Abstract
The goal of linkage mapping is to find the true order of loci from a chromosome. Since the number of possible orders is large even for a modest number of loci, the problem of finding the optimal solution is known as a NP-hard problem or traveling salesman problem (TSP). Although a number of algorithms are available, many either are low in the accuracy of recovering the true order of loci or require tremendous amounts of computational resources, thus making them difficult to use for reconstructing a large-scale map. We developed in this article a novel method called unidirectional growth (UG) to help solve this problem. The UG algorithm sequentially constructs the linkage map on the basis of novel results about additive distance. It not only is fast but also has a very high accuracy in recovering the true order of loci according to our simulation studies. Since the UG method requires n − 1 cycles to estimate the ordering of n loci, it is particularly useful for estimating linkage maps consisting of hundreds or even thousands of linked codominant loci on a chromosome.
ALTHOUGH more and more genomes are being sequenced, high-quality linkage maps for many organisms remain useful. For a majority of organisms, obtaining appropriate linkage maps will be a necessary step for understanding their genetic architecture. With the advance of technology as well as increasing demand of high-density linkage maps, the number of loci simultaneously examined in experiments is steadily growing, which presents a considerable computational challenge for estimating the underlying linkage map since the number of possible orders of loci increases rapidly with the number of loci (Olson and Boehnke 1990; Mester et al. 2003). The construction of linkage maps has been recognized as a special case of the traveling salesman problem (TSP) (Liu 1998). The TSP is a classical non-deterministic polynomial time (NP)-complete problem (Wilson 1988; Olson and Boehnke 1990; Falk 1992) that has attracted the attention of mathematicians and computer scientists. Currently, there are two approaches to tackle the problem. One is to find the answer by performing exhaustive searches and the other is to find approximations. Even for just 10 loci on a chromosome, there are 1,814,400 possible orders. Thus it is extremely time consuming (if not entirely impossible) to exhaustively search all possible orders when the number of loci on a chromosome is >30 (Mester et al. 2003). Therefore algorithms to obtain approximate optimal solutions are the only practical approach for large-scale linkage mapping (Liu 1998). To date, several approximation algorithms are available, including seriation (Buetow and Chakravarti (1987), simulated annealing (SA) (Thompson 1984; Weeks and Lange 1987), branch and bound (BB) (Lathrop et al. 1985), Lander–Green (LG) algorithm (Lander and Green 1987), and stepwise likelihood (Lathrop et al. 1984). Many of these algorithms have been implemented in software packages such as LINKAGE (Lathrop et al. 1984), MAPMAKER/EXP (Lander et al. 1987), LINKAGE MAP (Eppig and Eicher 1983), JoinMap (Stam 1993), LINKAGE-1 (Suiter et al. 1983), GMendel (Echt et al. 1992), and PGRI (Lu and Liu 1995). Recently Mester et al. (2003) proposed a promising genetic and evolutionary algorithm (GEA) for constructing large-scale genetic maps. The GEA searches for optimal solutions adaptively by mimicking the evolutionary process of a population that includes mutation, recombination, and selection. In addition to the GEA, Mester et al. (2003) also combined their GEA with the 2-Opt or 3-Opt (Lin and Kernighan 1973) to obtain a procedure called evolutionary strategy (ES).
All the approximate algorithms employ certain criteria to search for an optimal order of a given set of loci. Proposed criteria include Lalouel's least squares (Jensen and Jorgensen 1975; Weeks and Lange 1987), sum of adjacent recombination fractions (SARF) (Falk 1992), product of adjacent recombination fractions (PARF) (Wilson 1988), probability of double recombinants (PDR) (Knapp et al. 1989), sum of adjacent LOD score (SALOD) (Weeks and Lange 1987), and SALOD divided by the equivalent number of informative meioses (SALEQ) (Edwards 1971). Olson and Boehnke (1990) compared these criteria and concluded that SARF and SALEQ were the best overall criteria. These two criteria are derived on the basis of the assumption that the true order of a set of loci has a minimum SARF or maximum SALOD. Although the principle appears sound, their performance may be affected by experimental errors, sample size, interference of recombination, and double crossovers (Mester et al. 2003). In general, the computation of this type of algorithm remains very time consuming.
An alternative approach is to construct the linkage map sequentially. That is, start with a small map and add loci into it one at a time. Ellis (1997) proposed such an algorithm called neighbor mapping (NM), which used ideas similar to the neighbor-joining (NJ) method (Saitou and Nei 1987) for phylogeny reconstruction. One advantage of NM is its speed; unfortunately, its accuracy is not particularly high. The purpose of this article is to present a novel sequential approach called unidirectional growth (UG), which has all the advantages of the NM method but with much higher accuracy in recovering the true order of a given set of linked loci.
THEORY AND ALGORITHM
Consider a set of loci whose order in a chromosome is unknown. We are interested in estimating this unknown order of loci from distances defined between each pair of loci. A map of a given nonempty set of loci is defined throughout this article as an ordered list of some or all of the loci in the set. Given a set of loci, the smallest map has only one locus and the largest map includes all the loci available. The largest map is also called a complete map while a smaller map is called a partial map. Graphically a map is conveniently represented as a list of symbols separated by hyphens. For example, both x-y-z and y-z-x are maps of the three loci x, y, and z. A correct map of a set of loci is a map such that the order of the loci in the map is the same as the true order. Each of the original loci is regarded as a simple locus and a partial map is regarded as a composite locus.
The approach we advocate for estimating the map of a given set of loci requires measures of closeness between each pair of loci, which are referred to as distance. Let 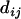 represent the distance between two loci i and j. Then the distance is said to be additive if
represent the distance between two loci i and j. Then the distance is said to be additive if 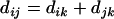 for every three loci (from the locus set) such that locus k lies between the two. The novel algorithm to be described stemmed from two theorems about additive distance.
for every three loci (from the locus set) such that locus k lies between the two. The novel algorithm to be described stemmed from two theorems about additive distance.
Theorem 1. For a set of n loci with distance dij between loci i and j, define
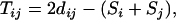 |
(1) |
where
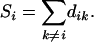 |
(2) |
Suppose that the true linkage map is 1-2-3-…-n; then
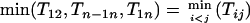 |
(3) |
if the distance is additive (see appendix a for proof).
This theorem indicates that for an additive distance at least one terminal locus of the map of a set of loci can be determined through Equation 3. Furthermore, we have
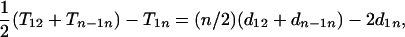 |
(4) |
which is clearly negative when loci are equally spaced. If the spacing between loci is random, then 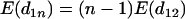 , so the expected value of the above expression is also negative. Therefore, there is a large chance that one of the
, so the expected value of the above expression is also negative. Therefore, there is a large chance that one of the 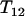 and
and 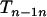 is smaller than
is smaller than 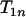 particularly with increasing n, which suggests that one terminal of the complete map (i.e., an end of the complete map) is likely found through Equation 3. When
particularly with increasing n, which suggests that one terminal of the complete map (i.e., an end of the complete map) is likely found through Equation 3. When 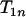 is the smallest among the three, it will lead to the identification through Equation 3 of both terminal loci.
is the smallest among the three, it will lead to the identification through Equation 3 of both terminal loci.
For a sequential algorithm for estimating a map on the basis of distance, it is necessary to update the distances during the process of reconstructing the complete map. It is highly desirable to maintain additivity for the updated distances. For a set L = {1, 2,…, n} of loci with additive distance 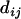 , if x-y is a terminal map, then fuse x and y into a composite locus (xy) and define its distance to a simple locus i as
, if x-y is a terminal map, then fuse x and y into a composite locus (xy) and define its distance to a simple locus i as
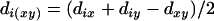 |
or
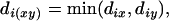 |
which will retain additivity for the updated distances, which can be proved as follows. Without loss of generality, assume that x is the terminal locus of the map. Then, because of additivity, 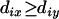 and
and 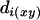 =
= 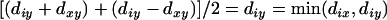 . Therefore, the updated distance defined on the set L-{x, y} + {(xy)} is the same as the original distance defined on the set L-{x}. Since x is the terminal locus, additivity is thus retained.
. Therefore, the updated distance defined on the set L-{x, y} + {(xy)} is the same as the original distance defined on the set L-{x}. Since x is the terminal locus, additivity is thus retained.
Taking advantage of the above results, a complete map can be reconstructed by first determining a terminal of the map and then growing the map sequentially by adding one locus at a time. The following theorem provides the basis for a strategy to extend a partial map (see appendix b for proof).
Theorem 2. Suppose that the true map of n loci is 1-2- …-n. Then for additive distance 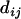 ,
,
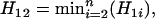 |
(5) |
where
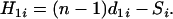 |
(6) |
The algorithm:
The above two theorems naturally lead to an algorithm for estimating the map of a given set of loci sequentially. In a nutshell, the algorithm first searches for one end of the map and then adds an adjacent locus to the map one at a time until it is completed. Because once a terminal is determined, the direction with which the map grows remains unchanged, we refer to this novel approach as the UG algorithm. The detailed steps of the UG algorithm are as follows:
- Step 1: Determine a terminal map and growth direction. Compute
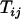 for all i < j. The pair of loci x and y that result in the smallest T-value is taken as a terminal map x-y, which is designated as locus
for all i < j. The pair of loci x and y that result in the smallest T-value is taken as a terminal map x-y, which is designated as locus 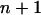 . The distance between locus
. The distance between locus 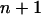 and a simple locus i is defined as
and a simple locus i is defined as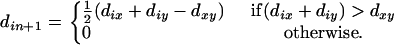
(7) - Compute with the newly defined distance
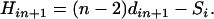
(8) The locus z that minimizes the value of
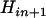 is chosen as the next locus to be added to the terminal map. The resulting partial map (the n + 2 locus) will be x-y-z if
is chosen as the next locus to be added to the terminal map. The resulting partial map (the n + 2 locus) will be x-y-z if 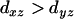 or z-x-y otherwise. The first situation indicates that all the subsequent growths will be from the left to the right and the second from the right to the left. Assign k = 2 and proceed to the following steps.
or z-x-y otherwise. The first situation indicates that all the subsequent growths will be from the left to the right and the second from the right to the left. Assign k = 2 and proceed to the following steps.- Step 2: Update distance. Compute the distance between composite locus
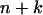 and a simple locus i as
and a simple locus i as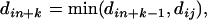
(9) where j is the simple locus that leads to the composite locus n + k by its inclusion to the n + k − 1 composite locus.
- Step 3: Grow map. Compute
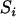 for each simple locus i with updated distance and
for each simple locus i with updated distance and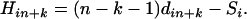
(10) The locus that gives the smallest H-value is then added to the partial map. The resulting new partial map is designated as locus n + k + 1.
Step 4: Repeat steps 2 and 3 until a complete map is obtained.
DEFINING DISTANCES BETWEEN LOCI FOR USE WITH THE UG ALGORITHM
The theory and algorithm established above calls for additive distance between loci. In general, distance between loci has to be estimated from relevant experimental data, which are often the frequencies of recombination between each pair of loci observed from comparing genotypes from the offspring to those from their parents. Estimate 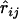 of the recombination fraction
of the recombination fraction 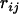 between loci i and j, which typically can be obtained by the EM algorithm (Liu 1998). Immediately a distance between a pair of loci can be defined as
between loci i and j, which typically can be obtained by the EM algorithm (Liu 1998). Immediately a distance between a pair of loci can be defined as 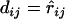 . Such a distance should be reasonable when the recombination fraction between the pair of loci is small. When the recombination fraction is sufficiently high between two loci, a double crossover may occur frequently, which typically leaves no trace in the data. As a result, the observed recombination frequencies between loci of high recombination rate may be downwardly biased. Therefore a correction is desirable to achieve a distance that is more addable.
. Such a distance should be reasonable when the recombination fraction between the pair of loci is small. When the recombination fraction is sufficiently high between two loci, a double crossover may occur frequently, which typically leaves no trace in the data. As a result, the observed recombination frequencies between loci of high recombination rate may be downwardly biased. Therefore a correction is desirable to achieve a distance that is more addable.
Suppose that 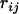 is the recombination frequency between loci i and j. If locus k lies between the two, then according to the three-point analysis (Kosambi 1944; Liu 1998),
is the recombination frequency between loci i and j. If locus k lies between the two, then according to the three-point analysis (Kosambi 1944; Liu 1998), 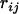 can be expressed as
can be expressed as
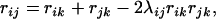 |
(11) |
where 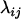 is a constant known as the coefficient of coincidence, which is defined as
is a constant known as the coefficient of coincidence, which is defined as 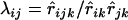 . Note that
. Note that 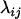 = 1 corresponds to the classical case of crossover independence (Haldane 1919) and
= 1 corresponds to the classical case of crossover independence (Haldane 1919) and 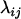 = 2
= 2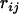 to the case of crossover interference (Kosambi 1944). One obvious corrected distance between loci i and j is defined as
to the case of crossover interference (Kosambi 1944). One obvious corrected distance between loci i and j is defined as
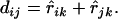 |
(12) |
The problem is that in general one does not know in advance which locus lies between loci i and j. Therefore to make use of Equation 12, we need to determine if a given locus should be considered as one between loci i and j. It appears that a minimal criterion is that 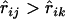 and
and 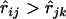 . Since there may be multiple loci lying between the loci i and j, and for each such locus k,
. Since there may be multiple loci lying between the loci i and j, and for each such locus k, 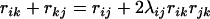 , we therefore define a distance between loci i and j as
, we therefore define a distance between loci i and j as
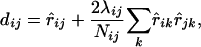 |
(13) |
where the summation is taken over all loci k, which satisfies 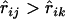 and
and 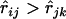 , and
, and 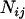 is the number of such loci. The definition implies that
is the number of such loci. The definition implies that 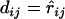 when there is no locus that appears to be between loci i and j.
when there is no locus that appears to be between loci i and j.
We found that letting 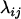 = 1 in Equation 13 works quite well so throughout this article
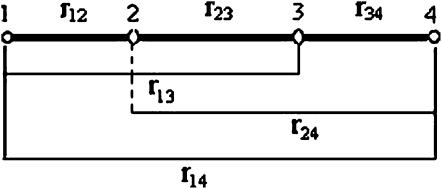
= 1 in Equation 13 works quite well so throughout this article 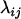 is assumed to be 1 for all loci considered. Figure 1 shows an example of the computation of distance
is assumed to be 1 for all loci considered. Figure 1 shows an example of the computation of distance 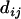 computed from Equation 13. In this example we have
computed from Equation 13. In this example we have 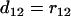 since no other locus satisfies the condition
since no other locus satisfies the condition 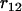 >
> 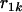 and
and 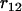 >
> 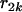 . For loci 1 and 3, we have
. For loci 1 and 3, we have 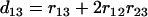 since
since 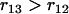 and
and 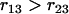 . Similarly, we have
. Similarly, we have 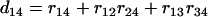 ,
, 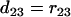 ,
, 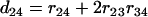 , and
, and 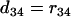 .
.
Figure 1.—
An example of a linkage map of four linked loci in which there are three adjacent intervals (1-2, 2-3, and 3-4) and three nonadjacent intervals (1-3, 2-4, and 1-4).
NUMERICAL EXAMPLES
We first illustrate the UG algorithm using a hypothetic data set (see Table 1) that was generated from the map (1-4-5-2-6-3-7) in which all the loci are codominant and the distances between adjacent loci are all 10 cM. As the algorithm indicates, the first step is to find a terminal map through Equation 3. It turns out that 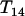 is (see Table 2) the smallest T-value (below the diagonal in Table 2). Therefore according to the algorithm the terminal map is 1-4, which is also designated as composite locus 8. After computing the distance between locus 8 and each of the remaining simple loci using Equation 7, it is found from Table 3 and Equation 8 that locus 5 is the next locus to be added to the map. Since
is (see Table 2) the smallest T-value (below the diagonal in Table 2). Therefore according to the algorithm the terminal map is 1-4, which is also designated as composite locus 8. After computing the distance between locus 8 and each of the remaining simple loci using Equation 7, it is found from Table 3 and Equation 8 that locus 5 is the next locus to be added to the map. Since 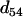 = 0.1 <
= 0.1 < 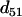 = 0.22, the map grows into 1-4-5, which is designated as locus 9. This completes step 1 of the algorithm. In the second step,
= 0.22, the map grows into 1-4-5, which is designated as locus 9. This completes step 1 of the algorithm. In the second step, 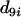 (i = 2, 3, 6, 7) is computed using Equation 9, and in the third step
(i = 2, 3, 6, 7) is computed using Equation 9, and in the third step 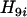 is computed using Equation 10. It turns out that
is computed using Equation 10. It turns out that 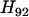 is the smallest (see Table 3), which leads to the partial map 1-4-5-2, which is designated as locus 10. Repeating steps 2 and 3, it is found in Table 3 that the next locus to be added is locus 6, then locus 3, and finally locus 7. The complete map is thus estimated to be 1-4-5-2-6-3-7, which is the same as the true map.
is the smallest (see Table 3), which leads to the partial map 1-4-5-2, which is designated as locus 10. Repeating steps 2 and 3, it is found in Table 3 that the next locus to be added is locus 6, then locus 3, and finally locus 7. The complete map is thus estimated to be 1-4-5-2-6-3-7, which is the same as the true map.
TABLE 1.
Recombination fractions between loci of the linkage map 1-4-5-2-6-3-7, where each of the adjacent intervals was assigned to be 10 cM
| Locus
|
||||||
|---|---|---|---|---|---|---|
| Locus | 1 | 2 | 3 | 4 | 5 | 6 |
| 2 | 0.30 | |||||
| 3 | 0.49 | 0.20 | ||||
| 4 | 0.10 | 0.20 | 0.40 | |||
| 5 | 0.20 | 0.10 | 0.30 | 0.10 | ||
| 6 | 0.40 | 0.10 | 0.10 | 0.30 | 0.20 | |
| 7 | 0.50 | 0.30 | 0.10 | 0.49 | 0.40 | 0.20 |
TABLE 2.
Distances (above diagonal) and T-values (below diagonal) for the seven loci specified in Table 1
| Locus
|
|||||||
|---|---|---|---|---|---|---|---|
| Locus | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 1 | 0.34 | 0.59 | 0.10 | 0.22 | 0.47 | 0.64 | |
| 2 | −2.9959 | 0.22 | 0.22 | 0.10 | 0.10 | 0.34 | |
| 3 | −2.9925 | −2.6967 | 0.47 | 0.34 | 0.10 | 0.10 | |
| 4 | −3.9725 | −2.6967 | −2.7000 | 0.10 | 0.34 | 0.59 | |
| 5 | −3.3625 | −2.5667 | −2.5833 | −3.0633 | 0.22 | 0.47 | |
| 6 | −2.8692 | −2.5667 | −3.0633 | −2.5833 | −2.4533 | 0.22 | |
| 7 | −3.4333 | −2.9959 | −3.9725 | −2.9925 | −2.8692 | −3.3625 | |
TABLE 3.
UG procedure for mapping the seven loci
| Linked locus (+) and remaining loci in cycle k
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Terminal locus | Growing point | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 8 | 1-4 | + | 0.16 | 1.16 | + | −0.69 | 0.78 | 1.13 |
| 9 | 8-5 | −0.36 | 0.60 | + | 0.24 | 0.75 | ||
| 10 | 9-2 | + | 0.24 | −0.12 | 0.36 | |||
| 11 | 10-6 | 0.0 | + | 0.12 | ||||
| 12 | 11-3 | + | 0.0 | |||||
| 12-7 | + | |||||||
To see how the UG algorithm performs in reality, we applied it to a real data set of 26 loci on barley chromosome I from the North American Barley Genome Mapping Project (see Liu 1998, p. 288). For the purpose of comparison, we applied both the UG and the NM algorithms to this data set, which yielded maps A and C, respectively, in Figure 2. Also included is the map (B) based on 1000 bootstrap samples from Liu (1998, p. 297) in statistical genomics-linkage, mapping, and QTL analysis. It is obvious that maps A and B are almost identical. The only notable difference is the positions of three pairs of loci. For the given observed data set, map A appears to be reasonable because the recombination fraction between loci BCD265B and Dhn6 is 7.97, which is larger than those between loci TubA1 and BCD265B (0.75) and between TubA1 and ABG3 (4.8). A similar situation also occurs among loci ABA3, ABG484, Pgk1, and ABR315. In comparison, there are considerable differences between maps B and C. It is noteworthy that sums of adjacent distances on linkage maps A, B, and C are, respectively, 132.8, 148.5, and 151 cM, which suggests that linkage map A is the best among these three linkage maps according to the principle of minimum SARF (Falk 1992; Liu 1998).
Figure 2.—
Comparison among three linkage maps. Maps A and C were estimated using the UG and NM methods, respectively, on the basis of a recombination fraction data set of 26 loci from Liu (1998) and map B was from Liu (1998), which was based on 1000 bootstrap samples.
PERFORMANCE OF THE UG ALGORITHM
Since few real data sets are available with known maps of the loci involved, we use computer simulation to generate data so that the performance of the UG algorithm can be compared to those of some widely used approaches such as NM, SA, SA-Opt2, and ES-2Opt (Mester et al. 2003). Although we carried out many comparisons, we present some representative results only for four numbers of codominant loci: 6, 30, 50, and 100. The latter two cases represent relatively large maps. For each number of loci, two types of map are considered. The first one is an equal distance (ED) map in which all recombination distances between adjacent loci are set to be 10 cM. The second one is a random distance (RD) map in which the distance between the adjacent loci is set to be a value randomly selected from the five possible values: 10, 15, 20, 25, and 30 cM.
The simulation starts with two isogenic lines, representing the paternal and maternal lineages, respectively. We employed the point process model by Foss et al. (1993) in our simulations. For each meiosis resulting in the F1 individuals, recombination events are assumed to occur between two adjacent loci with a certain probability that is proportional to the distance (in centimorgans). We considered both the presence and the absence of recombination interference. In the case of no interference, recombination between each pair of adjacent loci is independently simulated between any two nonsister chromatids (Weinstein 1936) with equal probability and proceeds without chromatid interference (Foss et al. 1993). For the case of interference, we considered only an extreme situation, which is a complete interference. In such a case, a crossover in a particular interval between two nonsister chromatids cannot occur when there is already a crossover in an interval within 30 cM.
The F2 generation is simulated by crossing the individuals of the F1 generation. The ratio of the paternal homozygotes, heterozygotes, and maternal homozygotes among F2 individuals is expected to be 1:2:1. To mimic the practice in a typical crossing experiment, we retain the sample of F2 individuals only when the ratio does not significantly deviate from 1:2:1. Once the sample of F2 individuals is obtained, the EM algorithm (see, for example, Liu 1998) is used to estimate the recombination fraction (r) between each pair of loci.
Since our main purpose is to recover the correct map, we measure the performance of a method by its success rate of recovering the true map to a given extent. Most important is whether a method is capable of completely recovering the true map. In all our results, we found that distance defined by Equation 13 performs slightly better than that defined by Equation 11; therefore, we report only the results based on the distance defined by Equation 13. Table 4 shows the results for the five methods for complete recovery of the map with six codominant loci. It is clear that the SA algorithm has almost no chance of recovering the true map, which agrees with the finding by Stam (1993). However, the combination of SA with the 2Opt optimization procedure (Lin and Kernighan 1973) improves it considerably, although it is still a poor performer. The ES-2Opt is overall more efficient than the SA-2Opt, particularly with large sample sizes. The performance of the NM algorithm is good overall except for the case of interference with equal distance between adjacent loci.
TABLE 4.
The efficiencies of different map-making methods in complete recovery of the true map with six codominants
| Distance status
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| ED
|
RD
|
||||||||
| Crossover status: | Independence
|
Interference
|
Independence
|
Interference
|
|||||
| Sample size: | 50 | 300 | 50 | 300 | 50 | 300 | 50 | 300 | |
| SA | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 2.0 | 0.0 | |
| SA-2Opt | 21.0 | 33.0 | 38.0 | 37.0 | 25.0 | 39.0 | 29.0 | 37.0 | |
| ES-2Opt | 36.0 | 58.0 | 41.0 | 63.0 | 46.0 | 60.0 | 49.0 | 62.0 | |
| NM | 31.0 | 79.0 | 20.0 | 25.0 | 39.0 | 93.0 | 36.0 | 89.0 | |
| UG | 87.0 | 100.0 | 84.0 | 100.0 | 82.0 | 100.0 | 76.0 | 100.0 | |
RD, random distances (10, 15, 20, 25, and 30 cM) scattered randomly in n − 1 adjacent intervals of n loci on a model linkage map. ED, equal distance (10 cM) for all adjacent intervals of n loci on a model linkage map.
Among the five methods compared, the UG algorithm is clearly the most efficient method; its improvement in efficiency over the other four methods is particularly profound when the sample size is small. For example, for a sample of 50 individuals, the UG algorithm has a 76–87% efficiency while the best of the other four methods achieves only 49% efficiency.
Since overall the NM is the best among the existing methods, we carried out more extensive comparisons between the performances of the NM and the UG. In addition to the complete recovery, it is also informative to see if a method can recover most of the true map, for example, recovering 90% of a true map. Table 5 shows the efficiencies of the NM method and the UG method in recovering several percentages of the true map in the cases of 30, 50, and 100 codominant loci. In the case of no interference, the efficiencies of both methods grow as sample size increases, but in any sample size the UG method greatly outperforms the NM method. In particular, the UG method is weakly sensitive to sample size. For example, in the samples of 100, 200, and 300 F2 individuals, the UG method has, respectively, 74, 86, and 88% accuracies to completely recover the true map of 100 1oci of RD and 82, 98, and 100% accuracies to completely recover the true map of 100 loci of ED. In comparison the NM method has only, respectively, 15, 30, and 31% accuracies for recovering the RD map and 22, 70, and 79% for recovering the ED map. On the other hand, it is also seen from Table 5 that, in comparison with the NM method, the UG method does not obviously tend to perform poorly as the number of linked loci increases and its mapping efficiency is not significantly affected by crossover interference.
TABLE 5.
Comparison between UG and EM methods in their accuracies of partially (80%, 90%) and completely (100%) recovering the model linkage maps of 30, 50, and 100 codominant loci
| Crossover status
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Independence
|
Interference
|
||||||||||
| Sample size: | 100
|
200
|
300
|
100: | 200: | 300: | |||||
| Loci | Recovering mode | Distance status: | ED | RD | ED | RD | ED | RD | RD | RD | RD |
| NM | |||||||||||
| 30 | 80% | 86.0 | 61.0 | 99.0 | 86.5 | 100.0 | 87.0 | 69.0 | 84.0 | 89.0 | |
| 90% | 86.0 | 60.0 | 98.0 | 86.5 | 100.0 | 87.0 | 66.0 | 84.0 | 89.0 | ||
| 100% | 85.0 | 59.0 | 98.0 | 86.0 | 100.0 | 86.0 | 63.0 | 84.0 | 89.0 | ||
| 50 | 80% | 74.0 | 39.0 | 95.0 | 64.0 | 100.0 | 70.0 | 42.0 | 63.3 | 73.0 | |
| 90% | 65.0 | 37.0 | 95.0 | 64.0 | 100.0 | 70.0 | 35.5 | 60.7 | 73.0 | ||
| 100% | 63.0 | 36.0 | 95.0 | 64.0 | 100.0 | 70.0 | 34.5 | 59.7 | 72.7 | ||
| 100 | 80% | 39.0 | 17.0 | 71.0 | 33.0 | 79.0 | 33.0 | 13.1 | 33.0 | 39.3 | |
| 90% | 32.0 | 16.0 | 71.0 | 31.0 | 79.0 | 32.0 | 10.7 | 32.0 | 39.3 | ||
| 100% | 22.0 | 15.0 | 70.0 | 30.0 | 79.0 | 31.0 | 7.1 | 32.0 | 39.3 | ||
| UG | |||||||||||
| 30 | 80% | 98.0 | 92.0 | 100.0 | 98.0 | 100.0 | 98.5 | 93.0 | 98.0 | 99.0 | |
| 90% | 98.0 | 92.0 | 100.0 | 98.0 | 100.0 | 98.5 | 92.0 | 98.0 | 99.0 | ||
| 100% | 95.0 | 92.0 | 100.0 | 98.0 | 100.0 | 98.5 | 92.0 | 98.0 | 99.0 | ||
| 50 | 80% | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 97.0 | 91.0 | 98.0 | 98.0 | |
| 90% | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 97.0 | 90.0 | 98.0 | 98.0 | ||
| 100% | 95.0 | 87.0 | 100.0 | 92.0 | 100.0 | 97.0 | 85.5 | 98.0 | 98.0 | ||
| 100 | 80% | 92.0 | 88.9 | 99.0 | 88.0 | 100.0 | 89.0 | 92.9 | 97.8 | 98.9 | |
| 90% | 92.0 | 85.6 | 99.0 | 86.0 | 100.0 | 89.0 | 89.3 | 97.8 | 98.9 | ||
| 100% | 82.0 | 74.4 | 98.0 | 86.0 | 100.0 | 88.0 | 81.0 | 97.8 | 97.8 | ||
RD, random distances (10, 15, 20, 25, and 30 cM) scattered randomly in n − 1 adjacent intervals of n loci on a model linkage map. ED, equal distance (10 cM) for all adjacent intervals of n loci on a model linkage map.
DISCUSSION
As pointed out in the Introduction, several principles can be used to reconstruct linkage maps. The SA, SA-2Opt, and ES-2Opt are methods aimed at minimizing the sum of adjacent recombination fractions or adjacent distances in the linkage map. This minimization principle is correct in theory but often does not work well in practice due to various types of random errors. Indeed, even with exactly the same linkage map for all the individuals subjected to the experiment, the observed recombination fractions may fluctuate widely from experiment to experiment due to sampling variation, ecological condition, sex, genotype, and age (Mester et al. 2003). Therefore, the estimated linkage map based on the minimization principle is often poor quality. The NM method is based on the neighbor joining of Saitou and Nei (1987). Since its search criteria closely mimic the minimization principle, the overall accuracy of the NM method is similar to the best in the class of methods based on the minimization principle.
The UG algorithm does not rely on the minimization principle, yet achieves higher accuracy in recovering the true map. Theorem 1 apparently is the foundation for its success, which indicates that there is a high probability that the terminal loci of the true map can be identified through utilization of distances among multiple loci simultaneously. Although the theorem is proven only for strictly additive distance, the fact that the UG algorithm works very well with distances defined either as the estimated recombination fractions or as the corrected estimates indicates that Equation 3 is robust against modest deviation by the distance from strict additivity. This pleasing result appears because the loci critical for determining the status of a particular locus are those nearby, whose distance to the given locus is likely more additive than that of those far away. In addition to its high accuracy, the UG algorithm also has the advantage of speed, particularly when the number of loci is large, compared with the NM algorithm. This is because it takes n − 1 cycles to complete while the NM algorithm takes n(n − 1)/2 cycles. Therefore, the UG algorithm should be a useful addition to the tools for large-scale linkage mapping and for evaluating the confidence of the estimated map by bootstrap or jackknife (Liu 1998; Mester et al. 2003).
Acknowledgments
We thank the High Performance Computer Center of Yunnan University for computational support and Sara Barton for editorial assistance. This work was partly supported by National Institutes of Health grant R01 GM50428 and funds from Yunnan University.
APPENDIX A
Proof of Theorem 1. For additive distance, 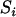 (i < n) can be written as
(i < n) can be written as
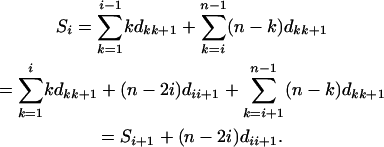 |
For 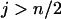 , i.e., the largest integer ≤n/2, we have
, i.e., the largest integer ≤n/2, we have
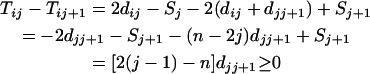 |
and for 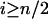
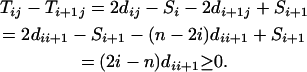 |
Similarly for 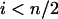 ,
, 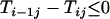 and for
and for 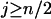 ,
, 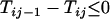 . It thus follows that when
. It thus follows that when 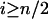 we have
we have 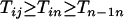 and when
and when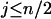 we have
we have 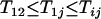 . Finally when
. Finally when 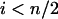 and
and 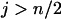 , we have
, we have 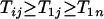 . ▪
. ▪
Furthermore we have
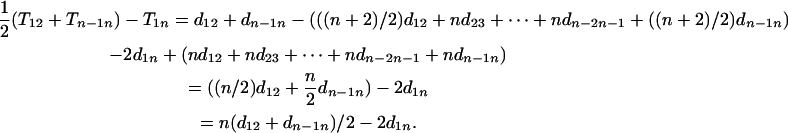 |
APPENDIX B
Proof of Theorem 2. Using the recurrence equation for 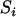 from appendix a, we have
from appendix a, we have
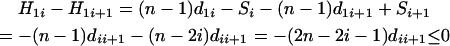 |
for 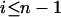 . It is clear that comparison between
. It is clear that comparison between 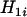 and
and 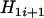 leads to
leads to
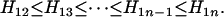 |
▪
References
- Buetow, K. N., and A. Chakravarti, 1987. Multipoint gene mapping using seriation. Am. J. Hum. Genet. 41: 189–201. [PMC free article] [PubMed] [Google Scholar]
- Echt, C., S. Knapp and B.-H. Liu, 1992. Genome mapping with non-inbred crosses using GMendel 2.0. Maize Genet. Coop. Newsl. 66: 27–29. [Google Scholar]
- Edwards, J. H., 1971. The analysis of X-linkage. Ann. Hum. Genet. 34: 229–250. [DOI] [PubMed] [Google Scholar]
- Ellis, T. H. N., 1997. Neighbor mapping as method for ordering genetic markers. Genet. Res. 69: 35–43. [Google Scholar]
- Eppig, J., and E. M. Eicher, 1983. The mouse linkage map. J. Hered. 74: 213–231. [DOI] [PubMed] [Google Scholar]
- Falk, C. T., 1992. Preliminary ordering of multiple linked loci using pairwise linkage data. Genet. Epidemiol. 9: 367–375. [DOI] [PubMed] [Google Scholar]
- Foss, E., R. Lande, F. W. Stahl and C. M. Steinberg, 1993. Chiasma interference as a function of genetic distance. Genetics 133: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Jensen, J., and J. H. Jorgensen, 1975. The barley chromosome 5 linkage map. Hereditas 80: 5–16. [Google Scholar]
- Knapp, M., M. Neugebauer, R. Fimmers, S. A. Seuchter and M. P. Baur, 1989. Preliminary ordering of multipoint linkage data, pp. 41–46 in Multipoint Mapping and Linkage Based Upon Affected Pedigree Members (Genetic Analysis Workshop), edited by R. C. Elston, M. A. Spence, S. E. Hodge and J. W. MacCluer. Alan R. Liss, New York.
- Kosambi, D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lander, E. S., and P. Green, 1987. Construction of multilocus linkage maps in human. Proc. Natl. Acad. Sci. USA 84: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MapMaker: an interactive computer package for constructing genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lathrop, G. M., J. M. Lalouel, C. Julier and J. Ott, 1984. Strategies for multilocus linkage analysis in human. Proc. Natl. Acad. Sci. USA 81: 3443–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop, G. M., J. M. Lalouel, C. Julier and J. Ott, 1985. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am. J. Hum. Genet. 37: 482–498. [PMC free article] [PubMed] [Google Scholar]
- Lin, S., and B. Kernighan, 1973. An effective heuristic algorithm for the TSP. Oper. Res. 21: 498–516. [Google Scholar]
- Liu, B.-H., 1998. Statistical Genomics: Linkage, Mapping, and QTL Analysis. CRC Press, New York.
- Lu, Y. Y., and B. H. Liu, 1995. PGRI, a software for plant genome research. Plant Genome III. Abstract, January, 1995, San Diego.
- Mester, D., Y. Ronin, D. Minkov, E. Nevo and A. Korol, 2003. Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics 165: 2269–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, J. M., and M. Boehnke, 1990. Monte Carlo comparison of preliminary methods of ordering multiple genetic loci. Am. J. Hum. Genet. 47: 470–482. [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Stam, P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3: 739–744. [Google Scholar]
- Suiter, K. A., J. F. Wendel and J. Case, 1983. SLINKAGE-1: a PASCAL computer program for the detection and analysis of genetic linkage. J. Hered. 74: 203–204. [DOI] [PubMed] [Google Scholar]
- Thompson, E. A., 1984. Information gain in joint linkage analysis. IMA J. Math. Appl. Med. Biol. 1: 31–49. [DOI] [PubMed] [Google Scholar]
- Weeks, D., and K. Lange, 1987. Preliminary ranking procedures for multilocus ordering. Genomics 1: 236–242. [DOI] [PubMed] [Google Scholar]
- Wilson, R. S., 1988. A major simplification in the preliminary ordering of linked loci. Genet. Epidemiol. 5: 75–80. [DOI] [PubMed] [Google Scholar]
- Weinstein, A., 1936. The theory of multiple-strand crossing over. Genetics 21: 155–199. [DOI] [PMC free article] [PubMed] [Google Scholar]