Abstract
We report that the Disc1 gene in all extant 129 mouse inbred substrains has a deletion, previously considered specific to the 129S6/SvEv substrain, which is predicted to abolish production of the full-length protein. This finding has implications for the study of knockout mice generated from 129-derived embryonic stem cells.
DISC1 (disrupted in schizophrenia 1) was directly implicated as a candidate gene for psychiatric disease by the identification of truncation mutations cosegregating with schizophrenia and affective disorder in two distinct Scottish (Millar et al. 2000) and American (Sachs et al. 2005) families. Other polymorphisms of DISC1 have shown association with these diseases in broader populations in the United States (Hodgkinson et al. 2004; Callicott et al. 2005) and northern Europe (Cannon et al. 2005; Zhang et al. 2006). DISC1 protein is present in multiple brain regions and in several other tissues, including heart and kidney (Millar et al. 2004). Although the functions of DISC1 are not understood in detail, it has been shown to interact with multiple proteins required for neuronal functions such as neurite extension, neurotransmission, signal transduction, and the cytoskeleton (Porteous and Millar 2006).
In the original Scottish DISC1 truncation family, Millar et al. (2005) found a 50% reduction of the normal level of DISC1 in cell lines derived from mutation hemizygotes, but could find no evidence for a stable, truncated DISC1 protein product. In an analogous finding in the mouse, Koike et al. (2006) reported that the 129S6/SvEv (129S6) inbred strain bred by Taconic Farms is homozygous for a 25-bp deletion in exon 6 of Disc1 that induces a frame shift in the reading frame of Disc1, resulting in 13 novel amino acids, followed by a premature stop codon. Neither full-length Disc1 nor the predicted C-terminally truncated protein was detectable in 129S6 brain tissue (Koike et al. 2006), suggesting that the deletion is a null allele. After backcrossing the deletion onto the C57BL/6J inbred strain to obtain a uniform genetic background, Koike et al. (2006) found that both heterozygous and homozygous mutants showed a consistent impairment in a working memory test, suggesting that Disc1 is haplo-insufficient in relation to this trait. Consistent with this haplo-insufficiency model, Kamiya et al. (2005) have shown that reduction of endogenous Disc1 expression in mice disturbs proper neuronal migration and arborization of dendritic neuronal processes in the developing cerebral cortex.
Fixation of spontaneous null mutations within mouse production colonies has been reported previously. C57BL/6JOlaHsd mice supplied by Harlan UK have a deletion of the α-synuclein locus (Specht and Schoepfer 2001), while C57BL/6J mice supplied by JAX Research Systems have a deletion in the nicotinamide nucleotide transhydrogenase gene (Huang et al. 2006). Neither deletion has been detected in other C57BL/6 substrains. As the Disc1 deletion was found to be absent in the AKR/J, BALB/cJ, C3H/HeJ, C57BL/6J, CBA/J, and DBA/2J strains, Koike et al. (2006) concluded that it represents a 129S6-specific polymorphism. However, the 129S6 is just one of several inbred substrains within the complex genealogy of the 129 strain family, which includes the parental, steel, Ter (teratoma susceptible), and X (genetically contaminated) lineages (Festing et al. 1999; Beck et al. 2000). As Koike et al. (2006) did not examine any other 129 substrains for the Disc1 deletion, they could not determine whether it arose de novo within the 129S6 colony at Taconic Farms.
As virtually all embryonic stem (ES) cell lines for gene targeting are derived from substrains of the 129 mouse (Table 1), the potential presence of a genetic ablation known to confer a cognitive impairment in the heterozygous state (Koike et al. 2006) deserves the attention of scientists using knockout mice to analyze brain function. Disc1 deficiency in these mice may have previously unforeseen consequent effects on the >50 putative Disc1 interacting proteins that have been reported (Porteous and Millar 2006). The objective of this study was thus to establish the prevalence of the Disc1 deletion among the other inbred substrains of 129, including the progenitors of 14 ES cell lines (Table 1).
TABLE 1.
129 mouse substrains assayed for the Disc1 deletion polymorphism
| Strain name | Abbreviation | Lineage | ES cell lines |
|---|---|---|---|
| 129P1/ReJ | 129P1 | Parental (P) | |
| 129P2/OlaHsd | 129P2 | Parental (P) | E14TG2a, HM-1 |
| 129P3/J | 129P3 | Parental (P) | mEMS32 |
| 129P4/RrRkJ | 129P4 | Parental (P) | |
| 129S1/SvImJ | 129S1 | Steel (S) | CJ7, W9.5 |
| 129S2/SvPas | 129S2 | Steel (S) | D3 |
| 129S4/SvJae | 129S4 | Steel (S) | AK7, J1 |
| 129S5/SvEvBrd | 129S5 | Steel (S) | Lex-1 |
| 129S7/SvEvBrd | 129S7 | Steel (S) | AB1, AB2.1 |
| 129S8/SvEv | 129S8 | Steel (S) | |
| 129T1/Sv | 129T1 | Ter (T) | |
| 129T2/SvEmsJ | 129T2 | Ter (T) | |
| 129X1/SvJ | 129X1 | Contaminated (X) | Pat5, PJ1-5, RW-4 |
We assayed all extant inbred substrains of 129 listed by Festing et al. (1999) and Beck et al. (2000). DNA samples from 129P1, 129P3, 129P4, 129S1, 129S4, 129S8, 129T1, 129T2, and C57BL/6J mice were obtained from the Mouse DNA Resource of The Jackson Laboratory (Bar Harbor, ME). DNA samples from 129S5 and 129S7 mice were obtained from Roswell Park Cancer Institute (Buffalo) and B&K Universal (Hull, Yorkshire, UK), respectively. DNA was isolated from tail biopsies from 129S6 and B6129F1 mice supplied by Taconic Farms (Germantown, NY), from 129P2 mice supplied by Harlan (Indianapolis), and from 129S2 mice supplied by Charles River (Saint-Constant, Quebec, Canada). ES cell line progenitor strains were identified using the Mouse Genome Database (2006). 129S1/SvImJ was previously named 129S3/SvImJ (Jackson Laboratory 2001). 129/Sv (JAX 000094), a cryopreserved strain that is heterozygous at many loci (Simpson et al. 1997), was excluded as it is clearly not inbred.
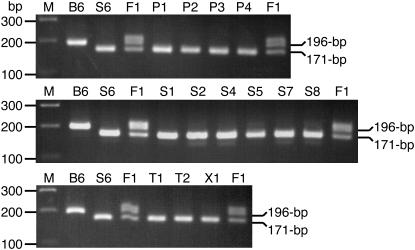
To simultaneously detect the 25-bp deletion of 129S6 and the wild-type allele of C57BL/6 mice, we devised a PCR-based genotyping assay (Figure 1). A trial PCR with control DNA of known genotype revealed that C57BL/6J had a single band of 196 bp, 129S6 had a single band of 171 bp, and the B6129F1 (C57BL/6N × 129S6) hybrid had two bands of 196 and 171 bp (Figure 2), which was consistent with the predicted action of the primers.
Figure 1.—
PCR genotyping assay to detect the Disc1 deletion. C57BL/6 mice have a wild-type Disc1 allele, while 129S6 mice have a 25-bp deletion (dots) in exon 6 of Disc1. The forward primer (5′-GCT GTG ACC TGA TGG CAC T-3′) within exon 6 corresponds to nucleotides 128,021,467–128,021,485, and the reverse primer (5′-CTT CTC ACC TGA GCA CAC CT-3′) within intron 6–7 is the reverse complement of nucleotides 128,021,642–128,021,662 of chromosome 8. Physical map distances between the primers were obtained using the UCSC Genome Browser (http://genome.ucsc.edu/; mouse assembly February 2006). The primers are designed to amplify a fragment of 171 bp from 129S6 and a fragment of 196 bp from C57BL/6. Each 25-μl PCR contains 17.7 μl nuclease-free water, 2.5 μl 10X PCR buffer [500 mm KCl, 100 mm Tris–HCl (pH 9.0 at 25°), 1% Triton X-100], 1.6 mm magnesium chloride, 0.2 mm each of dATP, dTTP, dCTP, and dGTP, 0.5 μm of each primer, 1.5 units Taq DNA polymerase, and 50 ng genomic DNA. Reactions are incubated at 94° for 5 min, followed by 35 cycles of 94° for 20 sec, 54° for 1 min, and 72° for 40 sec, followed by 72° for 10 min.
Figure 2.—
Disc1 deletion genotyping gel. M, 100-bp DNA ladder (1.0 μg; 70 ng per band). B6, C57BL/6J. S6, 129S6. F1, B6129F1. P1, 129P1. P2, 129P2. P3, 129P3. P4, 129P4. S1, 129S1. S2, 129S2. S4, 129S4. S5, 129S5. S7, 129S7. S8, 129S8. T1, 129T1. T2, 129T2. X1, 129X1. Following thermocycling, 20 μl of PCR product were electrophoresed at 8.5 V/cm through a 3% (w/v) agarose gel in 1X TAE buffer (40 mm tris-acetate, 2 mm EDTA) for 75 min. The sizes of the DNA ladder fragments and PCR products are indicated. The extra, upper band in the F1 (heterozygote) lanes is a heteroduplex of the wild-type and mutant strands, in which the 25-bp mismatch of the deletion forms a loop that slows migration through the gel.
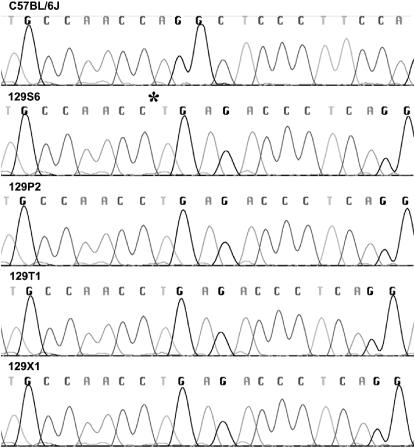
Application of the genotyping assay to 13 other substrains of 129 (Table 1) revealed that each had a single band of the same size as that of the 129S6 control (Figure 2). Sequence analysis of the fragments from single representatives of the parental (129P2), steel (129S6), Ter (129T1), and X (129X1) lineages confirmed the presence of the deletion, which was absent in the C57BL/6J control (Figure 3). This result indicates uniform homozygosity for the Disc1 deletion across all inbred lineages of 129.
Figure 3.—
Sequence chromatograms showing the 25-bp deletion of Disc1 in representatives of the steel (129S6), parental (129P2), Ter (129T1), and X (129X1) lineages of the 129 inbred strain family. An asterisk indicates the location of the deletion in the 129S6 sequence. Alignment and comparisons with the wild-type C57BL/6J sequence were performed using Sequencher software (Gene Codes, Ann Arbor, MI).
As the Disc1 deletion is common to all inbred substrains of 129, we conclude that it is not 129S6 specific. This is perhaps surprising given the substantial genetic heterogeneity between 129 substrains (Simpson et al. 1997; Threadgill et al. 1997), even within the same lineage (Sechler et al. 1997). The presence of the deletion in all lineages suggests that it was already fixed in the population of mice used to reestablish the Jackson Laboratory's 129 breeding colony in 1948, from which all the current substrains are descended (Simpson et al. 1997). Given the ancestral fixation of the Disc1 deletion in the 129 strain, the numerous 129 substrain-derived ES cell lines established within the past 20 years are also likely to harbor the deletion (Table 1). In light of our findings, studies of working memory in knockout mice with isogenic or hybrid 129 genetic backgrounds should perhaps be reevaluated (e.g., Bjorklund et al. 2001; Deller et al. 2003). To avoid potential confounds resulting from Disc1 deficiency, scientists using gene targeting to analyze brain function would be advised to genotype their knockout lines for the Disc1 deletion. Backcrossing to a wild-type Disc1 strain, such as C57BL/6, for several generations should remove the deletion if it is unlinked to the targeted gene. On a more positive note, investigators of psychiatric disease may welcome the news that Disc1-deficient mice can be obtained cheaply and easily by accessing any of the widely available 129 substrains.
Acknowledgments
We thank Y. Yu for 129S5/SvEvBrd DNA and D. Pomp for advice. This study was supported by grants-in-aid to J.C.R. from the Canadian Institutes of Health Research (GMH-79044) and the National Alliance for Research on Schizophrenia and Depression (NARSAD Distinguished Investigator Award). J.C.R. holds a Canada Research Chair. S.J.C. holds an Epilepsy Canada Research Fellowship.
References
- Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig et al., 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. [DOI] [PubMed] [Google Scholar]
- Bjorklund, M., I. Siverina, T. Heikkinen, H. Tanila, J. Sallinen et al., 2001. Spatial working memory improvement by an alpha2-adrenoceptor agonist dexmedetomidine is not mediated through alpha2C-adrenoceptor. Prog. Neuropsychopharmacol. Biol. Psychiatry 25: 1539–1554. [DOI] [PubMed] [Google Scholar]
- Callicott, J. H., R. E. Straub, L. Pezawas, M. F. Egan, V. S. Mattay et al., 2005. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl. Acad. Sci. USA 102: 8627–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, T. D., W. Hennah, T. G. M. van Erp, P. M. Thompson, J. Lonnqvist et al., 2005. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry 62: 1205–1213. [DOI] [PubMed] [Google Scholar]
- Deller, T., M. Korte, S. Chabanis, A. Drakew, H. Schwegler et al., 2003. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc. Natl. Acad. Sci. USA 100: 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing, M. F. W., E. M. Simpson, M. T. Davisson and L. E. Mobraaten, 1999. Revised nomenclature for strain 129 mice. Mamm. Genome 10: 836. [DOI] [PubMed] [Google Scholar]
- Hodgkinson, C. A., D. Goldman, J. Jaeger, S. Persaud, J. M. Kane et al., 2004. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 75: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T. T., M. Naeemuddin, S. Elchuri, M. Yamaguchi, H. M. Kozy et al., 2006. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxidase dismutase. Hum. Mol. Genet. 15: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Jackson Laboratory, 2001. New 129 nomenclature—revised. JAX Bulletin, No. 1 (http://jaxmice.jax.org/info/bulletin/bulletin01.html). The Jackson Laboratory, Bar Harbor, ME.
- Kamiya, A., K. Kubo, T. Tomoda, M. Takaki, R. Youn et al., 2005. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7: 1067–1078. [DOI] [PubMed] [Google Scholar]
- Koike, H., P. A. Arguello, M. Kvajo, M. Karayiorgou and J. A. Gogos, 2006. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc. Natl. Acad. Sci. USA 103: 3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, J. K., J. C. Wilson-Annan, S. Anderson, S. Christie, M. S. Taylor et al., 2000. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9: 1415–1423. [DOI] [PubMed] [Google Scholar]
- Millar, J. K., R. James, N. J. Brandon and P. A. Thomson, 2004. DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric disease. Ann. Med. 36: 1–12. [DOI] [PubMed] [Google Scholar]
- Millar, J. K., B. S. Pickard, S. Mackie, R. James, S. Christie et al., 2005. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 310: 1187–1190. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Database, 2006. ES cell line name and strain of origin. Mouse Genome Informatics (ftp://ftp.informatics.jax.org/pub/datasets/es_cell_lines.html). The Jackson Laboratory, Bar Harbor, ME.
- Porteous, D. J., and J. K. Millar, 2006. Disrupted in schizophrenia 1: building brains and memories. Trends Mol. Med. 12: 255–261. [DOI] [PubMed] [Google Scholar]
- Sachs, N. A., A. Sawa, S. E. Holmes, C. A. Ross, L. E. DeLisi et al., 2005. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol. Psychiatry 10: 758–764. [DOI] [PubMed] [Google Scholar]
- Sechler, J. M. G., J. C. Yip and A. S. Rosenberg, 1997. Genetic variation among 129 substrains: practical consequences. J. Immunol. 159: 5766–5768. [PubMed] [Google Scholar]
- Simpson, E. M., C. C. Linder, E. E. Sargent, M. T. Davisson, L. E. Mobraaten et al., 1997. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 16: 19–27. [DOI] [PubMed] [Google Scholar]
- Specht, C. G., and R. Schoepfer, 2001. Deletion of the alpha-synuclein locus in a sub-population of C57BL/6J inbred mice. BMC Neurosci. 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill, D. W., D. Yee, A. Matin, J. H. Nadeau and T. Magnuson, 1997. Genealogy of the 129 inbred strains: 129/SvJ is a contaminated inbred strain. Mamm. Genome 8: 390–393. [DOI] [PubMed] [Google Scholar]
- Zhang, F., J. Sarginson, C. Crombie, N. Walker, D. St. Clair et al., 2006. Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B: 155–159. [DOI] [PubMed] [Google Scholar]