clone pλBBg/A carries an additional 1097 bp in the 5′ region upstream from the start codon and displays heterokaryon incompatibility activity. Thus, a minimum region required for het-6PA incompatibility activity encompasses ∼1100 bp upstream from the start codon and ∼110 bp downstream from the stop codon (Figure 2A). In contrast, the additional noncoding sequences outside of the 6VPA3/6VPA4 primer sites are not required for het-6OR activity, accounting for the lack incompatibility activity reported earlier for het-6PA (Smith et al. 2000b). This observation also suggests that OR and PA forms of het-6 have distinct mechanisms of gene regulation since the additional 5′- and 3′-untranslated regions required for het-6PA activity do not appear to contain additional genes.
The het-6PA sequence was reported previously (Smith et al. 2000b). The OR and PA alleles at het-6 share 78% identity at the nucleotide level and only 68% identity at the amino-acid level. Nucleotide sequence differences are spread over the entire length of the het-6 open reading frame. As is the case for het-6OR, no intron consensus sequences occur in het-6PA (Bruschez et al. 1993; Edelman and Staben 1994). The gene putatively encodes a 680-amino-acid peptide with three regions of identity to at least 13 other putative N. crassa proteins of unknown function (Blast cutoff score >38, E < 1 × 10−4; Broad Institute). The three common regions of 10, 27, and 21 amino acids in length form the HET domain identified in other heterokaryon incompatibility-associated proteins from N. crassa (TOL) and Podospora anserina (HET-E) (Smith et al. 2000b). Genes similar to het-6 are also found in Aspergillus nidulans, Magnaporthe grisea, C. parasitica, and Fusarium graminearum sequence databases, for example, but not in the Saccharomyces cerevisiae genome, suggesting that this gene family may be restricted to filamentous fungi. In addition to the HET domain, there is a region bound by amino acids 434–516 in het-6 that displays amino acid similarity (∼50%) to a domain of unknown function in bacterial aconitases (data not shown).
un-24 and het-6 show evidence of diversifying selection:
The isolation of active PA alleles at un-24 and het-6 suggests that allelic interactions are responsible for incompatibility function, rather than nonallelic interactions as previously hypothesized (Smith et al. 2000b). Therefore, the allelic specificity domains of un-24 and het-6 may be recognized by comparing the respective OR and PA forms since nonself recognition loci are often subject to diversifying selection (Apanius et al. 1997; McDowell et al. 1998; Roalson and McCubbin 2003). Under diversifying selection, substitutions leading to amino-acid changes are favored to presumably allow for broader specificity of nonself recognition during protein interactions (Hughes 1999) as exemplified with the MHC in humans (Aguilar et al. 2004), the S locus in plants (Roalson and McCubbin 2003), and the het-c locus in N. crassa (Wu et al. 1998). This is in contrast to most genes undergoing neutral or directional selection, where the rate of synonymous substitutions typically exceeds that of nonsynonymous substitutions.
We tested for indicators of diversifying selection at un-24 and at het-6 by determining the number of synonymous and nonsynonymous nucleotide substitutions in the OR and PA alleles for each gene. For this analysis, DNA sequences corresponding to the entire coding region of OR and PA alleles of both genes were used (GenBank accession nos. AF206700, AF208542, AF171697, and DQ525966). Intron sequences were removed from un-24 sequences and pairwise alignments in all combinations were done using ClustalW (http://clustalw.genome.ad.jp/), adjusted by hand and analyzed for the distribution of synonymous and nonsynonymous substitutions.
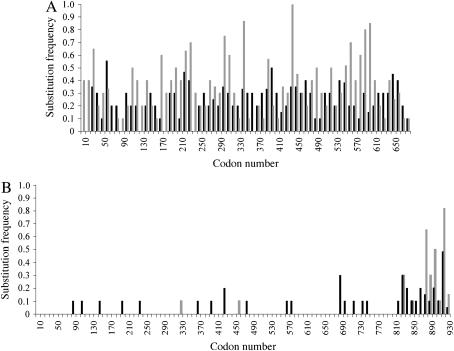
het-6 has a significantly greater frequency of nonsynonymous than synonymous substitutions throughout most of the coding region (overall dN/dS = 1.47; Figure 3A) on the basis of both parametric (two-tailed test, P < 0.001) and nonparametric (Kruskal–Wallis test, χ2 = 8.16, d.f. = 1, P = 0.004) statistics. Small islands with more synonymous than nonsynonymous substitutions are evident between codons 337 and 378, 389 and 403, and 638 and 670, and notably in the regions that correspond to part of the conserved HET domain (codons ∼53–95) and within the aconitase-like domain (codons 455–475). In contrast, two distinct patterns of nucleotide substitutions are apparent within un-24 (Figure 3B). un-24PA and un-24OR are very similar across the 5′ region of the open reading frame with only 7 of the 38 nucleotide substitutions in the first 871 codons resulting in significantly more synonymous than nonsynonymous substitutions across this region (dN/dS = 0.22; two-tailed test, P < 0.001; Kruskal–Wallis test, χ2 = 17.25, d.f. = 1, P < 0.001). This N-terminal region of un-24 is essential for ribonucleotide reductase function and is highly conserved in amino-acid sequence across taxa (Smith et al. 2000a). However, in the putative allelic-specificity domain located at the 3′-end of un-24 from codons 872 to 925, there is a strong bias toward nonsynonymous substitutions (dN/dS = 2.32; two-tailed test, P < 0.016; Kruskal–Wallis test, χ2 = 3.71, d.f. = 1, P = 0.05). Thus, both un-24 and het-6 display high levels of nonsynonymous substitutions in regions that are likely associated with heterokaryon incompatibility function, consistent with the view that diversifying selection is operating within both genes.
Figure 3.—
Frequencies of synonymous and nonsynonymous substitutions between alleles of het-6 and un-24. Plotted are the synonymous substitutions per synonymous site (dS, solid bars) and nonsynonymous substitutions per nonsynonymous site (dN, shaded bars) in comparisons between PA and OR alleles at het-6 (A) and un-24 (B) in N. crassa. dS and dN frequencies were calculated for nonoverlapping regions of 10 codons.
Fitness reduction is associated with un-24PA het-6OR and un-24OR het-6PA allelic combinations:
In addition to these patterns of nonsynonymous substitutions, un-24 and het-6 appear to be inherited as a block (either OR or PA) in population samples and across the species range (Mir-Rashed et al. 2000; Smith et al. 2000b). To explain this linkage disequilibrium, we explored the possibility that the missing allelic combinations of un-24PA het-6OR and un-24OR het-6PA may have a negative effect on fitness since vegetative incompatibility and/or sexual dysfunction have been associated with specific allelic combinations at unlinked het loci (i.e., nonallelic incompatibility) in P. anserina (Saupe 2000). We generated novel genetic combinations at un-24 and het-6 by transformation of un-24PA or het-6PA into the OR-haplotype strains C9-2 and C01(4)-2. As described above, self-incompatible transformants that are heteroallelic at un-24 or het-6 escape to wild-type growth rates within a few days after transfer to Vogel's medium. Escape from self-incompatibility results in the loss of either PA or OR incompatibility functionality and the recovery of some strains with the novel incompatibility specificities of un-24PA het-6OR and un-24OR het-6PA. The specificity of these escape strains can be inferred from heterokaryon incompatibility tests with OR and PA standard strains. In one such experiment, 15 escape strains were isolated from a transformation of strain C9-2 with un-24PA. To determine the heterokaryon incompatibility allele expressed at un-24, each escape strain was confronted in heterokaryon incompatibility tests to the OR strain C8c-32 (un-24 het-6OR). Eight of the escape strains formed vigorous heterokaryons with C8c-32 and were considered functionally un-24OR het-6OR whereas 7 escape strains produced spidery, self-incompatible heterokaryons and thus appeared to be functionally un-24PA het-6OR. All 15 escape strains were heteroallelic at un-24 on the basis of PCR analysis with primers 6JP11/6JP6 and digestion with MboI (Mir-Rashed et al. 2000). Thus, the functionally PA and OR escapes were all partial diploids with silenced incompatibility activity of either the endogenous (un-24OR) or the ectopic (un-24PA) copy, respectively. The silencing mechanism in these cases is not known. All escape strains displayed wild-type growth rates and conidiated, indicating that the novel un-24PA het-6OR allelic combination has no deleterious effect on vegetative growth. Each escape strain was subsequently mated with each of four mat-A strains C9-15 (un-24OR het-6OR), 6-13 (un-24PA het-6PA), C2(3)-25 (un-24PA het-6PA), and C2(2)-9 (un-24PA het-6PA). A majority of these crosses produced perithecia (90% of matings involving OR-compatible and 85% with OR-incompatible escape strains) and ascospores (89% matings involving OR-compatible and 87% OR-incompatible escape strains). However, ascospores derived from functionally un-24PA het-6OR parents showed a significantly lower germination frequency (22 ± 29%; 17 crosses analyzed) compared to progeny of functionally un-24OR het-6OR escapes (63 ± 17%; 9 crosses analyzed; P < 0.001, two-tailed t-test of arcsine-transformed frequencies) and this difference was observed whether or not the mating partner of the escape strain carried the OR or PA haplotype. Thus, N. crassa strains carrying the allelic combination un-24PA het-6OR are predicted to have reduced fitness, since they give rise to relatively few viable progeny.
In addition to mating capacity, we evaluated heterokaryon incompatibility characteristics of escape strains. If heterokaryon incompatibility has an adaptive function in maintaining individuality and in preventing the transmission of infectious elements, strong incompatibility reactions would presumably have a selective advantage. To assess the strength of heterokaryon incompatibility reactions, heterokaryon incompatibility tests were done with all possible allelic combinations at un-24 and het-6 (Table 3). For these experiments, functionally un-24PA het-6OR and un-24OR het-6PA strains were derived through escape as described above. The resulting heterokaryons were assessed for growth rate, colony morphology, and conidiation. As shown in Table 3, strains with the novel allelic combinations, un-24PA het-6OR and un-24OR het-6PA, displayed weak incompatibility reactions with strains that had differences at one or both het genes. These novel haplotypes are apparently compromised for nonself recognition and, in this context, individuals with these genetic combinations at un-24 and het-6 may be selected against in nature.
TABLE 3.
Relative strength of heterokaryon incompatibility reactions in strains with different allelic combinations of un-24 and het-6
| C01(4)-2 un-24 het-6OR | RLM58-18 un-24PAhet-6PA | C01-PA1 (un-24PA) het-6OR | C01-6PA un-24OR (het-6PA) | |
|---|---|---|---|---|
| C9-2 un-24ORhet-6OR | Nonea | Strongb | Weakc | Weak |
| C2(2)-5 un-24PAhet-6PA | Strong | None | Weak | Weak |
| C9-2 E18 (un-24PA) het-6OR | Weak | Weak | None | Weak |
| C9-2-6PA un-24OR (het-6PA) | Weak | Weak | Weak | None |
Escape strains C01-PA1 and C01-6PA were derived from C01(4)-2, C9-2 E18, and C9-2-6PA from C9-2. Allelic designation of the transgene expressed in the escape strain is in parentheses.
None, no heterokaryon incompatibility. Heterokaryons grow at wild-type rates of ∼30 mm/day and conidiate profusely.
Strong, pronounced incompatibility. No, or slight, growth of heterokaryons, no conidiation, escape infrequent even after ∼2 weeks.
Weak, weak incompatibility evident by slow growth rates (∼3 – 8 mm/day), abnormal “flat” morphology, irregular colony margins, and lack of conidiation. Escape to wild-type-like heterokaryons occurs rapidly, usually within ∼4 days.
Comparison of OR and PA un-24–het-6 regions:
Previously, it was suggested that linkage disequilibrium in the un-24–het-6 region may be due to sequence dissimilarity and low homologous recombination rates (Mir-Rashed et al. 2000). The isolation of pλB, encompassing both un-24 and het-6 PA alleles, allowed us to address this hypothesis through structural comparisons of OR and PA haplotypes. The cosmid G8:G1 (which spans the un-24OR–het-6OR region) and DNA sequence from contig B2A19 (MIPS, http://mips.gsf.de/proj/neurospora/) were used as reference points for the OR haplotype. λ-Clones from the FGSC1131 library were used to study the structure of the PA haplotype. pλB and pA3 (derived from FGSC1131 via FGSC2190; Smith et al. 1996; Mir-Rashed et al. 2000) were sequenced in the un-24PA–het-6PA region and used for RFLP analyses.
Southern hybridizations were done to compare the marker distribution and general structure of clones carrying PA- or OR-derived sequences (Figure 1). Hybridization membranes containing DNA from clones pA3, λB, λTLP131, and λ11-1 (PA haplotype) and G8:G1 (OR haplotype) were probed sequentially with radioactively labeled PCR products of primer pairs 6J-P11/6J-P6 (un-24OR) and cys3-P1/cys3-P2 (cys-3OR). Hybridization of the un-24OR probe to G8:G1, pA3, and λB was observed and RFLP patterns were consistent with previously published restriction maps (Mir-Rashed et al. 2000; Smith et al. 2000b). On the basis of hybridization to the cys-3OR probe, neither pA3 nor λB harbors this gene and this was verified by the absence of a PCR amplicon from either clone with cys-3-specific primers. However, cys-3 amplicons were obtained from G8:G1 (OR) and from the λTLP131 (PA) clone that maps centromere distal to un-24PA (Figure 1). Given that the intergenic distance between un-24 and het-6 is ∼5.2 kbp in the PA haplotype compared to ∼19 kbp in the OR haplotype, these results suggested the presence of an inversion in the region. This inversion, designated here as In(IIL)1131 het-6, for inversion associated with het-6, was verified by determining the DNA sequences of the termini of pA3. In pA3 (PA haplotype), un-24 and het-6 are transcribed from the same strand of DNA whereas in the OR haplotype (MIPS database) the genes are transcribed from opposite strands.
Characterization of inversion breakpoints brk-1 and brk-2:
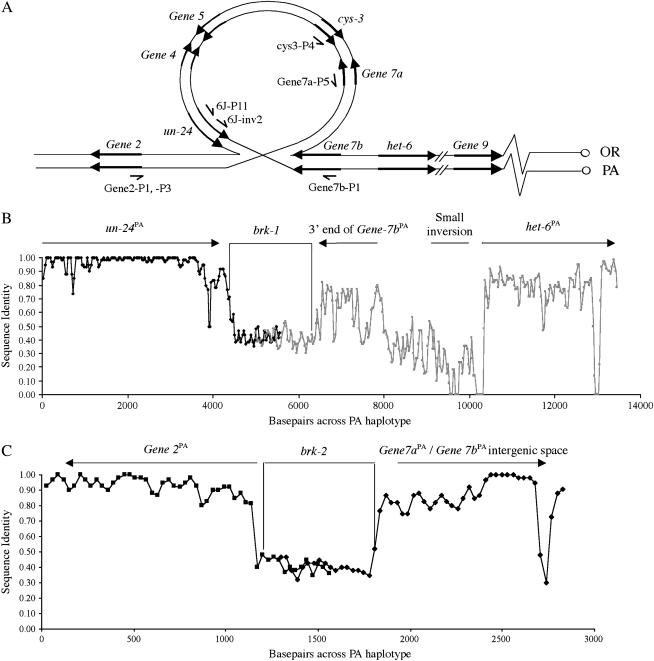
The location of the In(IIL)1131 het-6 [abbreviated as In(het-6) hereafter] breakpoints were determined by comparisons of the OR and PA haplotypes. On the basis of the map data (above), we surmised that brk-1, the centromere-proximal breakpoint, was located close to Gene7a or Gene7b with respect to the OR map (Figure 4A). The location of the distal breakpoint, brk-2, had to account for the shorter distance between un-24 and het-6 in PA relative to OR haplotypes and was inferred to be just downstream of un-24OR, likely between un-24OR and Gene2 relative to the OR map. We sought confirmation of these hypothetical maps by designing PCR primers based on the contig B2A19 sequence (MIPS) that would differentially amplify across the OR and PA configurations of brk-1 and brk-2 (Table 2 and Figure 4A). Genomic DNAs from RLM58-18 (PA) yielded amplicons with primer pairs Gene7b-P1/6J-P11 (brk-1PA) and Gene2-P3/Gene7a-P5 (brk-2PA), while 74-OR23-1V (OR) DNA produced amplicons with primers Gene7b-P1/cys3-P4 (brk-1OR) and Gene2-P3/6J-inv2 (brk-2OR). On the basis of these inversion breakpoint positions, In(het-6) spans an ∼18.6-kbp region from ∼4.2 kbp upstream of het-6 to ∼100 bp upstream of Gene 2 (based on the OR haplotype).
Figure 4.—
Sequence comparisons at In(het-6) breakpoints. Schematic (not to scale) of inversion loop expected during meiosis when In(het-6) is heterozygous (A) and sliding-window analyses of sequence identities between OR and PA haplotypes near brk-1 (B) and brk-2 (C). In A, sister chromatids are represented as a single line and primers are shown as one-sided arrows. In B and C, alignments were with ClustalX and sequence identity analysis was done with a window size of 60 bp and a window shift of 30 bp.
To characterize the inversion breakpoints in the PA haplotype, the entire pA3 clone was sequenced to cover brk-1PA, and brk-2PA was isolated by PCR from clone λG12 (Figure 1) using primers cys3-P4 and Gene2-P1, subcloned and sequenced. brk-1PA and brk-2PA sequences were compared to the OR genomic sequence available at MIPS. Sequence comparisons around the two inversion breakpoints are summarized in Figure 4 using sliding-window analyses. For the analyses, OR and PA sequences were aligned and sequence identity was calculated as the number of identical nucleotides in a window of 60 bp. Successive identity values were calculated as the window was incrementally shifted by 30 bp until the breakpoint region was covered. The inversion breakpoints are characterized by regions of sequence dissimilarity between OR and PA haplotypes of ∼2 kbp (brk-1) and 0.6 kbp (brk-2), flanked by regions of significant sequence identity. The sequence identity patterns to the right of brk-1 in Figure 4B suggested that an additional small inversion polymorphism just off the 5′-end of het-6 also differentiates the OR and PA haplotypes (Figure 4B).
A BLAST analysis of brk-1OR and brk-1PA revealed sequence identity to regions located elsewhere within the N. crassa genome. Notably, within brk-1PA, but not within brk-1OR, a 397-bp stretch of sequence shows ∼75% identity to at least 14 different regions within the N. crassa genome. This sequence also contains ∼53-bp inverted repeats that share 88% identity. Among these 14 regions, contigs 3.213 (mapping to LGVI), 3.622 (mapping to LGII), and 3.400 (LGVII) also carry similar inverted repeats (Figure 5). The remaining 11 sequences lack the inverted repeats but otherwise share significant sequence identity to the 397-bp internal sequence. In summary, this 397-bp sequence has several features that qualify it as a bona fide repetitive sequence (RS): inverted terminal repeats, short imperfect direct repeats (CCTAACC), and its presence in multiple (at least 14) copies in the genome of N. crassa. A BLAST survey indicated that neither the inverted repeats nor the internal repeated sequence matched any known mobile genetic elements or repeated sequences in the databases and that the region had no similarity to known transposases, which suggests that this is a novel RS element. Several RS elements and transposons have been implicated in chromosomal rearrangements in maize, Drosophila, and Anopheles species (Mathiopoulos et al. 1998; Cáceres et al. 1999; Zhang and Peterson 1999). In a majority of cases, inversion breakpoints were associated with mobile genetic elements, suggesting that recombination between the two homologous mobile genetic element sequences was responsible for the genetic rearrangement. The close spatial association of a repetitive genetic element, brk-1, and incompatibility genes under diversifying selection suggests a role for RS elements in the evolution of nonself recognition in N. crassa.
Figure 5.—
A 397-nucleotide sequence within brk-1PA has significant identity to ∼14 regions elsewhere in the N. crassa genome. Alignment shown is of the brk-1PA element and three sequences within contigs 3.213 (contig 7.12), 3.622 (contig 7.81), and 3.400 (contig 7.32) (Neurospora Genome Sequence Project, Broad Institute). Inverted repeats are indicated by arrows and direct repeats in the brk-1 region are underlined.
Evidence for recombination within In(het-6):
In(het-6) may partially account for some of the unusual characteristics of the un-24–het-6 region. For example, mutations may accumulate readily at inversion breakpoints due to low rates of recombination (Dobzhansky 1951) and could give rise to rapidly diverging sequences in the neighboring un-24 and het-6 sequences. Likewise, linkage disequilibrium of genes within inversions is predicted. Single crossover events between inverted/noninverted heterozygotes will cause dicentric bridges and acentric chromatid fragments during meiosis. Such aberrant chromatids are often lethal and result in an apparent absence of recombination within the inverted region (Perkins 1997). We surveyed the extent of genetic exchange between the inverted and noninverted haplotypes in PA and OR haplotype strains to test the hypothesis that In(het-6) blocks recombination in the region. In addition, we were interested in surveying the prevalence and correlation, if any, of inversion forms and genotypes at un-24 and het-6. If linkage disequilibrium at un-24 and het-6 is due to the In(het-6) polymorphism, inversion form and genotype at un-24 and het-6 are expected to be perfectly correlated. If either inversion form exists in both OR and PA haplotypes, then recombination within the un-24–het-6 region is possible as long as it occurs between identical inversion forms. Should this be the case, the inversion may not be a source of observed linkage disequilibrium at un-24 and het-6.
To explore this, 20 N. crassa strains (10 un-24OR het-6OR and 10 un-24PA het-6PA) were selected from different geographic origins to represent strains that have likely been separated by many generations (Figure 6). The het-6 and un-24 genotype of each strain was verified using a PCR–RFLP-based assay as described previously (Mir-Rashed et al. 2000). Inversion forms were determined using primers designed for the isolation of brk-1 and/or brk-2 and, in all strains tested, there was a perfect correlation between genotypes at het-6 and un-24 and inversion type (P < 0.001, Figure 6). This strongly supports the idea that the In(het-6) is responsible for linkage disequilibrium of the two incompatibility genes, het-6 and un-24.
To establish the extent of recombination within the inverted region, additional polymorphic markers (Table 4) were analyzed inside and outside of the inversion for each of the 20 strains, including the two standard strains. For all but one of these markers, Gene9, only two RFLP forms, either PA like or OR like, were detected. For Gene9, two adjacent PCR–RFLP markers identified five different variants, two corresponding to OR (O1 and OR) and three corresponding to PA (O2, O3, and PA) tester strains (Figure 6). The two Gene9 markers were congruent and are shown in a single column in Figure 6.
TABLE 4.
Diagnostic markers for allele typing in the un-24/het-6 region
| Approximate fragment size (kbp)
|
||||
|---|---|---|---|---|
| Genetic marker | Primer pair | Restriction enzyme | OR | PA |
| Gene2 | Gene2-P1 and Gene2-P2 | KpnI | 0.7, 0.2 | 0.9 |
| un-24 | 6J-P11 and 6J-P6 | PvuI | 0.75, 0.75 | 1.5 |
| Gene4-1 | Gene4-P1 and Gene4-P2 | XhoI | 0.8, 0.7 | 1.5 |
| Gene4-2 | Gene4-P1 and Gene4-P2 | KpnI | 1.1, 0.4 | 0.7, 0.4. 0.4 |
| Gene5 | Gene5-P1 and Gene5-P2 | NcoI | 2.3, 1.0, 0.3 | 2.3, 1.3, 0.4 |
| brk1-1 | 6V-P5 and 6J-P11 | None | — | 7.5 |
| brk1-2 | Gene7b-P1 and cys3-P4 | None | 4.5 | — |
| brk1-3 | Gene7b-P1 and 6J-P11 | None | — | 3.5 |
| brk2-1 | Gene2-P3 and 6J-inv2 | None | 1.8 | — |
| brk2-2 | Gene2-P3 and Gene7a-P5 | None | — | 2.3 |
| brk2-3 | Gene2-P1 and 6J-P11 | None | 5.6 | — |
| het-6 | 6V-P3 and 6V-P5 | MboI | 0.5, 0.5 | 1 |
| Gene9-1 | Gene9-P5 and Gene9-P6 | BglII | 1.9, 0.2 | 2.1 |
| Gene9-2 | Gene9-P5 and Gene9-P6 | HincII | 0.8, 0.7, 0.5 | 1.3, 0.7 |
To test whether there is a correlation between genotype at het-6 and genotype at each of the other markers, pairwise allelic associations were calculated using Fisher's exact test (Figure 6; http://www.physics.csbsju.edu/stats/exact.html; Jerrold 1984). Polymorphisms at het-6, un-24, and In(het-6) are perfectly correlated with no exceptions, as described previously. Other markers within the inversion breakpoints were significantly correlated to the allelic form of het-6 except for marker Gene4-2 (P = 0.152), which is situated toward the center of the inverted region. Markers outside the inverted region were not correlated to the het-6 genotype (P = 0.328 for Gene2, P = 0.328 for Gene9). Therefore, polymorphisms are more frequently shared between PA- and OR-haplotype strains toward the center of, and outside of, the inverted region. The presence of the shared polymorphisms at markers within In(het-6) is consistent with the view that gene conversion events and double crossovers are possible in the center of inversions, where chromosomal pairing is more efficient than at inversion breakpoints (Chovnik 1973). Apparently, as one approaches the breakpoints, and homologous pairing is not possible due to divergent sequences at each breakpoint, the correlation between markers increases to unity. Therefore, the proximity of the un-24 and het-6 incompatibility genes to the In(het-6) breakpoints, rather than the inversion per se, is responsible for linkage disequilibrium of un-24 and het-6 in N. crassa.
DISCUSSION
Significant linkage disequilibrium of loci associated with inversions is well documented in Drosophila (reviewed in Strobeck 1983), but not in Neurospora where only six pericentric inversions have been analyzed (Perkins 1997). Dobzhansky (1951) pointed out that inversions could have selective advantages in preserving gene complexes (called “supergenes” by Darlington and Mather 1949) since nondisjunction resulting from single crossovers between inverted and noninverted haplotypes will tend to suppress recombination and preserve allelic combinations of gene clusters within the inverted region. In N. crassa, a paracentric inversion, In(het-6), appears to be involved in the origin and/or maintenance of a nonself recognition gene complex consisting of un-24 and het-6. The two genes, although distinct in sequence, share a common incompatibility function and are inherited together as a functional incompatibility unit. The complete disequilibrium of these two genes is best explained by an absence of significant sequence identity in the regions between each gene and the inversion breakpoints. Thus, genes that are tightly linked to breakpoints of an inversion may be in severe linkage disequilibrium regardless of the physical distance between the genes. For example, we found that PA- and OR-specific polymorphisms at brk-1, brk-2, het-6, and un-24 are in complete linkage disequilibrium even though recombination in the central portion of In(het-6) has likely occurred.
Sequence differences in alleles of un-24 and het-6 are found mainly adjacent to the In(het-6) breakpoints and are characterized by high dN/dS ratios in un-24 and het-6, suggesting that diversifying selection is operating within the gene complex. The signature of diversifying selection is particularly apparent in the 3′ coding sequence of un-24, where the encoded protein sequences of OR and PA alleles are only 70% similar. Moving toward the 5′-end of the gene, an abrupt shift to high sequence similarity between the OR and PA alleles corresponds to the ribonucleotide reductase catalytic domain that is conserved across taxa (Smith et al. 2000a). Thus, the un-24 locus represents a battlefield between opposing selection pressures: diversifying selection acting in the incompatibility domain and purifying selection acting in the rest of the gene. Similarly, distinct selection pressures on various domains of nonself recognition loci were described in R-genes of plants and in MHC loci in vertebrates. In plants, protein domains involved in direct or indirect interactions with pathogen protein moieties [e.g., the ligand-binding leucine-rich repeat (LRR)] are hypervariable and carry the mark of diversifying selection, whereas structural domains and domains involved in host signaling are highly conserved (Noël et al. 1999; Bergelson et al. 2001). Given the conserved nature of the large subunit of ribonucleotide reductase across diverse taxa, it would appear that nonself recognition function by un-24 is a derived character in Neurospora.
The un-24–het-6 region also appears to be under balancing selection, which helps to maintain variability at nonself recognition loci over long time periods (Richman 2000). Two characteristics of balancing selection, which are not necessarily observed with diversifying selection, are balanced frequencies of alleles within populations and the preservation of specific polymorphisms across different species. Previous studies showed that the PA and OR haplotypes occur at about equal frequencies in N. crassa populations (Mir-Rashed et al. 2000; Smith et al. 2000b). In addition, evidence for linkage disequilibrium of the PA and OR haplotypes was also found in N. intermedia and N. tetrasperma (Mir-Rashed 1998; Powell 2002). These observations provide support for the trans-species maintenance of OR and PA alleles at un-24 and het-6 and for balancing selection acting on the gene complex. Significantly, the maintenance of linkage disequilibrium of the PA and OR haplotypes would suggest that In(het-6) also has a trans-species distribution.
That In(het-6) is old is suggested by two observations in addition to its inferred trans-species distribution. First is the extensive sequence divergence between PA and OR haplotypes immediately adjacent to In(het-6) breakpoints. While this may suggest an ancient haplotype split, strong diversifying selection could also result in rapid sequence divergence at the breakpoints. The second observation is that disequilibrium is limited to markers close to the inversion breakpoints. Since regional decay of linkage disequilibrium at neutral markers is expected to occur through time (Sabeti et al. 2002), the presence of shared polymorphisms in and around the inversion suggests that In(het-6) is old. A comprehensive taxonomic analysis of the distribution of In(het-6) would provide additional insight into the evolution of the incompatibility gene complex. In this context, it would be interesting to ask whether the association of un-24 with this inversion breakpoint marks the acquisition of heterokaryon incompatibility function or whether the two events are independent, with, perhaps, the inversion event locking a beneficial allelic combination of un-24 and het-6 into a supergene configuration in N. crassa. As for the former of these two possibilities, genetic rearrangements such as gene duplications, inversions, and somatic recombination are responsible for generating diversity at nonself recognition loci in other organisms. In tomato, for example, unequal crossing over between Cf-2 and Cf-5 R-genes can lead to deletions or duplications of open reading frames that affect plant disease resistance. In addition, intragenic recombination between repeated LRR domains within R-genes generates variation that directly affects R-gene function (Hammond-Kosack and Jones 1997). The MLA locus involved in powdery mildew resistance in barley is another example of a gene cluster that has undergone multiple rearrangements (Wei et al. 2002). A final extreme example is evident in somatic recombination within V, D, and J gene segments of B-lymphocytes (class switch recombination) to generate antibody diversity and thus increase nonself recognition potential in mammals (Li et al. 2004). Additional rearrangements around un-24–het-6, aside from the inversions described herein, are suggested by a general lack of synteny around the large subunit of ribonucleotide reductase among N. crassa and the other Sordariomycetes, F. graminearum, Chaetomium globosum, and M. grisea (S. Qadri and M. L. Smith, unpublished results).
An argument in favor of the second possibility—that the inversion locked together a beneficial combination of preexisting alleles—takes into consideration that a striking characteristic of the un-24–het-6 gene complex is its symmetric incompatibility function, as opposed to the asymmetry of activities by either gene alone. The molecular basis for this asymmetry is not known, but it seems to be a common feature of heterokaryon incompatibility genes (Williams and Wilson 1966; Biella et al. 2002), and a transition from asymmetric to symmetric incompatibility may have driven the establishment of the un-24–het-6 incompatibility gene complex. Recent studies support the idea that heterokaryon incompatibility is a gatekeeper of fungal individuality. Fungal heterokaryon incompatibility genes were shown to cause cell death in a structured, organized manner analogous to PCD (Biella et al. 2002; Dementhon et al. 2003; Glass and Kaneko 2003). Furthermore, incompatibility is negatively correlated to the probability of transmission of viruses in the chestnut blight fungus, C. parasitica (Biella et al. 2002). In this sense, cell death caused by heterokaryon incompatibility is analogous to the hypersensitive response in plants that limits pathogen infection and to the apoptotic response induced by killer T lymphocytes in response to viral infection (Kagi et al. 1996; Greenberg and Yao 2004). Here, however, heterokaryon incompatibility represents a preventive strategy in the battle against viral infection, rather than a direct response to pathogen attack. One would predict that strong incompatibility reactions would be more effective in limiting the transfer of infectious elements between incompatible individuals of Neurospora, as was observed for C. parasitica (Biella et al. 2002). Assuming that there is a selective advantage to incompatibility, then a gene complex that provides a strong, symmetric incompatibility reaction could provide the selective force to maintain In(het-6) as a balanced polymorphism.
The adaptive significance of the un-24–het-6 gene complex requires further investigation. In particular, the observation that escape strains that were functionally un-24PA het-6OR have low fitness, as measured by the frequency of viable ascospore progeny produced, suggests that specific interactions may have developed within the respective OR and PA gene complexes. These interactions apparently do not result in incompatibility of particular combinations of un-24 and het-6, as is observed with nonallelic heterokaryon incompatibility loci in P. anserina, for example (Saupe 2000), since, in this study, un-24PA het-6OR escape strains do not exhibit slow growth or reduced conidiation, two characteristics of self-incompatibility (Newmeyer and Galeazzi 1977; Smith et al. 1996). The low fecundity of un-24PA het-6OR escapes is also distinct from the barren phenotype observed when Neurospora strains with segmental duplications are mated to wild-type strains. In contrast to the barren condition, all un-24PA het-6OR escapes that we tested produced abundant perithecia and ascospores after mating. However, unlike functionally un-24OR het-6OR escape strains, in which the un-24PA copy is silenced, the un-24PA het-6OR escapes, which have the un-24OR copy silenced, produce a significantly lower frequency of spores that are viable. Furthermore, the inability of escape strains with either un-24PA het-6OR or un-24OR het-6PA phenotypes to undergo strong incompatibility reactions with any of the other haplotypes suggests that these combinations are also impaired in nonself recognition function. Such strains could provide a hub for the propagation of infectious elements in the population at large and may have been selected against.
Finally, the repetitive element noted in the vicinity of brk-1 has interesting implications for the evolution of incompatibility function. Similar elements are often associated with genome instability and rearrangement (Kazazian and Goodier 2002). For example, a variety of transposon types are associated with inversion breakpoints in Drosophila (Galileo, Cáceres et al. 1999; Hobo, Lyttle and Haymer 1993), mosquito (Odysseus, Mathiopoulos et al. 1998), and maize (Ac, Zhang and Peterson 1999). Inversions can occur by homologous recombination between two similar mobile elements situated on the same chromosome or may be the secondary consequence of a double-stranded break associated with a transposition event. It is therefore possible that the putative repetitive element associated with In(het-6) may have had a direct involvement in the inversion event. The incidence of a putative repetitive element between brk-1 and het-6, which itself is found in multiple copies in Neurospora and other genomes, is particularly interesting since it suggests that there may be a direct role for mobile elements in the origin of nonself recognition in fungi as is now hypothesized for mosquitoes (Sinkins et al. 2005).
Acknowledgments
The work described in this article was made possible through a Discovery Grant to M.L.S. from the Natural Sciences and Engineering Research Council of Canada.
References
- Aguilar, A., G. Roemer, S. Debenham, M. Binns, D. Garcelon et al., 2004. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl. Acad. Sci. USA 101: 3490–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanius, V., D. Penn, P. R. Slev, L. R. Ruff and W. K. Potts, 1997. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 17: 179–224. [DOI] [PubMed] [Google Scholar]
- Beadle, G. W., and V. I. Coonradt, 1944. Heterokaryosis in Neurospora crassa. Genetics 29: 291–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Biella, S., M. L. Smith, J. R. Aist, P. Cortesi and M. G. Milgroom, 2002. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. R. Soc. Lond. B Biol. Sci. 269: 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschez, J. J. P., J. Eberle and V. E. A. Russo, 1993. Regulatory sequences in the transcription of Neurospora crassa genes. Fungal Genet. Newsl. 40: 89–96. [Google Scholar]
- Cáceres, M., J. A. Ranz, A. Barbadilla, M. Long and A. Ruiz, 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418. [DOI] [PubMed] [Google Scholar]
- Carrol, A. M., J. A. Swiegard and B. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41: 22. [Google Scholar]
- Caten, C. E., 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72: 221–229. [DOI] [PubMed] [Google Scholar]
- Chovnik, A., 1973. Gene conversion and transfer of genetic information within the inverted region of inversion heterozygotes. Genetics 75: 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesi, P., C. E. McCulloch, H. Song, H. Lin and M.G. Milgroom, 2001. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 159: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington, C. D., and K. Mather, 1949. The Elements of Genetics. Macmillan, New York.
- Davis, R. H., and F. J. de Serres, 1970. Genetic and microbiological techniques for Neurospora crassa. Methods Enzymol. 17A: 79–143. [Google Scholar]
- Debets, A. J. M., and A. J. F. Griffiths, 1998. Polymorphism of het genes prevents resource plundering in Neurospora crassa. Mycol. Res. 102: 1343–1349. [Google Scholar]
- Dementhon, K., M. Paoletti, B. Pinan-Lucarré, N. Loubradou-Bourges, M. Sabourin et al., 2003. Rapamycin mimics the incompatibility reaction in the fungus Podospora anserina. Eukaryot. Cell 2: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1951. Genetics and the Origin of Species, Ed. 3, revised. Columbia University Press, New York.
- Edelman, S., and C. Staben, 1994. A statistical analysis of sequence features within genes from Neurospora crassa. Exp. Mycol. 18: 70–81. [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Glass, N. L., and I. Kaneko, 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 1: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, N. L., D. J. Jacobson and P. K. Shiu, 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34: 165–186. [DOI] [PubMed] [Google Scholar]
- Greenberg, J. T., and N. Yao, 2004. The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6: 201–211. [DOI] [PubMed] [Google Scholar]
- Grosberg, R. K., and J. F. Quinn, 1986. The genetic control and consequences of kin recognition by the larvae of a colonial marine invertebrate. Nature 322: 456–459. [Google Scholar]
- Hammond-Kosack, K. E., and J. D. G. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., E. R. Dempster and S. R. Brown, 1975. Adaptive significance of vegetative incompatibility in Neurospora crassa. Genetics 81: 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. L., 1999. Adaptive Evolution of Genes and Genomes. Oxford University Press, New York.
- Hughes, A. L., and M. Yeager, 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32: 415–435. [DOI] [PubMed] [Google Scholar]
- Jerrold, H. Z., 1984. Biostatistical Analysis, Ed. 2. Prentice-Hall, Engelwood Cliffs, NJ.
- Kagi, D., B. Ledermann, K. Burki, R. M. Zinkernagel and H. Hengartner, 1996. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu. Rev. Immunol. 14: 207–232. [DOI] [PubMed] [Google Scholar]
- Kazazian, H. H., and J. L. Goodier, 2002. LINE drive: retrotransposition and genome instability. Cell 110: 277–280. [DOI] [PubMed] [Google Scholar]
- Korber, B., 2000. HIV signature and sequence variation analysis, pp. 55–72 in Computational Analysis of HIV Molecular Sequences, edited by A. G. Rodrigo and G. H. Learn. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Li, Z., C. J. Woo, M. D. Iglesias-Ussel, D. Ronai and M. D. Scharff, 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18: 1–11. [DOI] [PubMed] [Google Scholar]
- Lyttle, T. W., and D. S. Haymer, 1993. The role of the transposable element hobo in the origin of endemic inversions in wild populations of Drosophila melanogaster, pp. 201–214 in Transposable Elements and Evolution, edited by J. F. McDonald. Kluwer Academic Publishers, Dordrecht, The Netherlands. [DOI] [PubMed]
- Mathiopoulos, K. D., A. Della Tore, V. Predazzi, V. Petrarca and M. Coluzzi, 1998. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc. Natl. Acad. Sci. USA 95: 12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J. M., M. Dhandaydham, T. A. Long, M. G. Aarts, S. Goff et al., 1998. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, O. C., and M. L. Smith, 2005. Biological concepts of vegetative self and nonself recognition in fungi, pp. 63–86 in Evolutionary Genetics of Fungi, edited by J. Xu. Horizon Bioscience, Norfolk, UK.
- Mir-Rashed, N., 1998. Molecular and functional analyses of incompatibility genes at het-6 as genetic markers in populations of Neurospora. M.Sc. Thesis, Carleton University, Ottawa. [DOI] [PubMed]
- Mir-Rashed, N., D. J. Jacobson, M. R. Dehghany, O. C. Micali and M. L. Smith, 2000. Incompatibility genes at het-6 as genetic markers in a population of Neurospora crassa. Fungal Genet. Biol. 30: 197–205. [DOI] [PubMed] [Google Scholar]
- Mylyk, O. M., 1975. Heterokaryon incompatibility genes in Neurospora crassa detected using duplication-producing chromosome rearrangements. Genetics 80: 107–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D., and D. R. Galeazzi, 1977. The instability of Neurospora duplication Dp(IL IR)H4250, and its genetic control. Genetics 56: 461–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L., T. L. Moores, E. A. van der Biezen, M. Parniske, M. J. Daniels et al., 1999. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Oakley, C. E., C. F. Weil, P. L. Kretz and R. B. Oakley, 1987. Cloning of the ribo B locus of Aspergillus nidulans. Gene 53: 293–298. [DOI] [PubMed] [Google Scholar]
- Paoletti, M., K. W. Buck and C. M. Brasier, 2006. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol. Ecol. 15: 249–262. [DOI] [PubMed] [Google Scholar]
- Perkins, D. D., 1988. Main features of vegetative incompatibility in Neurospora. Fungal Genet. Newsl. 35: 44–46. [Google Scholar]
- Perkins, D. D., 1997. Chromosome rearrangements in Neurospora and other filamentous fungi. Adv. Genet. 36: 239–398. [DOI] [PubMed] [Google Scholar]
- Powell, A. J., 2002. Non-self recognition and population biology in Neurospora tetrasperma. Ph.D. Thesis, University of New Mexico, Albuquerque, NM.
- Richman, A., 2000. Evolution of balanced genetic polymorphism. Mol. Ecol. 9: 1953–1963. [DOI] [PubMed] [Google Scholar]
- Roalson, E. H., and A. G. McCubbin, 2003. S-RNAses and sexual incompatibility: structure, functions and evolutionary perspectives. Mol. Phylogenet. Evol. 29: 490–506. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. P. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. E., F. Fritsch and T. Maniatis, 1989. Molecular Cloning, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saupe, S. J., 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64: 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S. J., G. A. Kuldau, M. L. Smith and N. L. Glass, 1996. The product of the het-c heterokaryon incompatibility gene of Neurospora crassa has characteristics of a glycine-rich cell wall protein. Genetics 143: 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte, U., I. Becker, H. W. Mewes and G. Mannhaupt, 2002. Large scale analysis of sequences from Neurospora crassa. J. Biotechnol. 94: 3–13. [DOI] [PubMed] [Google Scholar]
- Sinkins, S. P., T. Walker, A. R. Lynd, A. R. Steven, B. L. Makepeace et al., 2005. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 436: 257–260. [DOI] [PubMed] [Google Scholar]
- Smith, M. L., C. J. Yang, R. L. Metzenberg and N. L. Glass, 1996. Escape from het-6 incompatibility in Neurospora crassa partial diploids involves preferential deletion within the ectopic segment. Genetics 144: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. L., S. P. Hubbard, D. J. Jacobson, O. C. Micali and N. L. Glass, 2000. a An osmotic-remedial temperature-sensitive mutation in the allosteric activity site of ribonucleotide reductase in Neurospora crassa. Mol. Gen. Genet. 262: 1022–1035. [DOI] [PubMed] [Google Scholar]
- Smith, M. L., O. C. Micali, S. P. Hubbard, N. Mir-Rashed, D. J. Jacobson et al., 2000. b Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics 155: 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck, C., 1983. Expected linkage disequilibrium for a neutral locus linked to a chromosomal arrangement. Genetics 103: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F., R. A. Wing and R. P. Wise, 2002. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14: 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C. A., and J. F. Wilson, 1966. Cytoplasmic incompatibility reactions in Neurospora crassa. Ann. NY Acad. Sci. 129: 853–863. [Google Scholar]
- Wu, J., S. J. Saupe and N. L. Glass, 1998. Evidence of balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl. Acad. Sci. USA 95: 12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and T. Peterson, 1999. Genome rearrangement by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]