Abstract
Pleiotropy refers to the observation of a single gene influencing multiple phenotypic traits. Although pleiotropy is a common phenomenon with broad implications, its molecular basis is unclear. Using functional genomic data of the yeast Saccharomyces cerevisiae, here we show that, compared with genes of low pleiotropy, highly pleiotropic genes participate in more biological processes through distribution of the protein products in more cellular components and involvement in more protein–protein interactions. However, the two groups of genes do not differ in the number of molecular functions or the number of protein domains per gene. Thus, pleiotropy is generally caused by a single molecular function involved in multiple biological processes. We also provide genomewide evidence that the evolutionary conservation of genes and gene sequences positively correlates with the level of gene pleiotropy.
PLEIOTROPY refers to the observation that a single gene affects two or more distinct and seemingly unrelated traits. Pleiotropy is one of the most commonly observed attributes of genes, with broad implications in genetics, evolution, development, aging, disease, and drug discovery (Williams 1957; Wright 1968; Barton 1990; Hodgkin 1998; Waxman and Peck 1998; Brunner and van Driel 2004; Otto 2004; Promislow 2004; van de Peppel and Holstege 2005). Genes of high pleiotropy are expected to be under strong stabilizing selection because they affect multiple traits (Hodgkin 1998). Pleiotropy also causes compromises among adaptations of different traits, because a genetic change beneficial to one trait may be deleterious to another (Barton 1990; Otto 2004). It is believed that this property underlies many fundamental principles and phenomena in biology, including senescence, trade-off, and cooperation (Williams 1957; Foster et al. 2004; MacLean et al. 2004). For example, it was proposed that mutant genes advantageous to development and reproduction are deleterious after the reproductive age and cause senescence, which may explain why all species have a limited life span (Williams 1957). Social amoeba Dictyostelium discoideum can aggregate during starvation where some cells die to form a stalk that holds the other cells aloft as reproductive spores (Strassmann et al. 2000). A recent study showed that deleting the gene dimA in D. discoideum allows cells to avoid death, but leads to a great reduction in spore production (Foster et al. 2004). Hence, the pleiotropic effects of dimA stabilize the cooperation among amoeba. Pleiotropy also has important implications in human diseases. For instance, mutations in the homeobox gene ARX cause ambiguous genitalia and lissencephaly (whole or parts of the surface of the brain appear smooth) (OMIM at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM). Mutants of AMT, a phosphatidylinositol-3 kinase gene, show the symptoms of cerebellar ataxia, telangiectases (visibly dilated blood vessel on the skin or mucosal surface), immune defects, and a predisposition to malignancy (OMIM).
A central question about pleiotropy is whether the pleiotropic effects of a gene are conferred by multiple molecular functions of the gene or by multiple consequences of a single molecular function (Dudley et al. 2005; van de Peppel and Holstege 2005). A typical example of a pleiotropic gene with multiple molecular functions is the mammalian serum albumin, which is well known for binding fatty acids and toxic metabolites, but it is also involved in the oxidation of nitric oxide (Rafikova et al. 2002). By contrast, the yeast gene HIS7 encodes glutamine amidotransferase, yet this single catalytic activity is used in both histidine biosynthesis and purine nucleotide monophosphate biosynthesis. Although the molecular basis of pleiotropy may vary among genes, it is still important to ask whether one of the above two mechanisms explains the majority of pleiotropic genes in a genome. To address this question, we take advantage of a recently generated genomewide data set of gene pleiotropy from the yeast Saccharomyces cerevisiae (Dudley et al. 2005). We show that the pleiotropic effects of a gene are not usually conferred by multiple molecular functions of the gene, but by multiple consequences of a single molecular function.
MATERIALS AND METHODS
The S. cerevisiae gene pleiotropy data (Dudley et al. 2005) were downloaded from http://arep.med.harvard.edu/pheno/default.htm. The data set included information about the growth rate of 4710 yeast homozygous single-gene deletion mutants in 21 adverse conditions, compared to the growth rate under the control (YPD medium) condition. These 4710 genes were all nonessential, meaning that the homozygous deletion strains could grow on rich media. See Dudley et al. (2005) for detailed information about the 21 conditions. The high-confidence subset of the data was used. In other words, slow growth was inferred only when both replicates showed slow growth. We treated strong growth defect and moderate growth defect equally. In some cases, several mutants in which the same gene was deleted showed different phenotypes or deletion mutants showed higher growth rates under adverse conditions than under the control condition. Genes involved in either of the above two situations were excluded from our analyses. The final data set included 4494 genes, among which 741 genes exhibited growth defects in at least one of the 21 tested adverse conditions compared with the control condition. The analysis was repeated when the 21 conditions were classified into nine condition groups according to their differences from one another on the basis of their effects on gene phenotypes (Dudley et al. 2005). The nine condition groups were YPGly and YPLac; benomyl and MPA; FeLim, paraq, and YPRaff; Sorb, UV; CaCl2, cyclohex, and HU; CAD and EtOH; Caff and rap; lowPO4, pH 3, NaCl, and YPgal; and HygroB. A gene was considered to have a phenotypic effect in a condition group if it had a phenotypic effect in at least one of the constituents of the condition group.
The yeast Gene Ontology (GO) annotations were downloaded from ftp://genome-ftp.stanford.edu/pub/yeast/literature_curation. Genes with the “unknown” annotations (molecular function: 0005554; biological process: 0000004; cellular component: 0008372) were excluded and only nonredundant annotations were considered. Yeast Enzyme Commission (EC) codes were downloaded from ftp://ftpmips.gsf.de/yeast/catalogues/eccat/eccat_data_20062005. Only genes with at least one EC code were considered. The predicted functional domains of yeast proteins were obtained from Munich Information Center for Protein Sequences (MIPS) (ftp://ftpmips.gsf.de/yeast/catalogues/motifs/) and genes with no domain predictions were excluded. The number of genes directly regulated by each transcriptional factor was obtained from the ChIP–chip (chromatin immunoprecipitation followed by the identification of immunoprecipitated genomic fragments through the use of whole-genome DNA chips) experiments (Harbison et al. 2004). The yeast protein–protein interaction (PPI) data were compiled by Han and colleagues, who used stringent criteria to avoid false-positive data points (Han et al. 2004). Proteins with no known PPIs were excluded. The yeast stable protein complex data used here were downloaded from ftp://genome-ftp.stanford.edu/pub/yeast/literature_curation/go_protein_complex_slim.tab. We found that genes involved in stable protein complexes were more pleiotropic than genes not involved in protein complexes (P < 10−4, Mann–Whitney U-test).
For the evolutionary analysis, we conducted genomewide all-against-all BLASTP searches (E-value cutoff = 10−10) between 5773 yeast and 13,434 fruit fly (Drosophila melanogaster) proteins, which were downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org/) and ENSEMBL (http://www.ensembl.org), respectively. Similar BLASTP searches were also conducted between the yeast proteins and 19,873 nematode (Caenorhabditis elegans) proteins and between the yeast proteins and 4999 fission yeast (Schizosaccharomyces pombe) proteins, respectively. The nematode protein sequences were obtained from ENSEMBL, whereas the fission yeast protein sequences were downloaded from http://www.sanger.ac.uk/Projects/S_pombe. We then considered the 4494 yeast genes with pleiotropy information. Nonsynonymous nucleotide distances (dN) between orthologous genes of the yeasts S. cerevisiae and Saccharomyces bayanus were obtained from Zhang and He (2005). We measured the rank correlation between dN and gene pleiotropy. Furthermore, we measured the partial rank correlation between dN and gene pleiotropy when the expression level of the gene was controlled for.
RESULTS
Molecular functions:
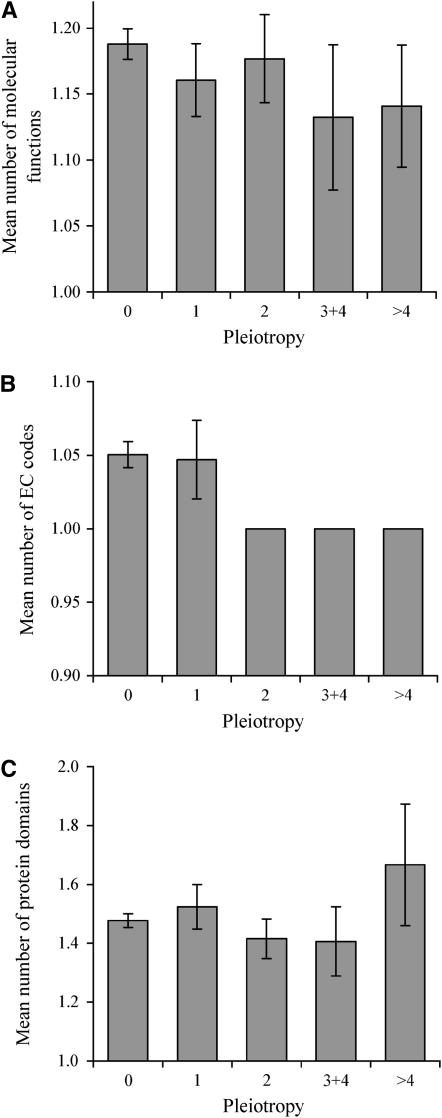
In the yeast gene pleiotropy data set that we use here (Dudley et al. 2005), the level of pleiotropy was measured for each yeast gene by the number of lab conditions (of the 21 conditions) under which the homozygous gene-deletion strain showed significantly slower growth than under the control condition. We first examine whether the level of gene pleiotropy is correlated with the number of molecular functions per gene. We use the molecular function annotation in GO (Ashburner et al. 2000), which describes activities, such as catalytic or binding activities, at the molecular level. A gene may possess one or more than one activity. For example, BMH1 shows both DNA-binding and protein-binding activities, thus having two molecular functions. Nevertheless, we find no correlation between the level of pleiotropy and the number of molecular functions among 2386 yeast genes for which both pleiotropy information and GO annotation are available (Spearman's rank correlation coefficient r = −0.01, two-tailed P = 0.57). We also group genes according to their level of pleiotropy, but find different groups to have similar mean numbers of molecular functions (Figure 1A). Next, we examine EC codes for all yeast enzymes. EC codes are a numerical classification scheme for enzymes based on the chemical reactions that they catalyze. Although the majority of enzymes have only one EC code, some have more than one code because they catalyze multiple different chemical reactions. For instance, THI6 is both a thiamine–phosphate diphosphorylase (EC 2.5.1.3) and a hydroxyethylthiazole kinase (EC 2.7.1.50). However, no significant correlation is observed between the number of EC codes and the level of pleiotropy among 917 genes for which both EC and pleiotropy information is available (r = −0.06, P = 0.09; see also Figure 1B). Furthermore, there is no correlation between the number of protein domains per gene and the level of gene pleiotropy (r = 0.01, P = 0.62; see Figure 1C). Thus, all three measures of molecular functions indicate that gene pleiotropy is not attributable to an excess of molecular functions.
Figure 1.—
Gene pleiotropy is not due to an excess of molecular functions per gene. Mean numbers of (A) GO-annotated molecular functions per gene, (B) EC codes per gene, and (C) protein domains per gene for genes of different levels of pleiotropy. Pleiotropy is measured by the number of adverse conditions (of 21 tested conditions) under which the homozygous gene-deletion strain shows significantly slower growth than under the control condition. For the unbinned data, the rank correlation coefficient is −0.01 (P = 0.57), −0.06 (P = 0.09), and 0.01 (P = 0.62) between pleiotropy and the numbers of molecular functions, EC codes, and protein domains, respectively. The numbers of genes in the five bins are 1890, 193, 164, 68, and 71, respectively, in A; 755, 64, 60, 18, and 20, respectively, in B; and 1213, 105, 89, 32, and 33, respectively, in C. Error bar shows one standard error of mean.
Biological processes:
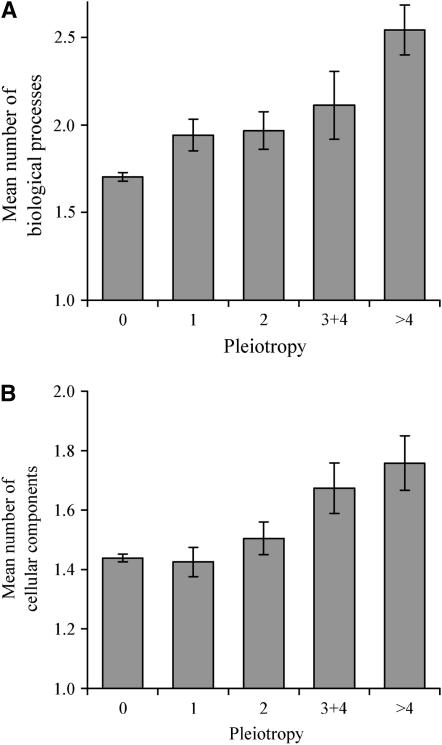
We then investigate the relationship between pleiotropy and the number of biological processes in which each gene participates, again using GO. A GO-annotated biological process is series of events accomplished by one or more ordered assemblies of molecular functions, such as pyrimidine metabolism or α-glucoside transport (Ashburner et al. 2000). A gene may participate in one or multiple biological processes. Not unexpectedly, a significant positive correlation exists between the level of gene pleiotropy and the number of biological processes in which the gene participates (r = 0.12, P < 10−10). When the genes are grouped by the level of pleiotropy, we observe a clear trend that the mean number of biological processes per gene increases with pleiotropy (Figure 2A). We also observe a weak, but significant, positive correlation between the number of cellular components where the product of a gene is located (as annotated by GO) and the level of gene pleiotropy (r = 0.05, P < 0.003; see Figure 2B). Here, a cellular component refers to a component of a cell but with the proviso that it is part of some larger object, which may be an anatomical structure or a gene product group (Ashburner et al. 2000). We hypothesize that the correlation between the number of cellular components and pleiotropy arises because gene products distributed among more cellular components have opportunities to participate in more biological processes. Indeed, the number of cellular components and the number of biological processes are significantly correlated (r = 0.15, P < 10−14). After we control for the number of biological processes, the correlation between the number of cellular components and pleiotropy is no longer significant (r = 0.03, P = 0.09). On the contrary, the correlation between the number of biological processes and pleiotropy is reduced only slightly by the control of the number of cellular components (r = 0.11, P < 10−8). These results indicate that gene pleiotropy is likely due to multiple biological processes in which the gene participates, and the multiple participations are realized in part by having the gene product distributed into multiple cellular components.
Figure 2.—
Gene pleiotropy correlates with (A) the number of GO-annotated biological processes and (B) the number of GO-annotated cellular components into which gene products are distributed. Pleiotropy is measured by the number of conditions under which the homozygous gene-deletion strain shows significantly slower growth than under the control condition. For the unbinned data, the rank correlation coefficient is 0.12 (P < 10−10) and 0.05 (P < 0.003) between pleiotropy and the numbers of biological processes and cellular components, respectively. The numbers of genes in the five bins are 2209, 244, 189, 88, and 92, respectively, in A and 2799, 268, 196, 92, and 95, respectively, in B. Error bar shows one standard error of mean.
Protein–protein interactions:
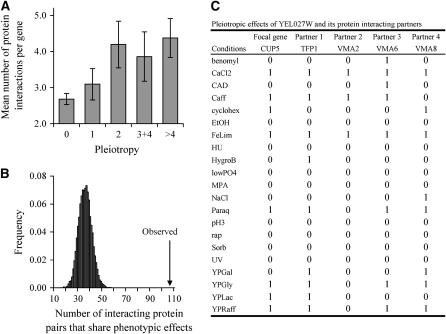
Genomewide studies showed that most genes function by PPIs (von Mering et al. 2002; He and Zhang 2006). We thus hypothesize that many pleiotropic genes participate in multiple biological processes through engaging in multiple PPIs. To test this hypothesis, we analyze a recently compiled yeast PPI data set (Han et al. 2004). We find that the number of PPIs that a gene has is positively correlated with its level of pleiotropy (r = 0.19, P < 10−6; see Figure 3A). Although pleiotropic genes also tend to be involved in stable protein complexes (see materials and methods), the correlation between the number of PPIs and the level of pleiotropy remains significant when proteins involved in protein complexes are removed (r = 0.16, P = 0.0015; see Figure S1 at http://www.genetics.org/supplemental/). Furthermore, from the yeast PPI network we identify 106 pairs (n) of interacting proteins that share at least one phenotype (i.e., condition under which slow growth is observed). The number n between random pairs of genes can be estimated by randomly rewiring the yeast PPI network while keeping the number of interactions constant for every protein. The average n from 10,000 randomly rewired PPI networks is 35.9, with none of the 10,000 n values ≥106 (Figure 3B). Thus, interacting proteins share phenotypic effects significantly more often than by chance (P < 10−4), suggesting that PPIs underlie some phenotypic effects of genes. We also identified 15 cases in which a focal gene shares different phenotypes with different interacting partners, indicative of pleiotropy arising from multiple PPIs. Figure 3C shows one example in which the seven phenotypes of the focal gene are shared with four different PPI partners. It should be pointed out that the different fitness effects of different gene deletions were controlled for when the yeast gene phenotypic data were generated (see materials and methods), so the observed relationship between pleiotropy and the number of PPIs is not due to the fact that genes with more PPIs tend to show detectable phenotypes upon deletions (Jeong et al. 2001; He and Zhang 2006).
Figure 3.—
Multiple PPIs underlie gene pleiotropy. (A) Gene pleiotropy correlates with the number of PPIs per gene. Pleiotropy is measured by the number of conditions under which the homozygous gene-deletion strain shows significantly slower growth than under the control condition. For the unbinned data, the rank correlation coefficient is 0.19 (P < 10−6) between pleiotropy and the number of PPIs per gene. The numbers of genes in the five bins are 501, 85, 61, 42, and 48, respectively. Error bar shows one standard error of mean. (B) Interacting proteins tend to share phenotypic effects. The arrow indicates the observed number of interacting protein pairs for which at least one phenotype (i.e., condition under which slow growth is found) is shared. The bars show the frequency distribution of the number of randomly paired proteins for which at least one phenotype is shared. The distribution is generated from 10,000 randomly rewired yeast PPI networks. (C) An example showing the phenotypes shared between a focal gene CUP5, also known as YEL027W, and all of its PPI partners. “0,” no phenotype; “1,” with phenotype. See Dudley et al. (2005) for the detailed information of the 21 conditions.
We reason that having multiple PPIs contributes to gene pleiotropy because a gene can participate in multiple biological processes through different PPIs. Because two randomly picked genes have only a negligible chance (1.56%) to share a biological process, biological processes shared between interacting proteins are likely dependent on their interaction, although the biological processes of some genes may have been inferred from the PPI information. We find a strong positive correlation between the number of PPIs that a gene has and the total number of nonredundant biological processes that the gene shares with its interacting partners (r = 0.33; P < 10−18). This result provides strong evidence that at least one of the molecular mechanisms by which highly pleiotropic genes participate in more biological processes is by multiple PPIs. Another potential mechanism is protein–DNA interaction. We find that although highly pleiotropic transcription factors do regulate more target genes on average, the correlation is not statistically significant (r = 0.14, P = 0.22). This result, however, could be due to the lack of statistical power, as only 83 transcriptional factor genes are included in our data set (see materials and methods). In an earlier study, Promislow (2004) found a positive correlation between the number of PPIs and gene pleiotropy. However, his gene pleiotropy was not directly measured, but was inferred from the number of functional classifications listed in MIPS. In effect, his finding was a correlation between the number of PPIs and the number of biological processes.
DISCUSSION
By conducting a genomewide analysis of the relationship between yeast gene pleiotropy and gene function, we discovered that gene pleiotropy is generally achieved by the use of a single molecular function in multiple biological processes, which is realized in part by the distribution of the gene product into multiple cellular components and by participation of the gene in different protein–protein interactions. Our analysis has several potential caveats. First, the gene pleiotropy data set analyzed here is not large. It includes only 21 conditions, and only 741 genes show phenotypic effects in at least one condition. Second, the 21 conditions tested for each gene-deletion strain may not be completely independent, which may lead to biased estimates of gene pleiotropy. This potential bias, however, should not affect our results as the bias applies to all genes equally. In fact, our main conclusion still holds when we merge the 21 conditions into nine highly independent condition groups and repeat the analyses (see materials and methods and Figures S2–S4 at http://www.genetics.org/supplemental/). Third, because yeast genes do not undergo alternative splicing, it is unknown whether alternative splicing is an important factor contributing to pleiotropy in species with prominent alternative splicing. Similarly, it is unknown whether pleiotropy could arise from gene expression in multiple tissues of multicellular organisms. Fourth, some of the GO biological processes may have been annotated by the yeast phenotypes upon gene deletion, although the exact overlap between the GO annotation and the current gene pleiotropy data (Dudley et al. 2005) is hard to assess. But, at any rate, the positive correlation between gene pleiotropy and the number of biological processes is what one should expect, even when GO annotation is completely independent from the gene pleiotropy data. The critical finding of our analysis is the lack of correlation between pleiotropy and the number of molecular functions, which is used to distinguish between the two competing hypotheses of the molecular basis of gene pleiotropy. Finally, although the functional annotations such as the GO terms, EC codes, and PPIs are much more complete and reliable for yeast than for other model organisms, false-positive and/or false-negative errors may still exist. These errors may in part explain why some of the statistically significant correlations are of small magnitudes. Furthermore, the errors could potentially limit our ability to discover true relationships. However, the lack of positive correlation between gene pleiotropy and the number of molecular functions is found for all three different measures of molecular functions and thus is likely to be real.
To test the robustness of our results, we analyzed another yeast gene pleiotropy data set (Parsons et al. 2004). In this data set, the growth rate of each single-gene deletion strain was measured in the presence and absence of 1 of 12 diverse inhibitory compounds. The pleiotropic level of a gene is defined by the number of compounds that inhibit the growth of the deletion strain. Similar to the above results, we do not find a significant correlation between the number of GO-defined molecular functions and the pleiotropic level of a gene (r = 0.03, two-tailed P = 0.13, n = 2386; see Figure S5a at http://www.genetics.org/supplemental/ for binned data). In contrast, a significant positive correlation between the number of GO-defined biological processes and pleiotropy is observed (r = 0.13, two-tailed P < 10−9, n = 2822; see Figure S5b at http://www.genetics.org/supplemental/ for binned data). Thus, our conclusion on the molecular basis of yeast gene pleiotropy appears robust for different data sets. At this time, however, no genomewide data sets of gene pleiotropy are available for other organisms, making it difficult to conclude whether the mechanisms revealed from the yeast apply to all organisms.
Revelation of the molecular basis of pleiotropy has several implications. Much effort has been employed to identify alleles of a pleiotropic gene, each of which affects one of the many phenotypic effects of the gene. This strategy deserves reevaluation because, if the pleiotropic effects of a gene are usually due to the same molecular function, it would be difficult or at least inefficient to isolate gene mutants that affect only one trait. This concern is particularly meaningful in human genetics where isolation of symptom-specific alleles is thought to be important for developing effective treatments (Dudley et al. 2005). Our finding suggests that developing drugs that target only one particular phenotypic effect of a pleiotropic gene is likely to be difficult. However, targeting a specific protein interaction or an interacting partner of the pleiotropic gene might be a useful strategy. Our results also show the universality that the same molecular function of a gene is repeatedly used in different biological processes.
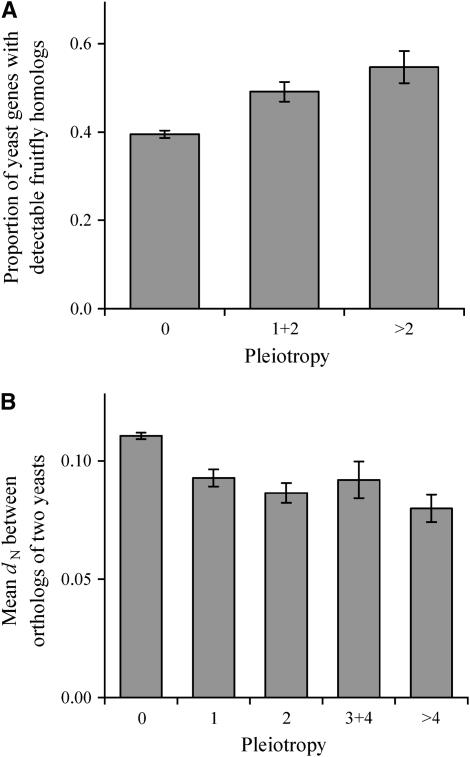
Pleiotropic genes are widely believed to be evolutionarily conserved because they are subject to purifying selection acting on multiple traits and are less likely to experience beneficial mutations (Fisher 1958; Hodgkin 1998). We find that 39.5 ± 0.8% of nonpleiotropic yeast genes (no phenotype in any condition) have detectable homologs in the fruit fly D. melanogaster (Figure 4A). In comparison, 49.2 ± 2.3% of low pleiotropic genes (with phenotypes in one to two conditions) and 54.7 ± 3.6% of high pleiotropic genes (with phenotypes in more than two conditions) have fruit fly homologs. Together, pleiotropic genes are significantly more likely to be retained in long-term evolution than nonpleiotropic genes (χ2 = 29, P < 10−7). Similarly, 52.6 ± 2.7% of pleiotropic yeast genes have detectable homologs in the nematode C. elegans, in comparison to 38.3 ± 1.1% of nonpleiotropic genes (χ2 = 20, P < 10−5). When the fungus S. pombe is compared, 71.7 ± 3.3% of pleiotropic yeast genes have detectable homologs, in comparison to 58.4 ± 1.3% of nonpleiotropic genes (χ2 = 41, P < 10−9). We also computed the nonsynonymous nucleotide distance (dN) between orthologous genes of S. cerevisiae and S. bayanus, two closely related yeast species, and observed a negative correlation between pleiotropy and dN (r = −0.12, P < 10−11), suggesting that pleiotropic genes tend to evolve more slowly at nonsynonymous sites (Figure 4B), consistent with a recent study that was based on fewer genes (Salathe et al. 2006). Our result is robust (r = −0.09, P < 10−7) even when we control for the level of gene expression, the most important determinant of dN in yeasts (Pal et al. 2001; Zhang and He 2005; Drummond et al. 2006). Thus, genomewide analyses demonstrate that pleiotropy leads to the evolutionary conservation of genes and gene sequences.
Figure 4.—
Evolutionary conservation of genes and gene sequences correlates with gene pleiotropy. (A) Proportion of yeast genes with detectable fruit fly homologs increases with the level of gene pleiotropy. The proportion is significantly greater among pleiotropic genes than among nonpleiotropic genes (χ2 = 29, P < 10−7). (B) The number of nonsynonymous substitutions per nonsynonymous site (dN) between orthologous genes of S. cerevisiae and S. bayanus decreases with gene pleiotropy. The numbers of yeast genes with fly homologs are 1317, 240, and 104, respectively, in the three bins of A. The numbers of genes are 2770, 259, 184, 86, and 87, respectively, in the five bins of B. Error bar shows one standard error of (A) the proportion estimate or (B) the mean dN estimate.
Acknowledgments
We thank Soochin Cho, Wendy Grus, Ondrej Podlaha, and two anonymous reviewers for valuable comments. This work was supported by research grants from the University of Michigan and the National Institutes of Health to J.Z.
References
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler et al., 2000. Gene ontology: tool for the unification of biology: the Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., 1990. Pleiotropic models of quantitative variation. Genetics 124: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, H. G., and M. A. van Driel, 2004. From syndrome families to functional genomics. Nat. Rev. Genet. 5: 545–551. [DOI] [PubMed] [Google Scholar]
- Drummond, D. A., A. Raval and C. O. Wilke, 2006. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 23: 327–337. [DOI] [PubMed] [Google Scholar]
- Dudley, A., D. Janse, A. Tanay, R. Shamir and G. Church, 2005. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 1: 2005.0001. [DOI] [PMC free article] [PubMed]
- Fisher, R. A., 1958. The Genetic Theory of Natural Selection. Dover, New York.
- Foster, K. R., G. Shaulsky, J. E. Strassmann, D. C. Queller and C. R. Thompson, 2004. Pleiotropy as a mechanism to stabilize cooperation. Nature 431: 693–696. [DOI] [PubMed] [Google Scholar]
- Han, J. D., N. Bertin, T. Hao, D. S. Goldberg, G. F. Berriz et al., 2004. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430: 88–93. [DOI] [PubMed] [Google Scholar]
- Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac et al., 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., and J. Zhang, 2006. Why do hubs tend to be essential? PLoS Genet. 2: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1998. Seven types of pleiotropy. Int. J. Dev. Biol. 42: 501–505. [PubMed] [Google Scholar]
- Jeong, H., S. P. Mason, A. L. Barabasi and Z. N. Oltvai, 2001. Lethality and centrality in protein networks. Nature 411: 41–42. [DOI] [PubMed] [Google Scholar]
- MacLean, R. C., G. Bell and P. B. Rainey, 2004. The evolution of a pleiotropic fitness tradeoff in Pseudomonas fluorescens. Proc. Natl. Acad. Sci. USA 101: 8072–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., 2004. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. Biol. Sci. 271: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, C., B. Papp and L. D. Hurst, 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, A. B., R. L. Brost, H. Ding, Z. Li, C. Zhang et al., 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22: 62–69. [DOI] [PubMed] [Google Scholar]
- Promislow, D. E., 2004. Protein networks, pleiotropy and the evolution of senescence. Proc. Biol. Sci. 271: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafikova, O., R. Rafikov and E. Nudler, 2002. Catalysis of S-nitrosothiols formation by serum albumin: the mechanism and implication in vascular control. Proc. Natl. Acad. Sci. USA 99: 5913–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathe, M., M. Ackermann and S. Bonhoeffer, 2006. The effect of multifunctionality on the rate of evolution in yeast. Mol. Biol. Evol. 23: 721–722. [DOI] [PubMed] [Google Scholar]
- Strassmann, J. E., Y. Zhu and D. C. Queller, 2000. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408: 965–967. [DOI] [PubMed] [Google Scholar]
- van de Peppel, J., and F. C. Holstege, 2005. Multifunctional genes. Mol. Syst. Biol. 1: 2005.0003. [DOI] [PMC free article] [PubMed]
- von Mering, C., R. Krause, B. Snel, M. Cornell, S. G. Oliver et al., 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- Waxman, D., and J. R. Peck, 1998. Pleiotropy and the preservation of perfection. Science 279: 1210–1213. [PubMed] [Google Scholar]
- Williams, G. C., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11: 398–411. [Google Scholar]
- Wright, S., 1968. Evolution and the Genetics of Populations. University of Chicago Press, Chicago.
- Zhang, J., and X. He, 2005. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol. Biol. Evol. 22: 1147–1155. [DOI] [PubMed] [Google Scholar]