Abstract
Protein kinases mediate much of the signal transduction in eukaryotic cells and defects in kinase function are associated with a variety of human diseases. To understand and correct these defects, we will need to identify the physiologically relevant substrates of these enzymes. The work presented here describes a novel approach to this identification process for the cAMP-dependent protein kinase (PKA) in Saccharomyces cerevisiae. This approach takes advantage of two catalytically inactive PKA variants, Tpk1K336A/H338A and Tpk1R324A, that exhibit a stable binding to their substrates. Most protein kinases, including the wild-type PKA, associate with substrates with a relatively low affinity. The binding observed here was specific to substrates and was dependent upon PKA residues known to be important for interactions with peptide substrates. The general utility of this approach was demonstrated by the ability to identify both previously described and novel PKA substrates in S. cerevisiae. Interestingly, the positions of the residues altered in these variants implicated a particular region within the PKA kinase domain, corresponding to subdomain XI, in the binding and/or release of protein substrates. Moreover, the high conservation of the residues altered and, in particular, the invariant nature of the R324 position suggest that this approach might be generally applicable to other protein kinases.
EUKARYOTIC cells utilize an elaborate network of signaling pathways to bring about the proper physiological response to changes in the extracellular environment. These pathways generally consist of a series of proteins that are responsible for amplifying the original signal and for moving it to different regions of the cell. Members of one particular family of enzymes, the protein kinases, are important mediators of much of this signal transduction (Hunter 2000; Manning et al. 2002). These enzymes catalyze the transfer of the terminal phosphate from ATP to a distinct set of protein targets. This phosphorylation results in specific changes in the activities associated with each of these target proteins. The net effect of activating any protein kinase therefore is due to the collective actions of the substrates of that kinase.
The cAMP-dependent protein kinase (PKA) is one of the best-characterized members of this protein family (Johnson et al. 2001). PKA was the first protein kinase structure to be described and much is known about the biochemical properties of this enzyme (Adams 2001; Taylor et al. 2004). PKA activity is found in all eukaryotes and influences a wide variety of biological processes, from basic glucose metabolism to the modulation of long-term memory (Picket-Gies and Walsh 1986; Arnsten et al. 2005). In the budding yeast Saccharomyces cerevisiae, PKA is a key regulator of cell growth and proliferation (Thevelein and de Winde 1999; Herman 2002). In particular, this enzyme appears to be a critical component of a central control mechanism that ensures that overall cell growth is properly coordinated with the available nutrient supply (Broach 1991; Herman 2002). These activities are mediated by three different PKA catalytic subunits, Tpk1, Tpk2, and Tpk3, that appear to have both overlapping and distinct cellular functions (Toda et al. 1987; Robertson et al. 2000; Ptacek et al. 2005). As with any protein kinase, a complete understanding of the biological role of the S. cerevisiae PKA will require the identification of the targets of these enzymes.
Over the years, a number of innovative strategies have been developed to identify protein kinase substrates (Shah et al. 1997; Zhu et al. 2001; Manning and Cantley 2002; Budovskaya et al. 2005). However, despite these advances, this process is still often a difficult and labor-intensive task, and we tend to know few, if any, of the physiologically relevant substrates of any given protein kinase (Manning and Cantley 2002; Johnson and Hunter 2005). Here, we describe a strategy for identifying PKA substrates that takes advantage of catalytically inactive variants that exhibit an increased binding to their in vivo targets. These Tpk1 variants were able to bind all substrates tested and showed no association with proteins that were not phosphorylated by PKA. In addition, no significant binding was observed between substrates and the wild-type Tpk1. This latter result was not unexpected as protein kinases typically interact with their substrates with relatively low affinity and few kinase targets have been identified by their ability to bind to these enzymes (Manning and Cantley 2002). The general utility of this substrate binding was demonstrated here by the identification of both previously described and novel PKA substrates in S. cerevisiae. Interestingly, our results suggest that a particular region at the C-terminus of the conserved protein kinase domain might be important for substrate interactions with PKA. Moreover, the conserved nature of the residues altered in this study suggests that the results obtained here might be generally applicable to other protein kinases.
MATERIALS AND METHODS
Yeast strains and growth conditions:
Standard yeast genetic methods and growth conditions were used throughout this study (Kaiser et al. 1994). The yeast strains used were PHY1220 (MATα his3-Δ200 leu2-3,112 lys2-801 trp1-101 ura3-52 suc2-Δ9), PJ69A-4A (MATa gal4Δ gal80Δ his3-Δ200 leu2-3,11 trp1-101 ura3-52 LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7∷lacZ), and TVY614 (PHY1220 prc1∷HIS3 pep4Δ∷LEU2 prb1Δ∷hisG) (James et al. 1996 ; Gerhardt et al. 1998; Chang et al. 2001). The yeast rich growth medium (YPAD), synthetic complete medium (SC), and yeast minimal medium (YM) have been described (Kaiser et al. 1994; Herman and Rine 1997).
Co-immunoprecipitation assays:
The yeast strain PJ69-4A was transformed with a low-copy plasmid encoding Tpk1 proteins tagged at their N-terminus with three copies of the hemagglutinin (HA) epitope. The strain also contained a high-copy plasmid encoding a particular substrate (or nonsubstrate) that had six copies of the myc epitope at its N-terminus. All of these constructs were under the control of the copper-inducible promoter from the CUP1 gene (Thiele and Hamer 1986). The proteins being tested for interaction included Cdc25 (codons 51–330), Cki1 (2–200), and Cki1-N (274–471). These strains were grown at 30° in SC-glucose medium to a density of 0.5 OD600 units/ml. CuSO4 was added to a concentration of 100 μm, and the cells were incubated for 2 additional hours. The cells were harvested, washed in TBS buffer containing 1 mm PMSF and 0.5% NP40, and then converted to spheroplasts and lysed as described (Budovskaya et al. 2002). The resulting protein extracts were incubated at 4° for 2 hr with 10 μl of an anti-HA affinity matrix (Roche). The beads were collected with a low-speed centrifugation and washed six times with 1 ml of TBS containing 1 mm PMSF and 0.5% NP40. The bound protein was eluted with 30 μl of SDS sample buffer, separated by SDS-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes. The relative amount of substrate present was assessed by Western immunoblotting with antibodies specific for the myc epitope (Cell Signaling). Following chemiluminescent development and exposure to X-ray film, the nitrocellulose membrane was stripped by incubating in a solution of 2% SDS, 0.0625 m Tris–HCl, pH 6.8, and 100 mm βME for 30 min at 50°. The blot was then washed and reprobed with anti-HA antibody (Roche) to assess the relative amount of Tpk1 protein present.
PKA in vitro kinase assays and Western immunoblot analyses:
PKA in vitro phosphorylation assays were performed as described (Chang et al. 2001). In general, these assays were performed with protein A fusion proteins that were constructed in the vector pPHY1044 that contains two repeats of the immunoglobulin-binding region of protein A from Staphylococcus aureus (Budovskaya et al. 2002). The protein fragments assayed included those listed in the above section and Dot6 (170–472). For the analysis of the in vivo phosphorylation levels with the anti-PKA substrate antibody (Cell Signaling), a glutathione S-transferase (GST)–Dot6 fusion protein was precipitated with an anti-GST antibody (Cell Signaling) and run out on an SDS-polyacrylamide gel (Zhu et al. 2001). The precipitated proteins were then examined by a Western immunoblot with either the anti-GST or anti-PKA substrate antibody used at a dilution of 1:5000. All site-directed mutageneses were performed as described (Kunkel 1985; Ausubel et al. 1995; Budovskaya et al. 2002).
Two-hybrid analyses:
For the two-hybrid assays shown, the Tpk1 variants were cloned into the Gal4 activation domain plasmid pGADT7 (Clontech) and the substrates into the Gal4 DNA-binding domain vector pGBKT7 (Clontech). The substrates tested included Rim15 (codons 1431–1671), Yak1 (193–362), Cki1 (2–200), Cdc25 (51–330), Kin28 (2–306), Ctk1 (2–528), Bur1 (2–657), Toa2 (2–122), and Pds1 (96–222). In general, the yeast strain PJ69-4A was transformed with derivatives of both two-hybrid plasmids and grown to midlog phase in YM-glucose minimal medium. The cells were then plated to media lacking either adenine or histidine and incubated for 2–3 days at 30° to assess the two-hybrid interaction. A modified two-hybrid assay system was used specifically for the experiments shown in Figure 2B. Since overexpression of the wild-type Tpk1 was found to produce a significant growth defect (see below), alleles encoding the wild-type Tpk1 and the Tpk1KH variant fused to the Gal4 activation domain were placed under the control of the CUP1 promoter in a single-copy plasmid. For this plasmid, nucleotides corresponding to −435 to −1 of the CUP1 locus (where +1 is the translation start site) were ligated into the pRS416 plasmid as an EcoRI–HindIII fragment and connected to the ATG of the Gal4 activation domain ORF via the linker TGCAAAG. Two-hybrid assays were then performed as described above except that all media were supplemented with 3 μm CuSO4.
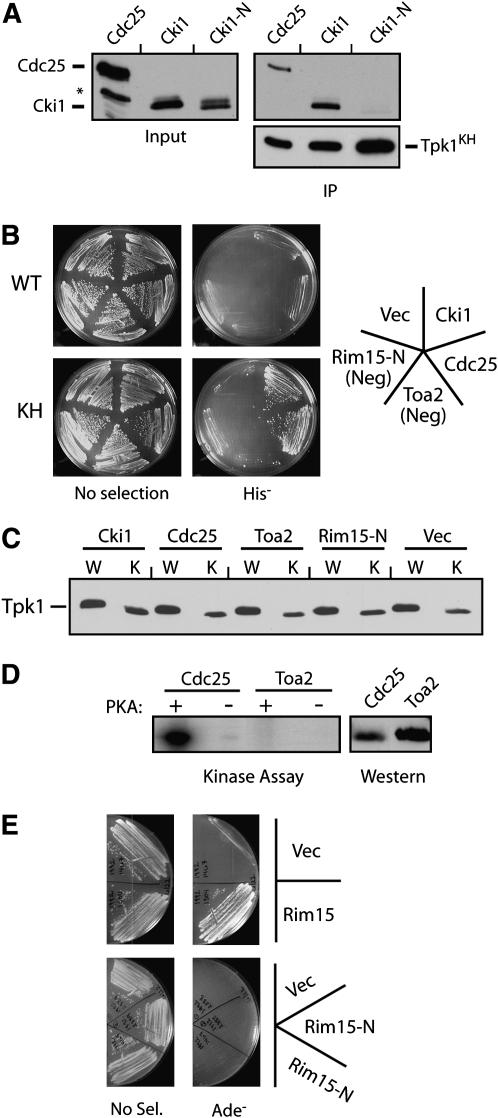
Figure 2.—
The interactions with the Tpk1KH variant were specific to PKA substrates. (A) The Tpk1KH variant did not associate with a fragment of Cki1 that lacks the sites of PKA phosphorylation. The relative amount of three different proteins, Cdc25 (codons 51–330), Cki1 (2–200), and Cki1-N (274–471), precipitating with the Tpk1KH variant was assessed as described in Figure 1A. The asterisk denotes a nonspecific protein band recognized by the α-myc epitope antibody. (B) A two-hybrid assay showing that the Tpk1KH variant interacted with the known substrates, Cdc25 and Cki1, but not with the nonsubstrates, Rim15-N and Toa2. The wild-type Tpk1 failed to interact with any of these test proteins. For these assays, the Tpk1 fusions to the Gal4 activation domain were under the control of the inducible promoter from the CUP1 gene (see materials and methods). Rim15-N (codons 361–657) was a negative control that was not phosphorylated by PKA. (C) A Western immunoblot showing the relative levels of the wild-type Tpk1 and Tpk1KH present in the two-hybrid strains shown in B. (D) Toa2 was not phosphorylated by PKA in vitro. Protein A fusions to Toa2 and a portion of Cdc25 (codons 51–330) were precipitated from yeast cell extracts and incubated with PKA and [γ-32P]ATP. The reaction products were separated on an SDS-polyacrylamide gel and the level of phosphorylation was assessed by autoradiography. (Right) A Western immunoblot showing the relative levels of the two substrates present in the kinase assays. (E) The Tpk1KH variant interacted with a Rim15 fragment containing the known sites of PKA phosphorylation (codons 1431–1671) but not with the Rim15-N fragment in a standard two-hybrid assay.
RESULTS
The catalytically inactive PKA variant Tpk1KH exhibited an increased binding to known substrates in S. cerevisiae:
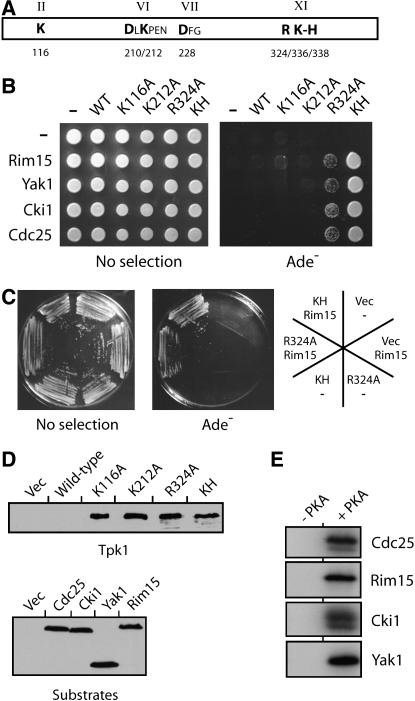
We are interested in defining the role of PKA in the control of S. cerevisiae growth, and thus have been attempting to identify the physiologically relevant substrates of this enzyme (Howard et al. 2002; Howard et al. 2003; Chang et al. 2004; Budovskaya et al. 2005). During the course of these studies, we found that a particular inactive variant of Tpk1 exhibited an increased binding to a number of known PKA substrates, including Cdc25 and Cki1 (Figure 1A). Cdc25 is the guanine nucleotide exchange factor for the Ras proteins in S. cerevisiae, and Cki1 is a choline kinase (Gross et al. 1992; Yu et al. 2002). In co-immunoprecipitation assays, the Tpk1KH variant bound significantly more of both of these substrates than did the wild-type protein (Figure 1, A and B). In general, very little, if any, substrate was found associated with the wild-type Tpk1 (Figure 1A). In this Tpk1KH variant, two relatively conserved residues important for catalytic activity, K336 and H338, were each replaced with an alanine. The K336 position is conserved within the AGC subfamily of protein kinases, whereas H338 is found in a somewhat broader spectrum of serine/threonine-specific kinases (Gibbs and Zoller 1991b; Hanks and Hunter 1995). Consistent with previous results with peptides (Gibbs and Zoller 1991b), we found that the Tpk1KH variant was ∼20-fold less active than the wild-type enzyme in an in vitro kinase assay with a protein substrate (Figure 1C). Both alterations in Tpk1KH were required for substrate binding as neither single mutant, K336A or H338A alone, exhibited any binding to Cdc25 or Cki1 (data not shown). Therefore, the Tpk1KH variant, unlike the wild-type Tpk1, was able to bind stably to a number of known PKA substrates.
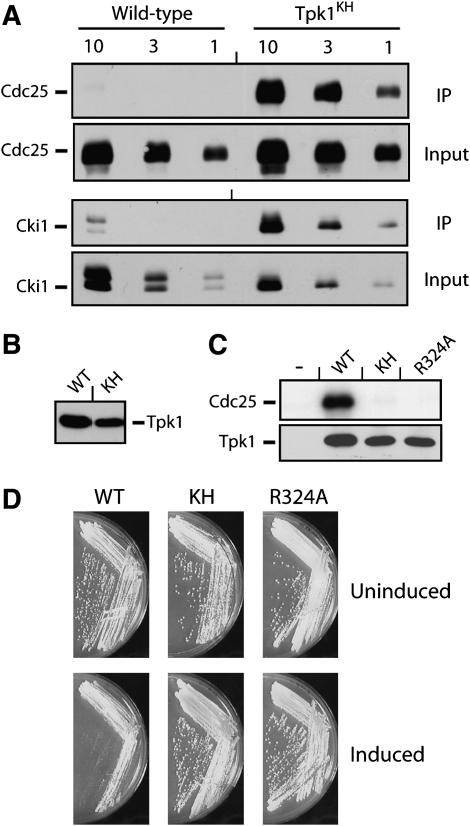
Figure 1.—
The Tpk1KH variant, but not the wild-type Tpk1, was physically associated with known PKA substrates. (A) The Tpk1KH variant was detected in immunoprecipitates with two PKA substrates, Cdc25 and Cki1. An HA epitope-tagged version of either the wild-type Tpk1 or the Tpk1KH variant was precipitated from cell extracts with an anti-HA affinity matrix. The bound substrate was eluted, separated on an SDS-polyacrylamide gel, and transferred to a nitrocellulose membrane. The relative amount of substrate present was then assessed by Western immunoblotting with antibodies specific for the myc epitope on the particular substrate present, either Cdc25 or Cki1. Three different amounts of the final immunoprecipitates and original inputs were loaded onto the gels, corresponding to 1, 3, and 10 μl of the 30 and 1000 μl present, respectively. (B) A Western immunoblot showing the relative levels of the wild-type Tpk1 and Tpk1KH present in the immunoprecipitates shown in A. (C) The Tpk1KH and Tpk1R324A variants were defective for PKA kinase activity in vitro. A myc epitope-tagged Cdc25 protein was incubated with [γ-32P]ATP and the wild-type Tpk1, Tpk1KH, or Tpk1R324A for 30 min at 25°. The reaction products were separated on an SDS-polyacrylamide gel and the extent of phosphorylation was assessed by autoradiography. (Bottom) A Western immunoblot showing the relative levels of the Tpk1 enzymes present in the kinase reactions. (D) The overexpression of the wild-type Tpk1 resulted in a growth defect in yeast cells. Wild-type cells were transformed with plasmids expressing the wild-type Tpk1, Tpk1KH, or Tpk1R324A from the CUP1 promoter. The cells were then plated to YM minimal media containing either 0 (Uninduced) or 100 μM (Induced) CuSO4 and incubated at 30° for 3 days.
TPK1 overexpression resulted in a severe growth defect:
During the course of the above studies, we found that we were not able to obtain stable expression of Tpk1 from clones that were under the control of constitutive promoters, like those from the ADH1 or TDH3 genes. One interpretation of these results was that the overexpression of TPK1 was detrimental to yeast cell growth and we tested this possibility with an inducible TPK1 construct. It should be pointed out that a previous study had shown that a TPK1 construct under the control of the galactose-inducible promoter from the GAL1 gene resulted in a growth defect specifically on galactose-containing media (Nehlin et al. 1992). However, the interpretation of these experiments is complicated by subsequent work showing that elevated levels of PKA activity inhibit GAL gene expression and can result in diminished growth on galactose media (Howard et al. 2002; Howard et al. 2006). Therefore, we tested whether the overexpression of TPK1 from the copper-inducible promoter from the CUP1 gene would also inhibit yeast cell growth. Previous work from our lab has shown that CUP1 gene expression is not significantly affected by changes in PKA activity (Budovskaya et al. 2004). We found that the elevated expression of the wild-type Tpk1, but not catalytically inactive versions like Tpk1KH, resulted in a severe growth defect on copper-containing media (Figure 1D). Thus, these results showed that elevated levels of Tpk1 are indeed deleterious to S. cerevisiae. Moreover, they indicated the importance of having inducible versions of the wild-type TPK1 for some of the experiments described here.
The binding to Tpk1KH was specific for PKA substrates:
The interactions with the Tpk1KH variant were specific for PKA substrates as no significant binding was detected with any protein that was not phosphorylated by Tpk1. For example, in the co-immunoprecipitation assay, no interaction was detected between this variant and Cki1-N, a control Cki1 fragment that lacks the two known sites of PKA phosphorylation (Figure 2A). Similar results were obtained with a two-hybrid assay system. Under conditions that produced equivalent levels of enzyme, a positive two-hybrid signal was observed with the Tpk1KH variant but not with the wild-type Tpk1 (Figure 2, B and C). Again, this signal was specific to substrates as two control proteins, Rim15-N and Toa2, did not interact with Tpk1KH (Figure 2B). Rim15 is a protein kinase important for stationary phase entry that has been shown to be a PKA target and Toa2 is a general transcription factor that is not a substrate for PKA (Reinders et al. 1998; Budovskaya et al. 2005) (Figure 2D). The Rim15-N fragment lacks the known PKA sites in this protein and is not phosphorylated by PKA (data not shown). In contrast, a Rim15 fragment that contains the known sites of PKA phosphorylation exhibited a robust interaction with the Tpk1KH variant (Figure 2E). Therefore, the Tpk1KH variant, but not the wild-type Tpk1, was able to interact specifically with known substrates of PKA.
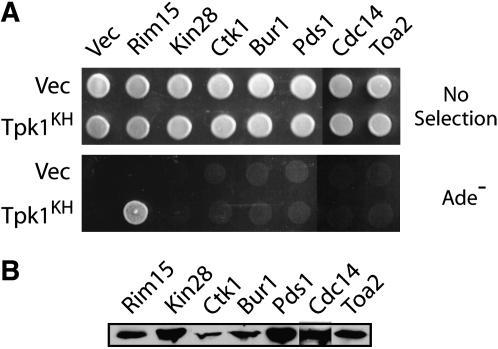
We further examined the specificity of this binding with a set of >40 proteins that were not substrates for PKA (Figure 3; data not shown). These negative controls included several proteins, such as Cdc14 and Toa2, that contain a reasonable match to the PKA consensus site of R−3-R−2-x−1-S/T-B+1, where x refers to any amino acid, B to a hydrophobic residue, and the S or T to the site of phosphorylation (Zetterqvist et al. 1976; Smith et al. 1999). Although many known PKA substrates are phosphorylated at a sequence that conforms to this consensus, most occurrences of this site in native proteins are not recognized by PKA (Shabb 2001; Budovskaya et al. 2005). We found that none of these nonsubstrate proteins exhibited any binding to the Tpk1KH variant (Figure 3, A and B). Therefore, Tpk1KH was not simply binding to proteins that contained a PKA consensus site but was instead recognizing bona fide substrates of this enzyme.
Figure 3.—
The Tpk1KH variant did not interact with proteins that were not substrates of PKA, including those that possessed sequences matching the PKA consensus phosphorylation site. (A) A two-hybrid assay examining the interactions between the Tpk1KH variant and a number of proteins that were not phosphorylated by PKA in vitro. Rim15 served as the positive control in these assays. (B) A Western immunoblot showing the relative levels of the different substrate proteins tested.
Substrate binding by the Tpk1KH variant was dependent upon PKA residues known to be important for the interaction with substrates:
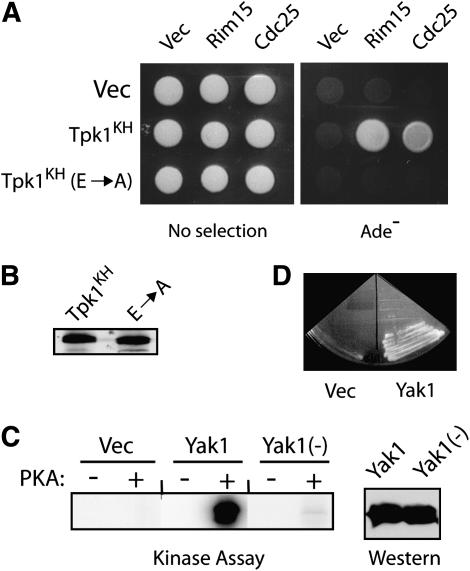
We tested whether the observed interactions with Tpk1KH were dependent on PKA residues previously shown to be important for substrate recognition. These studies took advantage of work that had identified amino acids within PKA that were critical for the association with a peptide substrate (Gibbs and Zoller 1991a; Kemp et al. 1994). For example, replacement of E171 and E274 with alanine in the wild-type Tpk1 resulted in a 40- to 80-fold increase in the apparent Km for this peptide (Gibbs and Zoller 1991a; Kemp et al. 1994). Here, we found that the introduction of these E171A and E274A alterations into the Tpk1KH variant completely abolished binding to two PKA substrates, Cdc25 and Rim15 (Figure 4A). Thus, the Tpk1KH variant may interact with substrates in a manner similar to that used by the wild-type enzyme.
Figure 4.—
Tpk1KH binding to substrates required amino acid residues known to be important for the interaction between the wild-type PKA and its substrates. (A) A two-hybrid assay assessing the interactions between Tpk1KH variants and two substrates, Rim15 and Cdc25. The Tpk1KH (E → A) variant has replaced both E171 and E274 with an alanine residue. These glutamic acid residues have been shown previously to be important for Tpk1 interactions with peptide substrates (Gibbs and Zoller 1991a; Kemp et al. 1994). (B) A Western immunoblot showing the relative levels of the two Tpk1 enzymes present in the two-hybrid strains with Cdc25 as substrate. (C) A Yak1 fragment containing codons 243–361 is phosphorylated at a single site by PKA in vitro. Protein A fusions to Yak1 with either a wild-type (Yak1) or altered PKA site [Yak1(−)] were precipitated from yeast cell extracts and incubated with PKA and [γ-32P]ATP. The reaction products were separated on an SDS-polyacrylamide gel and the level of phosphorylation was assessed by autoradiography. (Right) A Western immunoblot showing the relative levels of the two Yak1 proteins present in the kinase assays. (D) A Yak1 fragment containing only a single site of PKA phosphorylation (codons 243–361) was recognized by the Tpk1KH variant in a two-hybrid assay.
We tested the sensitivity of this binding assay with a substrate containing only a single site of PKA phosphorylation. Most of the targets used above are likely phosphorylated by PKA at more than one position (Gross et al. 1992; Reinders et al. 1998; Yu et al. 2002). For these experiments, we used a fragment of Yak1 that encompassed codons 243–361 and was phosphorylated by PKA at the serine residue at position 295 (Figure 4C). As with the other PKA substrates tested, we found that this Yak1 fragment exhibited a stable interaction specifically with the Tpk1KH variant (Figure 4D; data not shown). Thus, Tpk1KH was able to associate with targets containing only a single site of PKA phosphorylation.
A second alteration within the C-terminal region of the Tpk1 kinase domain resulted in an increased affinity for substrates:
We also tested whether the substrate binding observed with the Tpk1KH variant was a general property of catalytically inactive versions of PKA. For these studies, we analyzed several different Tpk1 variants, including Tpk1K116A, Tpk1D210A, Tpk1K212A, Tpk1D228A, and Tpk1R324A, that alter residues that are highly conserved in protein kinases (Hanks and Hunter 1995; Hanks 2003) (Figure 5A). A previous study had shown that each of these variants exhibited diminished activity toward a peptide substrate and we confirmed these results here with protein substrates (Gibbs and Zoller 1991b) (see Figure 1C; our unpublished data). We found that none of these inactive versions of Tpk1, except Tpk1R324A, exhibited any binding to the PKA substrates tested (Figure 5, B–E) (data not shown). In general, the two-hybrid signal with the Tpk1R324A variant was less than that observed with Tpk1KH (Figure 5, B and C). However, it should be pointed out that more subtle alterations that changed the R324 residue to either a lysine or a histidine residue resulted in a stronger binding with specific substrates (data not shown). In all, this Tpk1R324A variant is interesting for a couple of reasons. First, the residue altered, R324, is in the same C-terminal region of the protein kinase domain (i.e., within subdomain XI) as the K336 and H338 residues altered in Tpk1KH (Figure 5A). Thus, this region of PKA might play a specific role in the binding and/or release of protein substrates. Second, this arginine is conserved in all protein kinases and therefore it will be important to test if alterations of this position in other kinases result in a similar increase in substrate binding.
Figure 5.—
Tpk1KH and Tpk1R324A were the only inactive variants of Tpk1 that interacted with known substrates of PKA. (A) A schematic representing the protein kinase domain of Tpk1 (aa 90–390). The Tpk1 residues altered here are shown in boldface type; the relative positions of these residues in Tpk1 are indicated by the numbers below the diagram. The roman numerals above the diagram indicate the Hanks and Hunter subdomain designations for the indicated regions of the protein kinase domain (Hanks and Hunter 1995). (B) A two-hybrid analysis showing the interactions observed between different Tpk1 variants and four known PKA substrates, Rim15, Yak1, Cki1, and Cdc25. An interaction is indicated by growth on the YM minimal medium lacking adenine (Ade−). (C) Plates showing the relative two-hybrid interaction observed for the Tpk1KH and Tpk1R324A variants with the Rim15 substrate. (D) Western immunoblots showing the relative levels of the different Tpk1 variants and substrates present in the strains used for the two-hybrid analyses in B and C. Note that very low levels of the wild-type Tpk1 were found in these two-hybrid strains. (E) The substrates used in the above two-hybrid analysis were all efficient in vitro substrates for PKA. Each of the indicated substrate proteins was precipitated from yeast cell extracts and incubated with the yeast Tpk1 and [γ-32P]ATP as described in materials and methods. The reaction products were separated on an SDS-polyacrylamide gel and the level of phosphorylation was assessed by autoradiography.
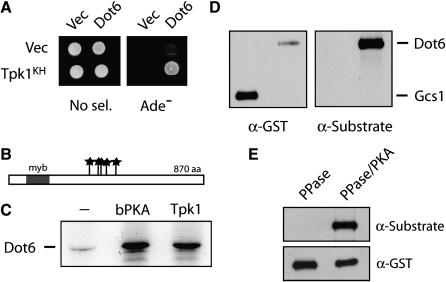
Dot6, a protein implicated in telomere function, is a substrate for PKA:
A preliminary screen with the Tpk1KH variant identified the Dot6 protein as a candidate substrate for Tpk1. As with other substrates, Dot6 was found to interact with the Tpk1KH and Tpk1R324 variants, but not with the wild-type Tpk1 (Figure 6A; data not shown). Although the precise biological role of Dot6 is unknown, previous work has shown that the overexpression of this protein disrupts the transcriptional silencing that normally occurs at yeast telomeres (Singer et al. 1998). Interestingly, the predicted Dot6 protein sequence contains five consensus PKA phosphorylation sites, the most sites present in any S. cerevisiae protein (Budovskaya et al. 2005) (Figure 6B). In addition, these sites are evolutionarily conserved as all five sites are found in the likely Dot6 ortholog present in the distantly related Saccharomyces species S. kluyveri (Budovskaya et al. 2005). Consistent with these observations, Dot6 was phosphorylated efficiently in vitro by both bovine and yeast PKA enzymes (Figure 6C). To assess whether Dot6 was also phosphorylated by PKA in vivo, we took advantage of a commercially available antibody that recognizes phosphorylated PKA sites. This antibody has been used to demonstrate that several proteins, including Srb9 and Cdc20, are phosphorylated by PKA in vivo (Chang et al. 2004; Searle et al. 2004). Here, we found that this antibody specifically recognized a GST–Dot6 protein that was precipitated from yeast cell extracts (Figure 6D). In contrast, this antibody failed to recognize several proteins, such as Gcs1, that were not substrates for PKA in vitro (see Figure 6D) (Budovskaya et al. 2005). This recognition of Dot6 was lost upon phosphatase treatment of the precipitated fusion protein and was subsequently restored following incubation with PKA and ATP (Figure 6E). In all, these data suggested that Dot6 was both an in vitro and an in vivo substrate for PKA in S. cerevisiae. Further work will be necessary to define precisely how PKA phosphorylation influences the normal functions of the Dot6 protein.
Figure 6.—
The Dot6 protein was identified as a candidate substrate for PKA. (A) Dot6 interacted with the Tpk1KH variant in a two-hybrid assay. (B) A schematic showing the positions of the five PKA consensus phosphorylation sites in the Dot6 protein; the sites are indicated by stars. A single myb domain, a DNA-binding motif originally identified in the c-myb oncoprotein, is present at the N-terminus of this protein (Singer et al. 1998). (C) The Dot6 protein was phosphorylated in vitro by the bovine PKA (bPKA) and the yeast Tpk1. (D) The Dot6 protein was recognized by an antibody specific for phosphorylated PKA sites. GST–Dot6 and GST–Gcs1 fusion proteins were precipitated with an anti-GST antibody, separated on an SDS-polyacrylamide gel, and transferred to a nitrocellulose membrane. The phosphorylation state of these proteins was then assessed by Western immunoblotting with an anti-PKA substrate antibody (Cell Signaling) that recognizes the phosphorylated forms of PKA target sites. The accompanying blot with an anti-GST antibody shows the relative levels of each fusion protein. (E) Dot6 recognition by the anti-PKA substrate antibody. The immunoprecipitated GST–Dot6 fusion was treated either with λ phosphatase only (PPase) or λ phosphatase and then bovine PKA and ATP (PPase/PKA). The reaction products were separated by SDS–PAGE electrophoresis, blotted to a nitrocellulose membrane, and analyzed by Western immunoblotting with either the anti-PKA substrate or anti-GST antibodies.
DISCUSSION
A complete understanding of the biological role of any protein kinase requires the identification of the relevant substrates of that enzyme. This study describes a novel approach to this identification process that takes advantage of protein kinase variants that bind stably to substrates. The two PKA variants identified here, Tpk1KH and Tpk1R324A, were able to bind to all substrates tested but exhibited no interactions with proteins that were not phosphorylated by PKA. Moreover, these interactions were dependent upon PKA residues previously shown to be required for the recognition of peptide substrates (Gibbs and Zoller 1991a; Kemp et al. 1994). Thus, these variants appear to interact specifically with substrates in a manner that may be similar to that used by the wild-type enzyme. The general utility of these variants was demonstrated by the identification of the Dot6 protein as a new substrate for PKA in S. cerevisiae.
In addition to identifying new substrates, the binding assay described here should also assist efforts to identify the substrate sequences that are required for a productive interaction with PKA. To date, most studies examining these elements have focused on the recognition and phosphorylation of peptide substrates (Songyang et al. 1994; Manning and Cantley 2002). The identification of the R-R-x-S/T-B consensus phosphorylation site discussed above is due, at least in part, to these efforts (Shabb 2001). However, one limitation of these types of studies is that they would miss any sequence elements in substrates that are distal to the actual site of PKA phosphorylation. In fact, although many PKA substrates are phosphorylated at sequences that generally conform to this consensus, most occurrences of these sequence elements in native proteins are not recognized by PKA (see, e.g., Budovskaya et al. 2005). Therefore, there must be additional sequence information present in substrates that governs the interaction with PKA. We feel that the use of the Tpk1 variants described here should facilitate the identification of these domains, and we are presently testing for the existence of such sequence elements in a number of PKA substrates from S. cerevisiae.
This assay may also provide complementary information about the sequences within PKA that are important for the interaction with substrates. In fact, the results presented here already implicate a small region at the C-terminal end of the PKA kinase domain in the binding and/or release of protein substrates. In general, all protein kinases possess a highly conserved, catalytic core domain of ∼250–300 amino acids that contains a number of signature residues critical for catalytic activity (Hanks and Hunter 1995; Manning et al. 2002; Johnson and Hunter 2005). This kinase domain has been further divided into 11 subdomains and all of the residues altered in the Tpk1KH and Tpk1R324A variants fall into subdomain XI (see Figure 5A). Inactivating alterations in other subdomains of the PKA catalytic core did not result in a concomitant increase in substrate binding. Interestingly, the arginine corresponding to position 324 in Tpk1 is thought to contribute to the stabilization of a peptide-binding loop in PKA that is in direct contact with peptide substrates (Johnson et al. 2001). Thus, this region of PKA may play an important role in coordinating the interactions between this enzyme and its in vivo substrates. This possibility could be tested directly if we could obtain co-crystals that contain the Tpk1KH variant with a relevant protein substrate.
An obvious question for the protein kinase field at large is whether the results obtained here might be generally applicable. Clearly, the key to success in this endeavor will be the ability to isolate kinase variants, analogous to Tpk1KH and Tpk1R324A, that can bind stably to their cognate substrates. In this regard, it is important to point out again that the arginine residue altered in the Tpk1R324A variant is found at an analogous position in all other protein kinases (Hanks and Hunter 1995). Thus, it will be important to test whether this invariant residue might play a role in regulating substrate interactions in other protein kinases and if alteration of this site might result in a similar increase in substrate binding. Alternatively, it might be necessary to alter distinct residues in other kinases to obtain an enzyme that binds stably to substrates. The important point is that the work presented here with PKA demonstrates the feasibility of this type of an approach and can serve as a framework for future studies with other protein kinases.
It is interesting also to speculate on the possibility that the substrate binding observed here could serve as a platform for the identification of a new class of protein kinase inhibitors. Most of the protein kinase inhibitors in use today target the conserved active site of these enzymes and act as competitive inhibitors of ATP (Cohen 2002). However, because of the highly conserved nature of the ATP-binding pocket, it can be difficult to obtain inhibitors of this type that have the specificity needed for clinical applications. In contrast, molecules that disrupt the substrate binding observed here would be interfering with the ability of the protein kinase to interact with its protein substrates. Since different families of protein kinases interact with substrates in quite distinct ways (Hardie and Hanks 1995), these latter types of inhibitors might exhibit a higher degree of specificity. In fact, this approach could potentially identify inhibitors that are substrate-specific, i.e., inhibitors that affect the phosphorylation of only one, or a subset, of the targets of a given protein kinase. The availability of such selective inhibitors would be invaluable in our attempts to understand which substrates are responsible for the particular actions of a protein kinase and could ultimately allow for a more precise treatment of the consequences of protein kinase malfunction associated with human disease.
In summary, this article describes how novel substrate-binding variants of protein kinases may be used to identify physiologically relevant targets of these enzymes. Although we presently do not know the mechanistic basis for the increased affinity toward PKA substrates observed here, previous studies with the Tpk1KH and Tpk1R324A variants suggested that these enzymes were specifically defective for catalytic activity (Gibbs and Zoller 1991b). The alterations examined resulted in a significant defect in the catalytic constant or kcat of the altered enzymes relative to the wild type, but had no effect on the observed Km for either ATP or a peptide substrate. One possibility suggested by these data is that these altered enzymes might bind normally to substrates but a specific defect prevents effective release of these targets. However, it is important to remember that Km values are not simply equivalent to dissociation constants and cannot be regarded as accurate gauges of enzyme/substrate affinity. Indeed, there are other possible explanations for the apparent increased substrate binding observed here. It may be especially important to emphasize that our studies were performed with protein substrates, whereas previous work utilized model peptides exclusively. Thus, it is possible that not yet identified aspects of the enzyme interaction with larger protein substrates, not reflected in the smaller peptides, might influence turnover. If this were the case, the kinetic parameters described previously for these Tpk1 variants might not apply. Further experimentation will therefore be necessary to determine the precise mechanisms underlying the increased substrate binding observed here.
Acknowledgments
We thank Joseph Heitman for the GST–Tpk1 plasmid; Michael Snyder for the yeast GST–Dot6 and GST–Gcs1 fusion proteins; Komudi Singh for the phosphorylation analysis of Toa2; Jack Dixon, Russell Hille, and Susan Taylor for helpful discussions and advice; and Joseph Stephan for comments on the manuscript. This study was supported by a grant from the National Institutes of Health (GM-65227) to P.K.H.
References
- Adams, J. A., 2001. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101: 2271–2290. [DOI] [PubMed] [Google Scholar]
- Arnsten, A. F., B. P. Ramos, S. G. Birnbaum and J. R. Taylor, 2005. Protein kinase A as a therapeutic target for memory disorders: rationale and challenges. Trends Mol. Med. 11: 121–128. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1995. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Broach, J. R., 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7: 28–33. [DOI] [PubMed] [Google Scholar]
- Budovskaya, Y. V., H. Hama, D. B. DeWald and P. K. Herman, 2002. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 277: 287–294. [DOI] [PubMed] [Google Scholar]
- Budovskaya, Y. V., J. S. Stephan, F. Reggiori, D. J. Klionsky and P. K. Herman, 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279: 20663–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya, Y. V., J. S. Stephan, S. J. Deminoff and P. K. Herman, 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102: 13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. W., S. C. Howard, Y. V. Budovskaya, J. Rine and P. K. Herman, 2001. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics 157: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. W., S. C. Howard and P. K. Herman, 2004. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell. 15: 107–116. [DOI] [PubMed] [Google Scholar]
- Cohen, P., 2002. Protein kinases—The major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 1: 309–315. [DOI] [PubMed] [Google Scholar]
- Gerhardt, B., T. J. Kordas, C. M. Thompson, P. Patel and T. Vida, 1998. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 273: 15818–15829. [DOI] [PubMed] [Google Scholar]
- Gibbs, C. S., and M. J. Zoller, 1991. a Identification of electrostatic interactions that determine the phosphorylation site specificity of the cAMP-dependent protein kinase. Biochemistry 30: 5329–5334. [DOI] [PubMed] [Google Scholar]
- Gibbs, C. S., and M. J. Zoller, 1991. b Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem. 266: 8923–8931. [PubMed] [Google Scholar]
- Gross, E., D. Goldberg and A. Levitzki, 1992. Phosphorylation of the S. cerevisiae Cdc25 in response to glucose results in its dissociation from Ras. Nature 360: 762–765. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., 2003. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S. K., and T. Hunter, 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596. [PubMed] [Google Scholar]
- Hardie, G., and S. Hanks (Editors), 1995. The Protein Kinase Factsbook: Protein-Serine Kinases. Academic Press, San Diego.
- Herman, P. K., 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5: 602–607. [DOI] [PubMed] [Google Scholar]
- Herman, P. K., and J. Rine, 1997. Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 16: 6171–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, S. C., Y. V. Budovskaya, Y. W. Chang and P. K. Herman, 2002. The C-terminal domain of the largest subunit of RNA polymerase II is required for stationary phase entry and functionally interacts with the Ras/PKA signaling pathway. J. Biol. Chem. 277: 19488–19497. [DOI] [PubMed] [Google Scholar]
- Howard, S. C., A. Hester and P. K. Herman, 2003. The Ras/PKA signaling pathway may control RNA polymerase II elongation via the Spt4p/Spt5p complex in Saccharomyces cerevisiae. Genetics 165: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, S. C., S. J. Deminoff and P. K. Herman, 2006. Increased phosphoglucomutase activity suppresses the galactose growth defect associated with elevated levels of Ras signaling in S. cerevisiae. Curr. Genet. 49: 1–6. [DOI] [PubMed] [Google Scholar]
- Hunter, T., 2000. Signaling—2000 and beyond. Cell 100: 113–127. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. A., P. Akamine, E. Radzio-Andzelm, M. Madhusudan and S. S. Taylor, 2001. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 101: 2243–2270. [DOI] [PubMed] [Google Scholar]
- Johnson, S. A., and T. Hunter, 2005. Kinomics: methods for deciphering the kinome. Nat. Methods 2: 17–25. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis and A. Mitchell, 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kemp, B. E., M. W. Parker, S. Hu, T. Tiganis and C. House, 1994. Substrate and pseudosubstrate interactions with protein kinases: determinants of specificity. Trends Biochem. Sci. 19: 440–444. [DOI] [PubMed] [Google Scholar]
- Kunkel, T. A., 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, B. D., and L. C. Cantley, 2002. Hitting the target: emerging technologies in the search for kinase substrates. Sci. STKE 162: 1–4. [DOI] [PubMed] [Google Scholar]
- Manning, G., G. D. Plowman, T. Hunter and S. Sudarsanam, 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27: 514–520. [DOI] [PubMed] [Google Scholar]
- Nehlin, J. O., M. Carlberg and H. Ronne, 1992. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 20: 5271–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picket-Gies, C. A., and D. A. Walsh, 1986. Phosphorylase kinase, pp. 395–459 in The Enzymes, edited by P. D. Boyer and E. G. Krebs. Academic Press, San Diego.
- Ptacek, J., G. Devgan, G. Michaud, H. Zhu, X. Zhu et al., 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684. [DOI] [PubMed] [Google Scholar]
- Reinders, A., N. Burckert, T. Boller, A. Wiemken and C. De Virgilio, 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12: 2943–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, L. S., H. C. Causton, R. A. Young and G. R. Fink, 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97: 5984–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, J. S., K. L. Schollaert, B. J. Wilkins and Y. Sanchez, 2004. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 6: 138–145. [DOI] [PubMed] [Google Scholar]
- Shabb, J. B., 2001. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 101: 2381–2411. [DOI] [PubMed] [Google Scholar]
- Shah, K., Y. Liu, C. Deirmengian and K. M. Shokat, 1997. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. USA 94: 3565–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson et al., 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. M., E. Radzio-Andzelm, K. R. Madhusudan, P. Akamine and S. S. Taylor, 1999. The catalytic subunit of cAMP-dependent protein kinase: prototype for an extended network of communication. Prog. Biophys. Mol. Biol. 71: 313–341. [DOI] [PubMed] [Google Scholar]
- Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms et al., 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4: 973–982. [DOI] [PubMed] [Google Scholar]
- Taylor, S. S., J. Yang, J. Wu, N. M. Haste, E. Radzio-Andzelm et al., 2004. PKA: a portrait of protein kinase dynamics. Biochim. Biophys. Acta 1697: 259–269. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., and J. H. de Winde, 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918. [DOI] [PubMed] [Google Scholar]
- Thiele, D. J., and D. H. Hamer, 1986. Tandemly duplicated upstream control sequences mediate copper-induced transcription of the Saccharomyces cerevisiae copper-metallothionein gene. Mol. Cell Biol. 6: 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., S. Cameron, P. Sass, M. Zoller and M. Wigler, 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50: 277–287. [DOI] [PubMed] [Google Scholar]
- Yu, Y., A. Sreenivas, D. B. Ostrander and G. M. Carman, 2002. Phosphorylation of Saccharomyces cerevisiae choline kinase on Ser30 and Ser85 by protein kinase A regulates phosphatidylcholine synthesis by the CDP-choline pathway. J. Biol. Chem. 277: 34978–34986. [DOI] [PubMed] [Google Scholar]
- Zetterqvist, O., U. Ragnarsson, E. Humble, L. Berglund and L. Engstrom, 1976. The minimum substrate of cyclic AMP-stimulated protein kinase, as studied by synthetic peptides representing the phosphorylatable site of pyruvate kinase (type L) of rat liver. Biochem. Biophys. Res. Commun. 70: 696–703. [DOI] [PubMed] [Google Scholar]
- Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor et al., 2001. Global analysis of protein activities using proteome chips. Science 293: 2101–2105. [DOI] [PubMed] [Google Scholar]