Abstract
When a double-strand break has a gap between the broken ends, the missing information can be restored through synthesis from a homologous template. Here we address the question of how long such a gap can be before this process fails. We measured the frequency of homologous repair in the Drosophila germ line following the creation of gaps of specific sizes ranging from 3.8 to 210 kb. We found that gaps of ≤11 kb can be repaired with approximately the same efficiency as breaks with no gap at all. However, a gap of 44 kb was repaired only rarely, and one of 210 kb was not repaired at a measurable frequency. We conclude that DNA gap repair is a length-limited process, but that this limitation is critical only for gaps ≫11 kb.
A cell faces a crisis with the occurrence of a double-strand break (DSB). It must either repair the break, perhaps imperfectly, or face cell cycle arrest. One indication of the importance of DSB repair is that defects in the process have been associated with elevated cancer risk in humans (Jackson 2002; Rothkamm and Lobrich 2002; Maser and DePinho 2003; Valerie and Povirk 2003; Bryant 2004).
The mechanisms for DSB repair are often classified according to whether or not a homologous template, usually the sister chromatid or the homolog, is used. Pathways that do not utilize a template, such as nonhomologous end joining (Moore and Haber 1996) or single-strand annealing (Fishman-Lobell et al. 1992; Preston et al. 2002), entail a greater risk of mutation but may be available in circumstances where templated repair is not. There are at least two pathways for homologous repair: one with the potential for crossing over and one without. Crossing over in meiosis is thought to depend on which of these two pathways is followed (Allers and Lichten 2001; Hunter and Kleckner 2001; Borner et al. 2004; Mazina et al. 2004). In mitotic cells there is evidence for competition among multiple pathways of homologous and nonhomologous mechanisms to repair the same pool of DSBs (Preston et al. 2006).
Homologous repair is necessary to restore the missing information when the DSB includes a gap. Studies in Drosophila (Nassif et al. 1994; Coveny et al. 2002) and yeast (Paques et al. 1998) have shown that gaps as long as 10 kb can be repaired with only a two- to fourfold reduction in efficiency relative to breaks with little or no missing sequence. Here we examine much larger gaps, up to 210 kb, to determine whether homologous repair is limited by the length of gap that must be filled in.
MATERIALS AND METHODS
Drosophila stocks and crosses:
Drosophila crosses were carried out with standard methods (Ashburner 1989). Genetic symbols can be found in FlyBase (Drysdale et al. 2005).
PCR tests and flanking DNA sequencing:
DNA extraction for all PCR tests was done with individual flies as described (Gloor and Engels 1992). Primers included D0 and G0 as described (Preston and Engels 1996). These primers span the P-insertion site at 50C and yield an amplicon of 145 bp if there is no insertion or deletion at this position. Gap lengths of the seven deletion chromosomes used here were determined by C. Flores, using thermal asymmetric interlaced PCR (Liu and Whittier 1995) to amplify a segment spanning the deletion and comparing its sequence with the Drosophila genome sequence.
RESULTS
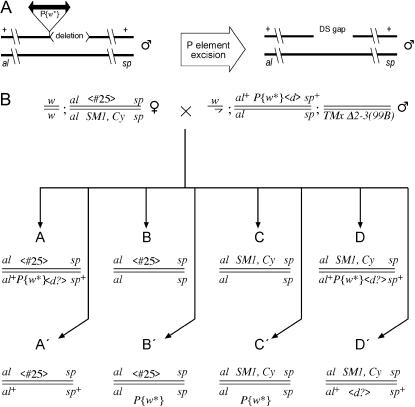
We made use of a series of chromosomes in which the same P-element insertion is precisely juxtaposed with deletions of various sizes (Preston et al. 1996). These chromosomes, listed in Table 1, were formed by a process called “hybrid element insertion”(HEI) (Gray et al. 1996; Preston et al. 1996), which involves two sister copies of a transposable element and an insertion site on the homologous chromosome. The result is a recombinant chromosome with the transposable element flanked by a duplication or deletion. Figure 1A shows how such a chromosome can be used to generate a gap relative to the homolog. Each transposition event represents an opportunity for gap repair, since the P element leaves behind a double-strand break (Engels et al. 1990; Beall and Rio 1997). This break can be repaired by copying from the sister chromatid, by copying from the homolog, or by end joining. Our experiment cannot detect homologous repair from the sister chromatid, but it can detect the other two events and distinguish between them. When the homolog is used, there is a gap that must be filled in. The size of the gap is equal to the length of the flanking deletion.
TABLE 1.
Progeny counts from the eight primary classes shown in Figure 1B
| Deletion size in bpa | IDb | Directionb | Single-male crosses | A | B | C | D | A′ | B′ | C′ | D′ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50C | None | 85 | 2,570 | 1,792 | 1,591 | 2,640 | 433 | 254 | 209 | 422 |
| 3,840 | 49 | Proximal | 79 | 1,510 | 1,532 | 1,480 | 2,040 | 91 | 37 | 35 | 78 |
| 6,848 | 81 | Proximal | 47 | 1,549 | 1,082 | 1,050 | 1,937 | 99 | 102 | 88 | 147 |
| 7,582 | 85 | Proximal | 75 | 1,256 | 1,074 | 1,179 | 1,630 | 99 | 118 | 100 | 108 |
| 10,160 | 102 | Proximal | 80 | 2c | 2,158 | 2,236 | 3,040 | 26 | 23 | 24 | 44 |
| 10,931 | 45 | Proximal | 47 | 28c | 827 | 439 | 1,089 | 25 | 88 | 57 | 125 |
| 42,645 | 36 | Distal | 114 | 4c | 2,915 | 2,503 | 3,690 | 4 | 357 | 305 | 195 |
| 210,463 | 38 | Distal | 239 | 0 | 7,205 | 6,030 | 9,067 | 0 | 666 | 500 | 394 |
Deletion lengths are based on a standard reference sequence and will vary slightly with stock.
Abbreviations for deletion chromosome names and directions are as reported (Preston et al. 1996). Standard nomenclature would be, e.g., Df(2R)50C-49.
The chromosomes with 10-, 11-, 43-, and 210-kb deletions are missing one or more essential genes encompassed by the deletion. Therefore, most individuals in category A do not survive from those crosses. A few exceptions were expected, however, resulting from coincidental occurrence of both gap repair and P-element transposition to place a copy of the P{w*} element on one of the other paternal chromosomes. We tested 20 of these exceptions by PCR similar to the test done for category A′ and confirmed that gap repair had occurred in each case.
Figure 1.—
(A) Gap formation in parental males. A P-transposable element at cytological position 50C on chromosome 2 is immediately adjacent to a deletion (Preston et al. 1996). Excision of this element in the male germ cells is activated by a P-transposase gene present on chromosome 3 (not included in the diagram). The result of this excision is a gap relative to the homolog equal to the length of the deletion. Males of this kind are used in the cross shown below to measure the frequencies of gap repair and other events. (B) Mating scheme for measuring repair frequency. See FlyBase (Drysdale et al. 2005) for all genetic symbols not explicitly defined. The male parent (top right) is equivalent to the males indicated in part A. The P element carrying a mini-white gene and designated P{w*} is located at cytological position 50C and is mobilized in the germ line by a third-chromosomal transposase source designated Δ2–3(99B) as described by Robertson et al. (1988). Deletions are denoted by angle brackets (“<d>,” etc.). In categories A, D, and D′ the deletion symbol is shown with a question mark to indicate that the presence of the deletion was not confirmed in those cases. The female parents are heterozygous for a specific deletion, Df(2R)50C-25, abbreviated <#25>, which was selected because it extends in both directions from the 50C insertion point and encompasses essential genes on both sides. It was obtained via two-step HEI as described by Preston et al. (1996). The symbol “TMx” refers to one of the balancer chromosomes TMS or TM2 (Drysdale et al. 2005). In categories B′ and C′ the P{w*} element is shown below the rest of the genotype to indicate that it represents a new insertion that can be on any of the paternal chromosomes. Otherwise, all genotypes are drawn with the maternal second chromosome above the paternal chromosome.
Figure 1B shows the detailed mating scheme and lists eight types of offspring classified according to the markers on chromosome 2. Gap formation and repair occur in the germ cells of males and are detected among the progeny. The top row (A–D) shows the expected genotypes if no repair, transposition, or excision events have occurred. Note that category A includes deletions on both homologs. Some of the deletions we tested—those of ≥10 kb—encompassed essential genes, thus rendering category A inviable. For that reason we used the average number of offspring in categories B, C, and D to estimate the Mendelian expectation for each class. Categories A′–D′ are similar to the corresponding genotypes in the top row except for the presence or absence of the P{w*} element, as indicated by eye color. These genotypes represent potential repair events (category A′), transposition events (categories B′ and C′), or excision events without complete templated repair (category D′). Offspring in category A′ were tested further by PCR as described (Table 2) to determine whether successful templated repair had occurred as opposed to end joining or other events that have been observed at this site (Preston and Engels 1996; Preston et al. 2002) or incomplete repair such as that described by McVey et al. (2004a,b). As described in Table 2, the observed ratio from these PCR results was then applied as a correction factor to determine the frequency of templated repair. Recombinants between al and sp were also scored, but these events were rare (203/72,601) and were not included in our calculations.
TABLE 2.
Estimates of P-element mobility and gap repair frequency
| Deletion size (kb) | No. of PCR testsa | PCR correction ± SDa | Transposition frequency ± SDb | Excision frequencyc | Repair frequency ± SDd | Transposition/repair ratio ± SDe |
|---|---|---|---|---|---|---|
| 0 | 405 | 0.24 ± 0.03 | 0.120 ± 0.006 | 0.138 | 0.045 ± 0.007 | 2.67 ± 0.46 |
| 4 | 79 | 0.56 ± 0.13 | 0.023 ± 0.003 | 0.037 | 0.029 ± 0.008 | 0.80± 0.24 |
| 7 | 85 | 0.46 ± 0.11 | 0.082 ± 0.009 | 0.071 | 0.031 ± 0.009 | 2.65 ± 0.82 |
| 8 | 91 | 0.56 ± 0.13 | 0.088 ± 0.008 | 0.062 | 0.040 ± 0.012 | 2.23 ± 0.69 |
| 10 | 23 | 1.00 ± 0.00 | 0.011 ± 0.002 | 0.014 | 0.010 ± 0.005 | 1.02 ± 0.51 |
| 11 | 14 | 1.00 ± 0.00 | 0.103 ± 0.008 | 0.103 | 0.029 ± 0.008 | 3.60 ± 1.04 |
| 43 | 3 | 1.00 ± 0.00 | 0.109 ± 0.005 | 0.050 | 0.001 ± 0.001 | 90.42 ± 44.59 |
| 210 | 0 | NA | 0.081 ± 0.003 | 0.042 | 0.000 ± 0.000 | 214.67 ± 124.21f |
DNA was extracted from gap repair candidates in phenotypic category A′ (Figure 1B) and amplified with primers D0 and G0 described previously (Preston et al. 1996). The correction factor was calculated as the proportion of PCR tests that yielded the 145-bp fragment expected from gap repair templated by the homolog. The standard deviation was obtained from the single-male independent replicates as described (Engels 1979). Note that in the case of the lethal-bearing deletions, all 40 PCR tests indicated complete homologous repair, as opposed to end joining or a partial repair of the kind observed in other systems (McVey et al. 2004a, McVey et al. 2004b).
Transposition frequency was estimated as 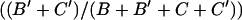 from the phenotypic categories shown in Figure 1B. Note that this estimate does not include a correction for transpositions to the al+ sp+ or X chromosomes. It can, therefore, be considered proportional to the absolute transposition rate. Its standard error was computed from the single-male independent crosses as described (Engels 1979).
from the phenotypic categories shown in Figure 1B. Note that this estimate does not include a correction for transpositions to the al+ sp+ or X chromosomes. It can, therefore, be considered proportional to the absolute transposition rate. Its standard error was computed from the single-male independent crosses as described (Engels 1979).
Excision frequency was computed from the phenotypic categories in Figure 1B as 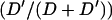 .
.
Repair frequency was estimated as 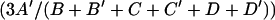 (PCR correction). The first term is the phenotypic repair frequency computed from the offspring counts shown in Table 1, and the second term comes from the third column of this table. Standard errors for the phenotypic repair frequency were computed from the single-male replicates as described (Engels 1979), and the standard errors for the corrected repair frequency were computed from the large-sample approximation for the standard error of a product of two random variables, x and y:
(PCR correction). The first term is the phenotypic repair frequency computed from the offspring counts shown in Table 1, and the second term comes from the third column of this table. Standard errors for the phenotypic repair frequency were computed from the single-male replicates as described (Engels 1979), and the standard errors for the corrected repair frequency were computed from the large-sample approximation for the standard error of a product of two random variables, x and y: 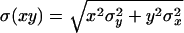 . In this case, x and y refer to the two factors in the expression for repair frequency, i.e., the phenotypic repair frequency and the PCR correction. This approximation requires that x and y not be strongly correlated. Inspection of a scatterplot of the phenotypic repair frequency vs. the PCR correction for individual male crosses showed no evidence of such a correlation.
. In this case, x and y refer to the two factors in the expression for repair frequency, i.e., the phenotypic repair frequency and the PCR correction. This approximation requires that x and y not be strongly correlated. Inspection of a scatterplot of the phenotypic repair frequency vs. the PCR correction for individual male crosses showed no evidence of such a correlation.
The transposition/repair ratio was computed from columns 4 and 6 of this table. The standard error was approximated by the large-sample method: 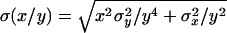 . In this case, x and y refer to columns 4 and 6.
. In this case, x and y refer to columns 4 and 6.
Since no repair events were observed for the 210-kb gap, we used as the denominator the upper limit of a 95% confidence interval. This procedure amounts to using the phenotypic repair frequency that would have been computed if we had observed –ln(0.05) = 3 repair events instead of zero. The resulting repair frequency and its (binomial) standard error were 0.00038 ± 0.00022.
Each cross in Figure 1 was set up with a single male and three to four females at room temperature, and offspring were scored up to day 21. The use of single-male crosses has the advantage of allowing accurate quantitative measurement of the frequency of gap repair and a robust statistical analysis. Thus, each individual male parent provides a statistically independent estimate of repair frequency.
Note that excision of the P{w*} element followed by end joining or incomplete repair can result in offspring phenotypically similar to category A′. To avoid error from these false positives, we extracted DNA from most of the A′ offspring and tested them by PCR as described in Table 2. Such false positives are expected primarily from the no-deletion controls and the three smaller deletions that do not encompass any essential genes. In the case of the four larger deletions, all end-joining events and most incomplete repair events will render genotype A′ inviable, and no false positive will occur. Therefore, as expected, the PCR correction given in Table 2 is substantial for the no-deletion controls and the three smaller deletions, but is nonexistent for the four largest deletions.
We tested eight chromosomes with deletions (i.e., gap lengths) ranging from 0 to 210 kb. An average of 96 individual male parents were testcrossed per chromosome for a total of 72,398 progeny scored in the experiment. The results are shown graphically in Figure 2 and are available numerically in more detail in Tables 1 and 2.
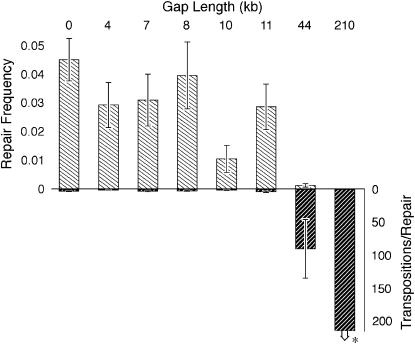
Figure 2.—
Frequency of homologous gap repair. Top bars show the frequency of homologous repair of gaps whose lengths are indicated above. The numerator for this measure is the number of progeny whose paternal second chromosome has undergone loss of the P element and restoration of the deleted sequence. Such progeny are recognized phenotypically as w−, al+, Cy+, sp+ as well as wild type for any essential genes encompassed by the deletion. The set of encompassed genes varies with the deletion size, but can include CP1 and several essential loci. The denominator is the (estimated) number of progeny to receive the upper paternal second chromosome. Many of the progeny in that class will be homozygous lethal. Therefore, we estimated the number using one-third of the other three second-chromosome classes, none of which can be homozygous lethal. Error bars show ±1 SD, computed as described (Table 2). The bottom bars make use of transposition as a proportional measure of DSB formation. The plotted values are proportional to the number of DSB events per successful homologous gap repair. Transposition is detected in progeny that receive the lower paternal second chromosome but carry a copy of the P element. (*) Since we found no cases of the 210-kb gap being restored, we used the upper limit of a 95% Poisson confidence interval to avoid dividing by zero. Otherwise, this bar would extend infinitely downward (see Table 2).
The top bars in Figure 2 show estimates of the frequency of gap formation and homologous repair among all gametes receiving the relevant (al+ sp+) chromosome from the males shown in Figure 1. Gap formation and repair can occur in the premeiotic germ cells, where a single event can yield multiple gametes. Our use of single-male crosses allowed us to take this clustering into account to calculate unbiased standard errors as described (Engels 1979; Preston et al. 2006).
The bottom bars show an alternative measure of gap repair that takes account of variation in P-element mobility among the deletion-bearing chromosomes. All of these chromosomes have the same P-element insertion, but its mobility can be affected by the flanking deletions that place one side of the element in a different DNA context. Since the P element carries a visible eye-color marker [mini-white (Drysdale et al. 2005), denoted w*], the same cross that yielded an estimate of the gap repair frequency also gave us an estimate of the P-element mobility. That is, the transposition rate is proportional to the number of al sp progeny carrying a copy of the P element, as described in more detail in Tables 1 and 2. Therefore, our alternative measure of repair efficiency is proportional to the number of transpositions (i.e., opportunities for gap repair) for each successful gap repair where the homolog serves as the template. Since it is a reciprocal measure of repair efficiency, the bars are drawn going downward in Figure 2.
We see from Figure 2 that gaps of length up to 11 kb were all repaired efficiently. In fact, only the 10-kb gap category had a repair frequency that differed significantly from any of the others in this group when comparisons were made using permutation tests with the single-male replicates. The chromosome carrying this 10-kb deletion was also unusual in having a transposition frequency of only 1%, which was less than half that of any of the other chromosomes in the study (Tables 1 and 2). If this chromosome is discounted, our results show no evidence that gaps up to 11 kb inhibit gap repair. The difference between gaps of up to 11 kb vs. the two larger gaps is even clearer when one compares the ratios of repair opportunities to successful templated repair events (Figure 2, bottom).
In contrast, the chromosome with a 43-kb deletion showed a markedly reduced repair frequency (Figure 2) even though its transposition rate of 11% was comparable to that of the other chromosomes (Tables 1 and 2). Even more striking was the complete lack of detectable homologous repair of 210-kb gaps. To maximize the power of our test with this chromosome, we performed 239 single-male crosses and scored 23,862 progeny but recovered no cases of homologous repair. We conclude that successful filling in of gaps is severely inhibited for gaps of 43 kb and essentially absent for gaps of 210 kb.
DISCUSSION
Several differences between this work and previous studies should be mentioned: We examined repair events in the germ cells of Drosophila males as opposed to the female germ line (Coveny et al. 2002) or yeast cells (Paques et al. 1998). In addition, there is evidence that the genomic position of a DSB can influence the relative usage of repair pathways (W. R. Engels and C. Preston, unpublished results). However, we believe the critical difference between the present study and previous investigations is that much larger gaps were examined: 210 kb vs. 10 kb. Only by testing repair of these large gaps were we able to detect the length limitation of DSB repair.
Our results provide strong evidence that homologous gap repair is a length-limited process, but other interpretations should be considered. In particular, note that the two chromosomes that produce gaps of 43 and 210 kb have deletions extending in the rightward direction from the original P insert whereas the other chromosomes in this study extend leftward (Table 1). If this directional difference were somehow affecting gap repair, it would offer an alternative explanation for our main results. However, two lines of evidence argue against a directional effect being an important factor in the observed repair differences. First, our data show that the 43-kb deletion was repaired significantly more often than the 210-kb deletion (P = 0.01), even though both are in the same direction. In addition, previous data showing symmetrical conversion tracts following excision of this same P insert (Preston and Engels 1996) argue against direction being crucial. That is, we saw no evidence of a directional effect, at least for the relatively short conversion tracts (<3 kb) observed in that study.
When a gap is too large to be filled in from the homolog, there are at least two other options that would allow continuation of the cell cycle. If the cell is in late S-phase or G2, templated repair from the sister chromatid is possible. In that case, only the length of the P{w*} element must be filled in (Figure 1). Another possibility is nonhomologous end joining (NHEJ), which does not depend on the stage of the cell cycle. In fact, several NHEJ products were observed in the present study among the offspring in category A′ in cases where the PCR test did not indicate templated repair. In some cases NHEJ was inferred from a PCR fragment larger or smaller than that expected from templated repair. In other cases, the mutant phenotype of the CP1 gene (Gray et al. 1998) implied loss of sequences flanking the insertion, as would be consistent with end joining. For example, the 4-kb deletion yielded 91 offspring of category A′ (Table 1). Of these, 39 had the CP1 mutant phenotype, suggesting an aberrant repair event of some kind, as was confirmed in 30/30 tested by PCR. In addition, the PCR tests showed that 5 of the 52 CP1+ offspring had no amplicon with primers D0 and G0, also indicating end joining or another aberrant event.
Why is homologous gap repair unable to handle longer gaps? Paques et al. (1998) suggested that the DNA polymerase used in DSB repair may lack processivity compared to the polymerase used for chromosome replication or break-induced replication (Kraus et al. 2001). A similar suggestion was made by McVey et al. (2004a) to explain partial repair events. Time constraints in the cell cycle might also prevent polymerase from completing extremely long tracts. Nonhomologous end joining is available as an alternative to homologous pathways for repair of the same breaks. Indeed, the relative usage of these pathways varies in a compensatory fashion (Preston et al. 2006). The inability of homologous gap repair to handle extremely long gaps provides another rationale for the existence of multiple, but not necessarily redundant, mechanisms for DSB repair.
Acknowledgments
Carlos Flores conceived the idea of using P inserts adjacent to very long deletions to test for very long gap repair. He also determined the lengths of the deletions by sequencing and provided comments on the manuscript. Christine Preston produced and characterized the deletion chromosomes and helped with the manuscript. This work was supported by the National Institutes of Health (GM30948).
References
- Allers, T., and M. Lichten, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Beall, E. L., and D. C. Rio, 1997. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 11: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Bryant, P. E., 2004. Repair and chromosomal damage. Radiother. Oncol. 72: 251–256. [DOI] [PubMed] [Google Scholar]
- Coveny, A. M., T. Dray and G. B. Gloor, 2002. The effect of heterologous insertions on gene conversion in mitotically dividing cells in Drosophila melanogaster. Genetics 161: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby, W. Gelbart, K. Campbell, D. Emmert et al., 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., 1979. The estimation of mutation rates when premeiotic events are involved. Environ. Mutagen. 1: 37–43. [DOI] [PubMed] [Google Scholar]
- Engels, W. R., D. M. Johnson-Schlitz, W. B. Eggleston and J. Sved, 1990. High-frequency P element loss in Drosophila is homolog-dependent. Cell 62: 515–525. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell, J., N. Rudin and J. E. Haber, 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G., and W. Engels, 1992. Single-fly DNA preps for PCR. Dros. Inf. Serv. 71: 148–149. [Google Scholar]
- Gray, Y. H., M. M. Tanaka and J. A. Sved, 1996. P-element-induced recombination in Drosophila melanogaster: hybrid element insertion. Genetics 144: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., J. A. Sved, C. R. Preston and W. R. Engels, 1998. Structure and associated mutational effects of the cysteine proteinase (CP1) gene of Drosophila melanogaster. Insect Mol. Biol. 7: 291–293. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jackson, S. P., 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23: 687–696. [DOI] [PubMed] [Google Scholar]
- Kraus, E., W. Y. Leung and J. E. Haber, 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98: 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. G., and R. F. Whittier, 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681. [DOI] [PubMed] [Google Scholar]
- Maser, R. S., and R. A. DePinho, 2003. Take care of your chromosomes lest cancer take care of you. Cancer Cell 3: 4–6. [DOI] [PubMed] [Google Scholar]
- Mazina, O. M., A. V. Mazin, T. Nakagawa, R. D. Kolodner and S. C. Kowalczykowski, 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- McVey, M., M. Adams, E. Staeva-Vieira and J. J. Sekelsky, 2004. a Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., D. Radut and J. J. Sekelsky, 2004. b End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif, N., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., W. Y. Leung and J. E. Haber, 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18: 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., and W. R. Engels, 1996. P-element-induced male recombination and gene conversion in Drosophila. Genetics 144: 1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., W. Engels and C. Flores, 2002. Efficient repair of DNA breaks in Drosophila: evidence for single-strand annealing and competition with other repair pathways. Genetics 161: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., C. C. Flores and W. R. Engels, 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm, K., and M. Lobrich, 2002. Misrepair of radiation-induced DNA double-strand breaks and its relevance for tumorigenesis and cancer treatment (review). Int. J. Oncol. 21: 433–440. [PubMed] [Google Scholar]
- Valerie, K., and L. F. Povirk, 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792–5812. [DOI] [PubMed] [Google Scholar]