Abstract
The establishment of transcriptional silencing in yeast requires cell-cycle progression, but the nature of this requirement is unknown. Sir2 is a protein deacetylase that is required for gene silencing in yeast. We have used temperature-sensitive alleles of the SIR2 gene to assess Sir2's contribution to silencing as a function of the cell cycle. When examined in vivo, these conditional alleles fall into two classes: one class exhibits a loss of silencing when raised to the nonpermissive temperature regardless of cell-cycle position, while the second class exhibits a mitosis-specific silencing defect. Alleles of the first class have a primary defect in protein deacetylase activity, while the alleles of the second class are specifically defective in Sir2–Sir4 interactions at nonpermissive temperatures. Using a SIR2 temperature-sensitive allele, we show that silencing can be established at the HML locus during progression through the G2/M–G1 interval. These results suggest that yeast heterochromatin undergoes structural transitions as a function of the cell cycle and support the existence of a critical assembly step for silent chromatin in mitosis.
IN Saccharomyces cerevisiae, transcriptional silencing is observed at the silent mating-type loci (HML and HMR), telomeres, and the ribosomal DNA repeats (for reviews see Huang 2002 and Rusche et al. 2002). At the HM loci and telomeres, silencing depends on the action of three Sir proteins, Sir2, Sir3, and Sir4. A Sir2–Sir4 complex is observed in vivo (Moazed et al. 1997), and this complex can also associate with Sir3 (Moazed et al. 1997; Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2003; Liou et al. 2005). The Sir protein complex can be recruited to telomeres and the HM loci via a specific interaction between Sir4 and the DNA-binding factor Rap1p (Moretti et al. 1994). Silencing at the rDNA locus does not require Sir3 or Sir4, but does depend on Sir2; here Sir2 is recruited as part of the RENT complex (Shou et al. 1999; Straight et al. 1999). Therefore, the Sir2 protein is central to all forms of transcriptional silencing in yeast. Sir2 is the founding member of a large family of NAD-dependent lysine deacetylases (Imai et al. 2000; Landry et al. 2000b; Smith et al. 2000) that includes members from bacteria to humans (Brachmann et al. 1995; Afshar 1999; Frye 2000; for Sir2 reviews see Moazed 2001; Smith et al. 2002). Yeast Sir2's likely substrate is histones. A relative lack of histone deacetylation is observed at all three silenced loci (Braunstein et al. 1993, 1996; Bryk et al. 2002; Buck et al. 2002), and Sir2's deacetylase activity is specifically required for silencing to be established (Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002) and maintained (Bedalov et al. 2001). Deacetylation of the histone H4 N-terminal tail may increase interactions with the Sir3 protein, promoting the spread of the Sir protein complex (Hecht et al. 1995; Carmen et al. 2002; Liou et al. 2005).
The establishment of silencing requires progression through the cell cycle (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001, 2006; Li et al. 2001; Lau et al. 2002; Martins-Taylor et al. 2004), but the nature of this constraint is unknown. Determining the requirement for cell-cycle dependence is likely to provide key insights into the mechanism of silencing in yeast and uncover general insights into the maintenance and propagation of transcriptional states. In this study we examine Sir2's specific contribution to silencing as a function of the cell cycle using conditional alleles of the SIR2 gene. Our data suggest that Sir2 makes distinct contributions to silencing and that a crucial step for the assembly of silent chromatin occurs in mitosis.
MATERIALS AND METHODS
Strains and plasmids:
To generate random mutations in the SIR2 gene, we used error-prone PCR followed by gap repair (Muhlrad et al. 1992). Error-prone PCR reactions included 5 units of Taq polymerase, 7 mm MgCl2, 0.5 mm MnCl2, 1 mm dCTP and dTTP, 0.2 mm dATP and dGTP, as well as the SIR2 template. Mutated SIR2 DNA fragments were cotransformed with SIR2 plasmid pAW2 that had been cut with SmaI into the HMLα MATa HMRα Δsir2 strain YSH311. Ura+ transformants were then screened for those able to promote mating at 23°, but not at 37°. All constructs were sequenced to identify or verify the existence of mutations. Temperature-sensitive alleles of SIR2 were cloned into pRS305 (Sikorski and Hieter 1989) to produce plasmids pMMi1 (bearing the sir2-604 allele), pMMi2 (sir2-614), and pMMi3 (sir2-620). These plasmids were then integrated into the leu2 locus of strain YSH501 by transforming plasmids cut with EcoRI. Yeast strains used in this study are listed in Table 1. SIR2 plasmids used in silencing assays are based on pR415 (Sikorski and Hieter 1989) and include pKAM1 (SIR2), pMM80 (sir2-604), pMM81 (sir2-614), and pMM82 (sir2-620). All plasmid-borne SIR2 alleles described in this work have identical flanking sequences and include 300 bp of wild-type sequences 5′ to the open reading frame and 320 bp 3′ to the open reading frame. A complete description and characterization of SIR2 temperature-sensitive alleles will be presented elsewhere (M. Hickman and S. Holmes, unpublished results).
TABLE 1.
Description of strains
| Strain | Genotype | Sourcea |
|---|---|---|
| YSH189 (DMY1) | MATaura3 ade2 lys1 his5 leu2 can1 | Mahoney and Broach (1989) |
| YSH501 | MATaura3 ade2 lys1 his5 leu2 can1 Δsir2∷KAN | |
| YSH502 | YSH501; leu2∷sir2-604-LEU2 | |
| YSH498 | YSH501; leu2∷sir2-614-LEU2 | |
| YSH499 | YSH501; leu2∷sir2-620-LEU2 | |
| PJ69-4a | MATatrp1-901 leu2-3,112 ura3-52 his3-200 Δgal4 Δgal80 LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ | James et al. (1996) |
| YSH563 | PJ69-4α; Δsir3∷NAT1MX Δsir4∷URA3 | |
| YSH625 | PJ69-4α; Δsir2∷HYG Δsir3∷NAT1MX Δsir4∷URA3 |
Unless noted, strains were constructed during the course of this work.
Silencing assays:
To measure silencing at HML using a pheromone response assay, cultures were grown in YPD (1% bacto yeast extract, 2% bactopeptone, 2% dextrose) to early log phase (∼2 × 106 cells/ml) when α-factor was added at 10 μg/ml. Cultures were further incubated with shaking at the indicated temperatures for 5 hr. Cell-cycle arrest was determined by microscopic examination of cell morphology. Unbudded cells were assumed to be in G1 phase. Unbudded cells with obvious growth projections were further designated as “shmoos.” Cells with buds composing <50% of the volume of the mother cell were designated as small-budded cells, while cells with buds composing >50% of the volume of the mother cells were designated as large-budded cells. A minimum of 100 cells was assayed for each determination. For pedigree analysis of α-factor sensitivity, cultures were applied directly to YPD media containing α-factor. Mating assays were performed as described (Dula and Holmes 2000). Reverse-transcriptase PCR (RT–PCR) measurements of ACT1 and α1 message were performed exactly as described (Martins-Taylor et al. 2004) except that PCR products were run on 8% acrylamide gels, stained using sybr gold dye (Invitrogen, San Diego), and the gels were converted to tif files using a Storm 840 PhosphorImager. Identical results were achieved in at least two independent experiments and in repeated determinations from RNA collected from individual experiments.
Western blots:
Protein extraction and detection by Western blotting was performed as described (Tanny et al. 1999). For cycling cells, protein was extracted from log-phase cultures (cell density was ∼1.5 × 107 cells/ml) grown at either 23° or 37°. For cultures blocked in the cell cycle, cells were grown at 23° to an OD600 of ∼0.2. Cell-cycle blocking agents were added and cells were allowed to block for ∼4–5 hr until >90% of the cells were arrested. For G1-S experiments, α-factor was added to a final concentration of 10 μg/ml. Once cells were >90% arrested in G1, hydroxyurea was added to a final concentration of 30 mg/ml. Cultures were incubated at 23° for an additional 30 min when the culture was divided, half remaining at 23° and half shifted to 37°. Following an additional incubation of 3 hr, cells were pelleted and protein was extracted. For G2/M-blocked cultures, nocodazole was added to a final concentration of 15 μg/ml. Once >90% of the cells were arrested, the cultures were divided; half of the culture remained at 23°, while the other half was shifted to 37° for 3 hr when cells were pelleted and protein was extracted. Bradford assays were performed on each sample to equalize protein loadings on the gel. Duplicate gels were run and stained with Coomassie to confirm consistent loading from lane to lane. Blots were probed with an antibody directed to the N terminus of Sir2 (Santa Cruz Biotechnology), used at a dilution of 1:2000. Secondary detection was performed using either the Renaissance enhanced chemiluminescence detection system (New England Nuclear, Boston) or a mouse anti-goat IgG–biotin secondary antibody from Santa Cruz used at a 1:500 dilution, followed by addition of streptavidin–HRP (Bio-Rad, Hercules, CA) at a dilution of 1:2500 and the Western Lightning chemiluminescence reagent (Perkin-Elmer, Norwalk, CT). Western blots run with varying dilutions of protein extracts indicated that the assay was sensitive to small differences in protein levels. Similar results were obtained in three independent experiments.
Cell-cycle blocks:
Cell-cycle blocks and interval experiments were performed as described (Martins-Taylor et al. 2004). α-Factor (10 μg/ml), nocodazole (15 μg/ml), or hydroxyurea (20 mg/ml) was used to block cells in G1, G2/M, or early S phase, respectively. Unless noted, cells exhibited at least a 90% arrest in the cell cycle. Cell-cycle arrest was determined by microscopic examination of cell morphology. For all interval experiments, log-phase cells were incubated in the initial blocking agent until >90% of cells were arrested in the cell cycle. Media were then removed by filtration, and cells were washed with several volumes of water and resuspended in media containing the second blocking agent until at least 90% of cells exhibited cell-cycle arrest. Experiments were initiated when cultures were at early log phase (∼2–3 × 106 cells/ml).
Two-hybrid assays:
Vectors and methods for performing two-hybrid screens have been described (James et al. 1996; Uetz et al. 2000). SIR2 alleles were amplified using primers SP280 (ACCCCACCAAACCCAAAAAAAGAGATCGAATTCCAGCTGACCACCATGACCATCC CACATATG) and SP281 (CTACGATTCATAGATCTCTGCAGGTCGACGGATCCCCGGGAATT GCCATGTTAGAGGGTTTTGGGATG). The sir4Δ730N allele was amplified using primers SP278 (ACCCCACCAAACCCAAAAAAAGAGATCGAATTCCAGCTGACCACCATGCC AAATGACAATAAGAC) and SP279 (CTACGATTCATAGATCTCTGCAGGTCGACGGATCCC CGGGAATTGCCATGTCAATACGGTTTTATCTCCT). These fragments were cotransformed with cut pOBD2 or pOAD; gap repair of the cut plasmids created plasmids expressing binding domain or activation domain fusions (Uetz et al. 2000). Deletions of SIR2 and SIR3 in strain PJ69-4α were made via PCR-mediated gene deletion using pAG25 and pAG32 as templates (Goldstein and McCusker 1999). SIR4 was deleted by transforming PJ69α with pCTC77 cut with HindIII (Stone et al. 1991).
NAD hydrolysis assays:
Sir2 enzymatic assays were performed on Sir2 protein complexes affinity purified from yeast (Tanny et al. 2004). SIR2 alleles fused to an N-terminal tandem affinity purification (TAP) tag (Rigaut et al. 1999; Puig et al. 2001) were constructed by gap repair; this addition did not affect the mating behavior of these SIR2 alleles (not shown). The TAP tag was amplified from plasmid pBS1761 (Rigaut et al. 1999) using PCR primers SP259 (CGCTAGTCTTTGATACGGCGTATTTCATATGTGGGATGGTCATCTTATCGTCATCATCAAGTG) and SP269 (GTAGACACATTCAAACCATTTTTCCCTCATCGGCACATT AAAGCTGGATGGCAGGCCTTGCGCAACA). These primers amplify the TAP affinity tag with overhangs identical to sequences flanking the start of the SIR2 open reading frame. This fragment was transformed into yeast along with plasmids containing alleles of the SIR2 gene partially digested with NdaI, an enzyme that cuts near the start of the SIR2 open reading frame. Successful gap repair creates plasmids expressing SIR2 alleles from their own promoter, altered solely by the addition of the TAP tag to the N terminus. Plasmids were recovered from yeast and verified by sequencing. The TAP–SIR2 plasmids that were created included pMM84 (SIR2), pMM87 (sir2-604), pMM85 (sir2-614), and pMM86 (sir2-620).
Purification and assay of Sir2 was essentially as described (Tanny et al. 2004). Cultures were grown in glucose medium lacking leucine. Fifty-milliliter cultures were grown to OD600 1.5; cells were washed with ice-cold water and pelleted by centrifugation. Cell pellets were frozen using liquid nitrogen and stored at −70°. Each of the remaining steps was performed at 4°. The frozen cell pellets were resuspended in 400 μl of lysis buffer (50 mm HEPES KOH, pH 7.6, 10 mm MgOAc, 0.5 m KOAc, 5 mm EGTA, 0.1 mm EDTA, 0.25% NP-40, 5% glycerol, 1 mm DTT). Freshly made PMSF (1 mm) and the protease inhibitors leupeptin, bestatin, and pepstatin (2 μg/ml each) were added to lysis buffer immediately before use. Cells were disrupted by grinding with glass beads on a bead beater (2 × 30 sec with 5 min rest on ice in between). Eppendorf tubes were punctured at the bottom and lysates were collected in centrifuge tubes by centrifugation at 2 krpm for 1 min at 4°. The lysates were transferred to new tubes and microcentrifuged at 13 krpm for 10 min at 4°. After centrifugation, the amount of total protein in each lysate was determined using the Bradford assay. Equal amounts of the total protein from each lysate (10–15 mg) were added to 10 μl IgG sepharose beads (Pharmacia 17-0969-01) that had been washed with lysis buffer. After 2 hr of nutation at 4°, beads were collected by centrifugation at 2 krpm for 1 min at 4° and washed three times with wash buffer (50 mm HEPES KOH, pH 7.6, 150 mm NaOAc, 5 mm MgOAc, 5% glycerol, 1 mm DTT, 1 mm PMSF). For Western detection, the washed beads were resuspended in 1.5× sample buffer (75 m Tris–HCl, pH 6.8, 3% SDS, 15% glycerol, 0.15% bromophenol blue, 7.5% β-mercaptoethanol, 0.1 mm PMSF, and 1 mm DTT), boiled at 65° for 10 min, and run on an 8% PAGE gel. The proteins were transferred to nitrocellulose and incubated with Sir2-specific antibodies (used at 1:5000 dilution); protein bands were visualized using the New England Nuclear–Renaissance system.
For the enzymatic activity assay, TAP–Sir2 was affinity purified as described above. After the second wash, the beads were split. One-third of the beads were washed with wash buffer one more time and stored at −20° in 1.5× sample buffer to be used in the Western blot assay. The other two-thirds of the beads were washed with reaction buffer (50 mm Tris 7.5, 100 mm NaCl, 1 mm DTT), divided, and used in the assay for Sir2 activity by nicotinamide release. Sir2-bound beads were incubated in 8.5 μl of reaction buffer, 0.05 μCi [carbonyl-14C]NAD (Amersham Pharmacia; 51 mCi/mmol; label on the nicotinamide ring), and 10 μg of synthetic N-terminal H4 peptides with either acetylated or deacetylated lysines. Samples were incubated for 1 hr at either 23° or 37°. The reactions ware stopped with 0.5 m sodium borate, pH 8.0, and the released nicotinamide was extracted by ethyl acetate. After addition of ethyl acetate, the samples were vortexed for 5 sec and centrifuged for 1 min at 13 krpm in a microcentrifuge. The ethyl acetate phase was removed and counted in 2 ml of scintillation fluid. The NAD breakdown activity for each mutant protein was expressed as percentage of activity measured for the wild type in a concurrent experiment. The amount of Sir2 protein in individual experiments was normalized to the amount of wild-type Sir2 used in the same experiment. We performed quantitative Western blotting to ensure that equal amounts of Sir2 protein were added to each reaction. Samples from individual experiments were analyzed by Western detection, the autoradiography film was scanned, and the Sir2 bands were digitized using UN-SCAN-IT software (Silk Scientific, Orem, UT). Serial dilutions of wild-type Sir2 samples were included to generate a standard curve. The data shown in Table 5 are the results of four independent experiments.
TABLE 5.
NADase activity of Sir2 proteins
| Sir2 protein | 23° | 37° |
|---|---|---|
| sir2-604 | 55 ± 29 | 29 ± 13 |
| sir2-614 | 60 ± 22 | 6 ± 3 |
| sir2-620 | 60 ± 39 | 23 ± 6 |
| sir2-H364Y | 3 ± 2 | 1.6 ± 1.5 |
The ability of the indicated Sir2 proteins to hydrolyze NAD in the presence of a substrate bearing an acetylated lysine was measured. Values are expressed as a percentage of a wild-type control purified and tested at the same time. The sir2-H364Y protein has been previously shown to have minimal activity in in vitro deacetylase assays (Tanny et al. 1999; Imai et al. 2000). In these assays, wild-type Sir2 had ∼5% higher activity at 37° vs. 23°.
RESULTS
A mitosis-specific function of Sir2?
We assessed the contributions of the Sir2 protein to silencing as a function of the cell cycle using conditional alleles of SIR2. For these studies, we focused on three temperature-sensitive alleles (Table 2; also see materials and methods). Full characterization of these and other temperature-sensitive alleles of SIR2 will be described elsewhere (M. Hickman and S. Holmes, unpublished results).
TABLE 2.
SIR2 alleles
| SIR2 allele | Amino acid changes |
|---|---|
| sir2-604 | D515Y Δ30C |
| sir2-614 | E174G N299I L438H K523E K542R |
| sir2-620 | S204P K414R K475E V500A Δ24C |
A shorthand description for mutations present in each allele is used. For amino acid substitutions, the wild-type amino acid (in its one-letter code) and position are listed, followed by the amino acid replacing it in the mutated allele. For deletions caused by premature stop codons, the number of amino acids removed is listed after the “Δ,” followed by a “C” for carboxy-terminal deletion. Amino acid changes known to be sufficient to confer the temperature-sensitive mating phenotype are in italics (M. Hickman and S. Holmes, unpublished results). Both N299I and L438H mutations of sir2-614 are necessary to observe the temperature-sensitive phenotype. SIR2 alleles containing only the Δ24C, K475E, or K414R/K475E mutations found in sir2-620 exhibit no mating phenotypes.
We created strains with integrated copies of temperature-sensitive SIR2 alleles in a MATa strain lacking the wild-type SIR2 gene. Silencing was initially examined using an α-factor sensitivity assay (Table 3). Cultures were grown to steady state (at least 10 cell doublings) at the permissive or nonpermissive temperature and then challenged with α-factor. Cells that lose silencing at HMLα in these MATa strains will lose sensitivity to α-factor and progress through the cell cycle, while cells that retain silencing will block in G1 and adopt an altered (“shmoo”) morphology. Strains with these alleles were found to retain near wild-type levels of silencing at the permissive temperature and exhibited essentially no silencing at the nonpermissive temperature by this assay (Table 3).
TABLE 3.
Temperature-sensitive silencing mediated by conditional SIR2 alleles
| % α-factor arrested
|
||
|---|---|---|
| SIR2 allele | 23° | 37° |
| SIR2 | 91 | 96 |
| sir3-8 | 96 | 1 |
| sir2-604 | 96 | 2 |
| sir2-614 | 98 | 2 |
| sir2-620 | 96 | 0 |
Yeast strains bearing different SIR2 alleles were grown to log phase at 23° or 37° and then challenged with 10 μg/ml α-factor. The percentage of unbudded (G1 blocked) cells after 5 hr incubation in α-factor is listed. A strain bearing the temperature-sensitive sir3-8 allele was assayed as a control. A minimum of 200 cells was assayed for each determination.
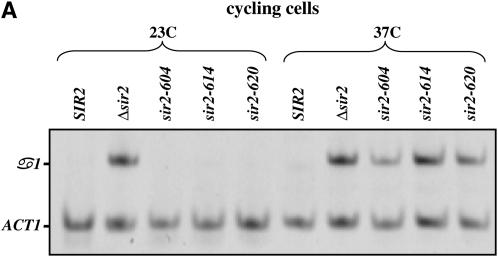
As an independent measure of silencing we used RT–PCR to measure the α1 message transcribed from the HMLα locus (see Figure 1). Loss of silencing in MATa strains allows expression of the α1 and α2 genes from HML. α1 and, to a lesser extent, α2 are subsequently subject to repression by the action of the a1/α2 heterodimer (Klar et al. 1981; Nasmyth et al. 1981; Siliciano and Tatchell 1984). However, α1 message remains detectable in MATa strains lacking Sir2 (Chi and Shore 1996; Wyrick et al. 1999). In our experiments we find that assaying α1 expression provides the most consistent and quantitative measure of HML expression. Similar to the prior experiment, cultures were grown to steady state at the permissive or nonpermissive temperature, RNA was collected, and the levels of α1 message were determined. As a control for these experiments message from the ACT1 gene was also measured. Consistent with the α-factor sensitivity assay, each of the strains bearing temperature-sensitive SIR2 alleles exhibits wild-type silencing at the permissive temperature and is defective for silencing at the nonpermissive temperature (Figure 1A).
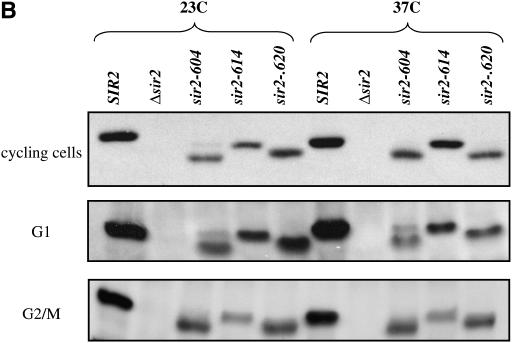
Figure 1.—
Conditional alleles of SIR2. (A) Levels of α1 and ACT1 mRNA were measured by RT–PCR in strains bearing different alleles of SIR2. Cultures were maintained in log phase for >10 generations at the listed temperatures, and then RNA was collected and analyzed as described in materials and methods. α1 indicates steady-state levels of message transcribed from the HML locus; ACT1 message is shown as a control. RNA was analyzed from a wild-type strain, a strain lacking the SIR2 gene, and strains bearing the indicated alleles of SIR2. (B) Protein extracts made from cells bearing the indicated SIR2 alleles were subjected to a Western blot using an antibody specific for the Sir2 protein, as described in materials and methods. For the cycling cells, parallel cultures were grown to log phase at either 23° or 37°. G1/early S-blocked cells were grown to early log phase at 23° and then blocked with α-factor. Half the culture was maintained at 23°, while the other half was shifted to 37° for 3 hr. G2/M-blocked cells were grown to early log phase at 23° and then blocked with nocodazole. Half the culture was maintained at 23°, while the other half was shifted to 37° for 3 hr. (C) For each of the experiments shown in B, duplicate gels were run and stained with Coomassie to confirm consistent loading of samples from lane to lane.
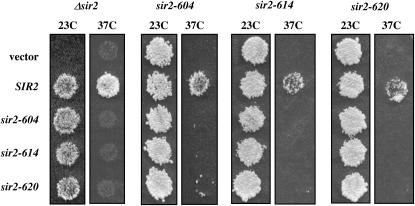
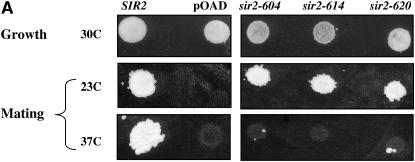
The absence of silencing at nonpermissive temperatures could be due to a loss of Sir2 function or to a temperature-dependent defect in the synthesis or stability of Sir2. To examine the steady-state levels of Sir2 protein coded for by these alleles, we performed Western blotting on strains grown at permissive and nonpermissive temperatures. As shown in Figure 1B, we find that strains bearing mutated alleles of Sir2 have lower steady-state levels of Sir2 protein than a strain expressing wild-type Sir2, but in each of the strains Sir2 protein levels either do not change, or appear to increase (see sir2-614p in cycling cells), at the nonpermissive temperature. When strains bearing specific SIR2 alleles are transformed with low-copy plasmids bearing the same SIR2 allele, the mating behavior of the strain does not change (Figure 5). Thus, the Sir2 proteins coded for by these alleles are defective for function at elevated temperatures, and these defects are not likely due to limiting Sir2 protein levels.
Figure 5.—
Complementation of SIR2 alleles. Plasmids bearing the SIR2 allele listed at the left were introduced into strains bearing integrated alleles of the SIR2 alleles listed at the top. Mating assays were performed at 23° and 37° as described in the Figure 4 legend.
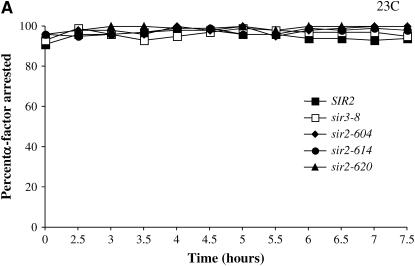
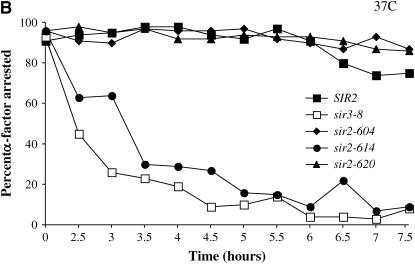
To test for the requirement for Sir2 in G1 phase, we grew cultures bearing conditional SIR2 alleles to log phase at permissive temperature, blocked them with α-factor, and then shifted the culture to the nonpermissive temperature. This design allowed us to observe the immediate effects of Sir2 inactivation vs. the cumulative effects of the steady-state experiments. Cultures were monitored at several time points following the temperature shift using the α-factor sensitivity assay. As a control we conducted the same experiment with a strain bearing the temperature-sensitive sir3-8 allele, known to lose silencing under these conditions (Miller and Nasmyth 1984; Holmes and Broach 1996). Results from these experiments are shown in Figure 2. We find that at the permissive temperature all alleles promote efficient silencing and remain arrested in G1 phase (Figure 2A). However, when raised to the nonpermissive temperature, our strains bearing temperature-sensitive alleles of SIR2 exhibit distinct phenotypes in this assay: the sir-604 and sir-620 alleles promote silencing at 37°, while a strain with the sir2-614 allele rapidly loses silencing (Figure 2B). Therefore, the sir2-604 and sir2-620 alleles may be defective for a function of Sir2 that is dispensable in G1 phase. Alternatively, the sir2-604 and sir2-620 alleles may code for Sir2 proteins that are slower than the sir2-614 protein to lose function at the nonpermissive temperature.
Figure 2.—
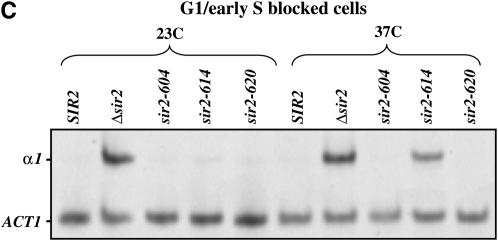
Sir2 function in G1 phase. (A and B) SIR2 alleles with cell-cycle-specific silencing defects. Cells grown to log phase at 23° were arrested with α-factor (10 μg/ml) in G1 phase. When at least 95% of cells exhibited an unbudded morphology, each culture was divided. One half was maintained at 23°, while the other was shifted to 37°. Silencing at HML was assessed by the bud morphology of the culture; unsilenced cells failed to arrest and form buds. The percentage of unbudded cells (α-factor-arrested cells) is plotted vs. time measured from the division of the culture. HML is efficiently silenced in all cultures maintained at 23° (Figure 2A). Silencing is lost in G1 at the nonpermissive temperature (37°) only in strains bearing the sir2-614 allele or the sir3-8 allele (Figure 2B). (C) α1 mRNA levels in G1-arrested strains. Cells grown to log phase at 23° were arrested with α-factor (10 μg/ml) in G1 phase. When at least 95% of cells exhibited an unbudded morphology, hydroxyurea (20 mg/ml) was added and each culture was divided. One-half was maintained at 23°, while the other was shifted to 37°. Following 4 hr of incubation, RNA was collected and RT–PCR was used to assay α1 and ACT1 message levels. RNA was analyzed from a wild-type strain, a strain lacking the SIR2 gene, and strains bearing the indicated alleles of SIR2.
We conducted a similar experiment using RT–PCR to assay silencing of HML. Cultures were blocked in G1 at the permissive temperature and then shifted to the nonpermissive temperature. In this experiment, cells that lose silencing escape the G1 arrest and progress through the cell cycle. To restrict their progress, hydroxyurea was added to each culture prior to the temperature shift. Hydroxyurea blocks cells early in S phase, soon after the initiation of DNA replication; thus, cells escaping from G1 were assayed in this short interval. Following 4 hr at the nonpermissive temperature, RNA was collected and α1 message levels were determined. The RT–PCR experiment produced results identical to the α-factor sensitivity assay; no message was detected from strains bearing the sir2-604 or sir2-620 alleles, while message was readily detected from the strain bearing the sir2-614 allele (see Figure 2C).
We next used the same set of strains to examine the requirement for Sir2 function in mitosis. For our first experiment, we used α-factor sensitivity to examine the role of Sir2 during the G2/M–G1 interval. Cells were blocked in G2/M with nocodazole at the permissive temperature, where they arrest as large-budded cells. Cultures were then shifted to the nonpermissive temperature, held at the nonpermissive temperature for 1 hr, and then released from the nocodazole block into medium containing α-factor. In this medium, cells will complete mitosis and form two unbudded cells. If cells maintain silencing at HML, these cells will block in G1 and remain unbudded. However, cells that lose silencing will be insensitive to α-factor and continue through G1 into S phase, resulting in cells with small buds. Results from a block-and-release experiment are shown in Table 4. When performed entirely at the permissive temperature, we find that nearly all cells that progress out of the nocodazole block are sensitive to α-factor and are therefore silenced (note that in this experiment cultures with SIR2 mutations fail to escape from the G2/M arrest as efficiently as wild-type cells, more frequently persisting as large-budded cells). In contrast, when cells are shifted to the nonpermissive temperature, released from the nocodazole block, and allowed to progress through mitosis, we find that all strains bearing temperature-sensitive alleles of SIR2 have lost silencing. The total amount of time at the nonpermissive temperature is similar in the G1 block and mitosis interval experiments, indicating that cells bearing the sir2-604 and sir2-620 alleles are more sensitive to a shift to the nonpermissive temperature in mitosis than in G1 phase. Thus, the differences in the alleles cannot be attributed solely to a difference in the time that it takes to manifest their temperature-sensitive phenotypes.
TABLE 4.
Sir2 function is required in mitosis
| 23° → 23°
|
23° → 37°
|
|||||||
|---|---|---|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
|
| SIR2 allele | Small budded | Large budded | Unbudded | Shmoo | Small budded | Large budded | Unbudded | Shmoo |
| SIR2 | 0 | 2 | 98 | 92 | 1 | 5 | 94 | 80 |
| sir2-604 | 10 | 14 | 76 | 66 | 52 | 11 | 37 | 8 |
| sir2-614 | 16 | 25 | 59 | 50 | 59 | 24 | 17 | 0 |
| sir2-620 | 3 | 9 | 88 | 80 | 60 | 29 | 11 | 1 |
Cultures bearing the indicated SIR2 alleles were grown to log phase at 23° and then blocked in G2/M phase with nocodazole (15 μg/ml). When >90% of cells exhibited large buds, the cultures were divided. Half was maintained at 23° for 1 hr and then released from the nocodazole block into media containing α-factor and incubated at 23° (23° → 23° columns). The other half was shifted to 37° for 1 hr and then released into media containing α-factor and incubated at 37° (23° → 37° columns). Budding morphology was monitored at various times and is reported here 4 hr after release from nocodazole. Values shown are the percentage of cells in each morphological class; shmoos are a subset of unbudded cells.
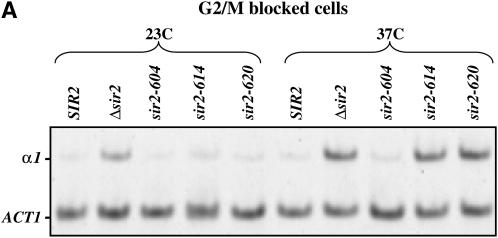
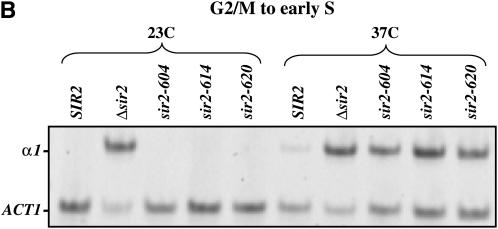
We next used RT–PCR measurements to more narrowly define the point at which silencing is lost in the mitotic interval. First, we blocked cells in G2/M with nocodazole at the permissive temperature and then shifted the culture to the nonpermissive temperature for 5 hr. RNA was collected and analyzed for expression of α1 message. Figure 3A shows that the strain bearing the sir2-604 allele remains substantially repressed despite the temperature shift, while strains bearing the sir2-614 and sir2-620 alleles are derepressed at HML. Finally, we measured α1 message in cells progressing from the G2/M boundary to early S phase. Cells were blocked with nocodazole, shifted to the nonpermissive temperature, and then released from the nocodazole block into media containing hydroxyurea. Figure 3B shows that all strains bearing conditional SIR2 alleles lose repression of HML when traversing this interval at the nonpermissive temperature.
Figure 3.—
Sir2 function during mitosis. (A) α1 mRNA levels in nocodazole-arrested strains. Log-phase cultures grown at 23° were blocked at the G2/M boundary with nocodazole. Each culture was then divided; half was maintained at 23°, and the other half was shifted to 37°. After maintaining cultures at their respective temperatures for 5 hr, RNA was collected and RT–PCR was used to assay α1 and ACT1 message, as described in materials and methods. (B) α1 mRNA levels in strains progressing from G2/M to early S phase. Log-phase cultures grown at 23° were blocked in G2/M with nocodazole. Each culture was then divided; half was maintained at 23°, and the other half was shifted to 37°. Cultures were retained at the nocodazole block for 4 hr and then released at their respective temperatures into media containing hydroxyurea. Eight hours following release from the nocodazole block, RNA was collected and RT–PCR was used to assay α1 and ACT1 message.
Sir2–Sir4 interactions:
These results suggest that the mutations conferring the temperature-sensitive phenotype define distinct functions for Sir2. Silencing is sensitive to disruptions in the function defined by the sir2-614 allele at all points in the cell cycle that we have tested, while silencing is preferentially sensitive to disruptions in the function or functions defined by the sir2-604 and sir2-620 alleles during metaphase and mitosis. To determine the specific functions compromised in the Sir2 proteins produced from these alleles, we examined two known functions of Sir2 likely to be essential for silencing at HML: the ability to interact with Sir4 (Moazed et al. 1997; Cockell et al. 2000) and the ability to deacetylate histone substrates (Imai et al. 2000; Landry et al. 2000b; Smith et al. 2000).
To examine Sir2's ability to interact with Sir4, we performed two-hybrid assays (Toby and Golemis 2001). For bait we used the C-terminal 629 amino acids of Sir4 fused to the DNA-binding domain of the Gal4 protein. Prior studies found that truncations of Sir4's N terminus increased the ability to detect interactions with Sir2 (Moazed et al. 1997; Chang et al. 2003). The two-hybrid reporter strain that we used includes a HIS3 reporter gene with binding sites for the Gal4 DNA-binding domain in its promoter (James et al. 1996). Interaction of the Sir4-binding domain fusion protein with a Sir2–Gal4 activation domain fusion protein allows expression of the HIS3 gene, permitting growth on media lacking histidine. To determine whether the Sir2–Gal4AD fusion proteins used in this study retained their silencing function, we performed the control experiment shown in Figure 4A. Plasmids expressing fusion proteins were introduced into a strain lacking the wild-type SIR2 gene and a mating assay was performed. In each case, the activation domain fusions recapitulated the temperature-sensitive silencing phenotypes seen with the unfused SIR2 alleles.
Figure 4.—
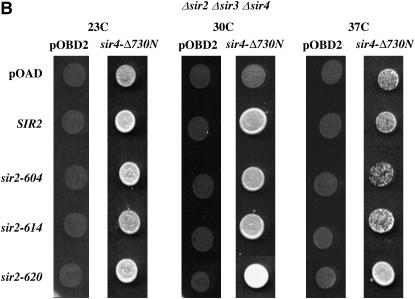
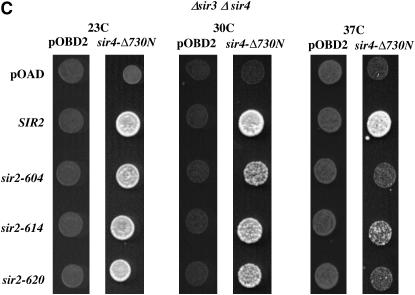
Two-hybrid assays of Sir2–Sir4 interactions. (A) Sir2 fusion proteins maintain function. A mating assay was performed on strains expressing Sir2 proteins fused at their N terminus to the Gal4 activation domain. An equal number of cells from each culture were spotted onto plates spread with a MATα haploid strain. These plates were incubated overnight at 23° or 37° to allow mating, replica plated to media selecting for diploids, and then incubated an additional night at their respective temperature, when the photos shown were taken. “Growth” shows the same haploid cultures spotted onto nonselective plates lacking a mating partner. Equivalent growth controls were done at 23° and 37° with identical results. pOAD is the base vector expressing only the Gal4-activation domain. (B) Sir2–Sir4 interactions. Each row is labeled with the activation domain fusion used; pOAD is the vector control expressing only the Gal4-activation domain. Each column lists the binding domain fusion used; pOBD2 is the vector control expressing only the Gal4-binding domain. Cultures were grown and plated at the temperatures indicated. This experiment was conducted in a strain lacking the endogenous SIR2, SIR3, and SIR4 genes. Equal numbers of cells were applied to media selecting for expression of a HIS3 reporter gene containing Gal4-binding sites in its promoter. Duplicate platings on media selecting only for the plasmids bearing the binding domain and activation domain fusions ensured that platings were equal (not shown). The Sir4–Δ730N binding domain fusion lacks the N-terminal 730 amino acids of Sir4. (C) An experiment was conducted exactly as described in B in a strain lacking the SIR3 and SIR4 genes.
To decrease the possibility of observing signals dependent on indirect interactions between the fusion proteins and endogenous Sir proteins, we initially performed our assay in a strain deleted for the endogenous SIR2, SIR3, and SIR4 genes. Assays were performed at 23°, 30°, and 37° to detect temperature-dependent effects. As shown in Figure 4B, at 23° and 30° all Sir2 proteins tested were able to interact with Sir4, while at 37° the sir2-604 protein has a significantly reduced interaction with Sir4. We also performed this assay in a strain containing the wild-type SIR2 gene. In this experiment, we observed that both the sir2-604 and the sir2-620 protein are unable to interact with the Sir4–BD fusion protein at the nonpermissive temperature, while the sir2-614 protein shows a milder defect (Figure 4C). Competition between the wild-type and Sir2–AD fusion proteins in the SIR2+ strain may have helped to reveal defects in the ability of sir2-620 and sir2-614 to interact with Sir4. Differences in the two strains could also be explained by more complex models invoking interactions between wild-type and mutant Sir2 proteins (Cubizolles et al. 2006).
Deacetylase activity of Sir2 mutants:
Silencing likely requires a Sir2-mediated deacetylation of histones at silenced locations (Imai et al. 2000; Hoppe et al. 2002; Luo et al. 2002; Matecic et al. 2002; Rusche et al. 2002). Removal of acetyl groups from lysines by Sir2 is coupled to the hydrolysis of NAD (Landry et al. 2000a; Tanner et al. 2000; Tanny and Moazed 2001). To examine whether Sir2 enzymatic activity was altered by these SIR2 alleles, we measured its ability to hydrolyze NAD in the presence of a peptide mimicking an acetylated histone tail. Sir2-containing protein complexes were purified from yeast and then assayed for NADase activity (Tanny et al. 2004 and materials and methods). Even at the permissive temperature, each of the alleles has reduced activity compared to wild-type Sir2 (Table 5). At the nonpermissive temperature, the activity of each allele is further reduced; this decrease is most pronounced for the sir2-614 allele. Thus, while these assays do not allow an unambiguous assignment of the specific defect causing the loss of silencing in strains bearing these SIR2 alleles, they do distinguish the alleles in a way that correlates with their phenotypes: one class of alleles, represented by the sir2-614 allele, is particularly defective in enzymatic activity, and silencing is lost at all cell-cycle positions tested. A second class, composed of the sir2-604 and sir2-620 alleles, has primary defects in Sir2–Sir4 interactions and exhibits a cell-cycle-specific defect in silencing.
Complementation of SIR2 alleles:
In some cases, distinct alleles of the same gene are able to complement each other. For instance, coexpression of two distinct, nonfunctional alleles of SIR4 restores silencing to cells lacking the SIR4 gene (Marshall et al. 1987). The observation of transcomplementation can suggest distinct functional domains in the protein. To test the ability of our SIR2 alleles to complement each other, plasmids bearing SIR2 alleles were introduced into strains containing integrated alleles of SIR2 (Figure 5). We used a mating assay to assess silencing at HML. As shown in Figure 5, these plasmid-borne alleles support mating at 23°, but not at 37°, in a strain lacking a chromosomal copy of SIR2. The remaining experiments show that (1) each of the temperature-sensitive alleles is recessive to wild type, (2) increasing the dosage of individual alleles does not significantly affect silencing by this assay, and (3) these sir2 temperature-sensitive alleles do not complement each other. This could mean that the mutations do not affect distinct functions of the protein or that independent functions must reside in a single molecule of Sir2 to promote silencing.
Strains bearing these SIR2 alleles exhibit cell-cycle silencing defects. To examine the possibility that Sir2 levels exhibit cell-cycle dependent changes in stability, we performed Western blots on extracts from cells blocked at G1 or G2/M, at permissive and nonpermissive temperatures (Figure 1B). We found that the levels of each temperature-sensitive protein decreases relative to the wild-type Sir2 protein. However, the level of individual proteins is not affected by temperature, and the ability to promote silencing does not correlate with Sir2 protein levels within the range that we observed. Coupled with our observation that increases in gene dosage do not affect the temperature-sensitive phenotype, we consider it unlikely that alterations in protein levels are responsible for the phenotypes that we see.
Sir2 and the establishment of silencing:
In all contexts reported thus far, the establishment of silencing requires progression through the cell cycle. Our experiments using conditional SIR2 alleles suggest that silencing is particularly sensitive to decreases in the ability of the Sir2 and Sir4 proteins to interact during mitosis, possibly indicating a crucial assembly step for silent chromatin at this point of the cell cycle. To determine if silencing can be established in this interval, we performed a pedigree assay on a yeast strain bearing the sir2-614 allele (Figure 6). Cells grown at the permissive temperature were blocked at the G2/M boundary with nocodazole, subjected to a temperature shift to inactivate Sir2, and then applied to solid media containing α-factor. G2/M-blocked cells were then monitored as they resumed progress through the cell cycle. If cells restore silencing in this interval, they will be sensitive to α-factor, blocked in the subsequent G1 phase of the cell cycle, and eventually adopt a shmoo morphology. Unsilenced cells will be insensitive to α-factor and enter a new cell cycle. Control cells maintained at the permissive temperature throughout the experiment efficiently blocked in α-factor following release from the G2/M block (87% shmoo; see Figure 6), while cells that were maintained at the nonpermissive temperature following release from G2/M showed little response to α-factor (7% shmoo). When cells blocked at G2/M were shifted to the nonpermissive temperature for 3 hr, shifted back to the permissive temperature for 2 hr, and then applied to media containing α-factor, a significant fraction were sensitive to α-factor (66% shmoo). We can compare this number to cells that were shifted to the nonpermissive temperature for 3 hr and then directly transferred out of the nocodazole-containing media to a plate containing α-factor (40% shmoo). Although the pedigree experiment does not allow us to measure the fraction of cells that were unsilenced when released from the nocodazole block, comparison of the last two conditions indicates that significant repression (χ2, P < 0.001) is established when cells are permitted more time at the permissive temperature in this interval.
Figure 6.—
Establishment of silencing in M phase. A strain bearing the sir2-614 allele was grown at 23° and blocked at G2/M with nocodazole. After >90% of the cells in the culture exhibited a large-bud morphology, the culture was divided and subjected to the indicated temperature shifts. At the release point, cells were applied to solid media containing α-factor. Released from the nocodazole-induced block, large-budded cells continued through the cell cycle and were either sensitive to α-factor, forming shmoos, or not sensitive to α-factor, forming buds (cells that neither budded nor formed shmoos, always <10% of the total, were not counted). The percentage of large-budded cells in which at least one of the cell–cell pair exhibited sensitivity to α-factor by forming a shmoo is indicated. Data shown are the cumulative results of two independent experiments. At least 90 large-budded cells were assayed for each condition. The difference between 40 and 66% is significant (χ2, P < 0.001), given the number of events assayed for each condition.
DISCUSSION
Sir2 and mitosis:
Analysis of our conditional alleles of SIR2 suggests a cell-cycle-dependent requirement for Sir2 function. When shifted to the nonpermissive temperature, cells bearing the sir2-614 allele lose silencing in G1 phase and during mitosis. We observed a distinct phenotype when analyzing the sir2-604 and sir2-620 alleles. Strains bearing these alleles did not lose silencing when raised to the nonpermissive temperature in G1 phase, but did during progression from the G2/M boundary to G1. There are two general possibilities for the different phenotypes exhibited by these alleles. First, mutations in SIR2 could affect a single function of the protein, with the sir-614 allele having a more pronounced effect at high temperature than the sir-604 or sir-620 mutations. In this model, cells in mitosis are more sensitive to this deficit, and sir2-614p is more deficient in this function than sir2-604p or sir2-620p. However, if the copy number of the SIR2 temperature-sensitive alleles is increased in haploid strains, there is no change in mating behavior, suggesting that the silencing defects are not simply due to dosage effects. Alternatively, the mutations could affect independent functions of Sir2, with the sir2-604 and sir2-620 alleles defining a cell-cycle-specific function.
In our conditional alleles of SIR2, the mutations that are sufficient to confer the temperature-sensitive mating defect cause alterations of the conserved region of Sir2 known to be sufficient for enzymatic function, within the carboxy-terminal region required for interacting with Sir4, or both (see Table 2). The sir2-614 and sir2-620 alleles introduce amino acid substitutions within the enzymatic core, while the mutations in sir2-604 and sir2-620 introduce a premature stop codon resulting in the deletion of 30 or 24 amino acids, respectively, from the carboxy terminus. None of these mutations alter amino acids highly conserved in the Sir2 family. However, this C-terminal region has been shown to be important for Sir2's interaction with Sir4 (Cockell et al. 2000). Our experiments suggest that the Sir2-614 protein has a temperature-dependent defect in enzymatic activity. This result is consistent with the prior observation that a small molecule inhibitor of Sir2's catalytic activity causes loss of silencing in log-phase cells or in cells arrested in G1 phase (Bedalov et al. 2001). While we cannot rule out the possibility that the Sir2-614 protein has defects in addition to impaired enzymatic function, these results are consistent with a requirement for Sir2's enzymatic activity throughout the cell cycle. In cycling cells, the levels of the sir2-614 protein appear to increase at the nonpermissive temperature (Figure 1B). A recent report indicates that Sir2 levels are subject to autoregulation (Michel et al. 2005); our results suggest that Sir2's enzymatic activity may be required for feedback inhibition.
The Sir2-604 and Sir2-620 proteins exhibit a decreased ability to interact with Sir4 and also a more minor impairment in enzymatic activity. Again, while these alleles could have additional defects not yet revealed by our assays, these results may reflect a greater sensitivity to the Sir2–Sir4 interaction at specific points of the cell cycle, including mitosis. Interaction with Sir4 increases the catalytic activity of Sir2 (Tanny et al. 2004); thus, this decrease in Sir4 interaction could contribute to the decrease in catalytic activity that we measured in these proteins.
There is no evidence that silencing fails at any point in the cell cycle. However, our results suggest that the structure maintaining transcriptional repression is not static; decreasing the function of Sir2 has differential effects depending on cell-cycle position. A conclusion that silent chromatin is dynamic is supported by prior observations. First, expression of a transactivator is able to promote expression of a telomere-linked URA3 gene when cells are blocked at the G2/M boundary, but is unable to do so in cells blocked in G1 phase (Aparicio and Gottschling 1994), suggesting that a transition in silencing occurs in mitosis. A silenced HML locus uncoupled from the cis-acting silencer sequences is derepressed specifically by passage through mitosis (Martins-Taylor et al. 2004), supporting the view that silent chromatin is particularly vulnerable to disruption at this point. Several studies have demonstrated that the establishment of silencing requires cell-cycle progression. The cell-cycle interval necessary for the establishment of silencing may depend upon the experimental system. In specific instances, S-phase passage has been shown to be necessary and sufficient to establish silencing (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Kirchmaier and Rine 2006). However, establishment of silencing following inactivation and reactivation of a conditional SIR3 allele occurs primarily in M phase (Lau et al. 2002; Martins-Taylor et al. 2004) while M-phase passage is necessary and sufficient to silence a telomere-linked gene following induced elevation of Sir3p levels (Martins-Taylor et al. 2004). Here we show that passage through the G2/M–G1 interval is sufficient to restore silencing in a strain bearing the sir2-614 allele. Our identification of SIR2 alleles with a mitosis-specific defect is consistent with a critical assembly step coinciding with mitosis.
In wild-type cells, silencing rarely fails and thus does not require frequent reestablishment. The significance of establishment experiments is in identifying intervals of dynamic reassembly or alteration of silent chromatin. In particular, these experiments suggest that a transition from a tentatively silenced state to a more stable state occurs during mitosis. What is the crucial event in mitosis promoting the assembly of silent chromatin? The identification of the Scc1 cohesin as a modifier of silencing (Lau et al. 2002; Papacs et al. 2004; Suter et al. 2004) is consistent with a hypothesis that silencing is tied to the dynamic changes in chromosome structure that accompany cell-cycle progression.
While several proteins necessary for silencing have been identified and characterized, the specific mechanism of silencing and the physical state of chromatin sufficient to repress gene transcription are not clear. Detailed characterization of these dynamic transitions will likely continue to provide valuable insights into the silencing mechanism.
Acknowledgments
We thank members of the Holmes Lab for valuable discussions, Tania Rozario for assistance in data analysis, Katrina Catron for comments on the manuscript, and the University of Washington Yeast Resource Center for strains and plasmids. M.H. gratefully acknowledges support from a Howard Hughes Foundation Undergraduate Research Grant to Wesleyan University. This work was supported by grants from the American Cancer Society (RPG-98-351-01-MGO) and the National Science Foundation (MCB-0096561) to S.G.H.
References
- Afshar, G., and J. P. Murnane, 1999. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene 234: 161–168. [DOI] [PubMed] [Google Scholar]
- Aparicio, O. M., and D. E. Gottschling, 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Bedalov, A., T. Gatbonton, W. P. Irvine, D. E. Gottschling and J. A. Simon, 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98: 15113–15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus et al., 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9: 2888–2902. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis and J. R. Broach, 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7: 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., R. E. Sobel, C. D. Allis, B. M. Turner and J. R. Broach, 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16: 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis et al., 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12: 165–170. [DOI] [PubMed] [Google Scholar]
- Buck, S. W., J. J. Sandmeier and J. S. Smith, 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111: 1003–1014. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., L. Milne and M. Grunstein, 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277: 4778–4781. [DOI] [PubMed] [Google Scholar]
- Chang, J. F., B. E. Hall, J. C. Tanny, D. Moazed, D. Filman et al., 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11: 637–649. [DOI] [PubMed] [Google Scholar]
- Chi, M.-H., and D. Shore, 1996. SUM1–1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16: 4281–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell, M. M., S. Perrod and S. M. Gasser, 2000. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154: 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubizolles, F., F. Martino, S. Perrod and S. M. Gasser, 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21: 825–836. [DOI] [PubMed] [Google Scholar]
- Dula, M. L., and S. G. Holmes, 2000. MGA2 and SPT23 are modifiers of transcriptional silencing in yeast. Genetics 156: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C., A. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547–1551. [DOI] [PubMed] [Google Scholar]
- Frye, R. A., 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273: 793–798. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser and M. Grunstein, 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., and J. R. Broach, 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30: 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, S., C. M. Armstrong, M. Kaeberlein and L. Guarente, 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291: 646–650. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2006. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar, A. J., J. N. Strathern, J. R. Broach and J. B. Hicks, 1981. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature 289: 239–244. [DOI] [PubMed] [Google Scholar]
- Landry, J., J. T. Slama and R. Sternglanz, 2000. a Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278: 685–690. [DOI] [PubMed] [Google Scholar]
- Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins et al., 2000. b The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16: 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650–653. [DOI] [PubMed] [Google Scholar]
- Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz and D. Moazed, 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527. [DOI] [PubMed] [Google Scholar]
- Luo, K., M. A. Vega-Palas and M. Grunstein, 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, D. J., and J. R. Broach, 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9: 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, M., D. Mahoney, A. Rose, J. B. Hicks and J. R. Broach, 1987. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor, K., M. L. Dula and S. G. Holmes, 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic, M., S. Stuart and S. G. Holmes, 2002. SIR2-induced inviability is suppressed by histone H4 overexpression. Genetics 162: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, A. H., B. Kornmann, K. Dubrana and D. Shore, 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19: 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A., and K. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312: 247–251. [DOI] [PubMed] [Google Scholar]
- Moazed, D., 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13: 232–238. [DOI] [PubMed] [Google Scholar]
- Moazed, D., A. Kistler, A. Axelrod, J. Rine and A. Johnson, 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94: 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti, P., K. Freeman, L. Coodly and D. Shore, 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8: 2257–2269. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., R. Hunter and R. Parker, 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8: 79–82. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. A., K. Tatchell, B. D. Hall, C. Astell and M. Smith, 1981. A position effect in the control of transcription at yeast mating type loci. Nature 289: 244–250. [DOI] [PubMed] [Google Scholar]
- Papacs, L. A., Y. Sun, E. L. Anderson, J. Sun and S. G. Holmes, 2004. REP3-mediated silencing in Saccharomyces cerevisiae. Genetics 166: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret et al., 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann et al., 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 27: 481–516. [DOI] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244. [DOI] [PubMed] [Google Scholar]
- Sikorski, R., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, P. G., and K. Tatchell, 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37: 969–978. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad et al., 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97: 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., J. Avalos, I. Celic, S. Muhammad, C. Wolberger et al., 2002. SIR2 family of NAD(+)-dependent protein deacetylases. Methods Enzymol. 353: 282–300. [DOI] [PubMed] [Google Scholar]
- Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks and R. Sternglanz, 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11: 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies et al., 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256. [DOI] [PubMed] [Google Scholar]
- Suter, B., A. Tong, M. Chang, L. Yu, G. W. Brown et al., 2004. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, K. G., J. Landry, R. Sternglanz and J. M. Denu, 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97: 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., and D. Moazed, 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz and D. Moazed, 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745. [DOI] [PubMed] [Google Scholar]
- Tanny, J. C., D. S. Kirkpatrick, S. A. Gerber, S. P. Gygi and D. Moazed, 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24: 6931–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby, G. G., and E. A. Golemis, 2001. Using the yeast interaction trap and other two-hybrid-based approaches to study protein-protein interactions. Methods 24: 201–217. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Wyrick, J. J., F. C. P. Holstege, E. G. Jennings, H. C. Causton, D. Shore et al., 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]