Abstract
UPC2 and ECM22 belong to a Zn(2)–Cys(6) family of fungal transcription factors and have been implicated in the regulation of sterol synthesis in Saccharomyces cerevisiae and Candida albicans. Previous reports suggest that double deletion of these genes in S. cerevisiae is lethal depending on the genetic background of the strain. In this investigation we demonstrate that lethality of upc2Δ ecm22Δ in the S288c genetic background is attributable to a mutation in the HAP1 transcription factor. In addition we demonstrate that strains containing upc2Δ ecm22Δ are also inviable when carrying deletions of ERG6 and ERG28 but not when carrying deletions of ERG3, ERG4, or ERG5. It has previously been demonstrated that UPC2 and ECM22 regulate S. cerevisiae ERG2 and ERG3 and that the erg2Δ upc2Δ ecm22Δ triple mutant is also synthetically lethal. We used transposon mutagenesis to isolate viable suppressors of hap1Δ, erg2Δ, erg6Δ, and erg28Δ in the upc2Δ ecm22Δ genetic background. Mutations in two genes (YND1 and GDA1) encoding apyrases were found to suppress the synthetic lethality of three of these triple mutants but not erg2Δ upc2Δ ecm22Δ. We show that deletion of YND1, like deletion of GDA1, alters the sphingolipid profiles, suggesting that changes in sphingolipids compensate for lethality produced by changes in sterol composition and abundance.
THE yeast Saccharomyces cerevisiae has provided a powerful model system to study the biochemistry of lipid biosynthesis. Many genes encoding the enzymes for sterol, fatty acid, phospholipid, and sphingolipid synthesis have been isolated, their products characterized, and the orthologous genes in human cells identified (Daum et al. 1998; Kelley and Herman 2001). However, less is known about the regulation of lipid synthesis and transport between organelles. Recently, two transcription factors, Upc2p and Ecm22p, were implicated in the coordination of these processes in this model organism (Vik and Rine 2001; Wilcox et al. 2002).
Upc2p and Ecm22p are members of the Zn[2]–CyS[6] binuclear cluster family of transcription factors (Todd and Andrianopoulos 1997) and share significant amino acid sequence identity (45%). A semidominant allele of UPC2 (upc2-1) was demonstrated to confer aerobic sterol influx, a process normally restricted to anaerobiosis (Lewis et al. 1988; Crowley et al. 1998), while a lesser role of Ecm22p in sterol transport has also been reported (Shianna et al. 2001). Microarray analyses of the upc2-1 mutant identified novel genes involved in sterol influx, including AUS1 and PDR11, which encode ABC transporters required for anaerobic sterol influx (Wilcox et al. 2002).
In addition to a role in sterol transport, an involvement of Upc2p and Ecm22p in regulating sterol biosynthesis has been suggested (Vik and Rine 2000). Both transcription factors have been referred to as sterol regulatory element (SRE)binding proteins (SREBPs), responsible for regulating the transcription of the ergosterol biosynthetic genes ERG2 and ERG3 via binding to an 11-bp SRE (Vik and Rine 2001). However, these proteins lack sequence conservation with the analogous mammalian or recently identified Schizosaccharomyces pombe SREBPs (Brown and Goldstein 1997; Hughes et al. 2005); whether they exhibit functional similarity remains to be established. Despite this, a large number of ergosterol biosynthetic genes, such as ERG1, ERG6, ERG8, ERG12, ERG13, and ERG25 contain this SRE and are likely under UPC2/ECM22 transcriptional control. In Candida albicans, loss of UPC2 resulted in an inability to upregulate ERG2 and ERG11 in response to azole treatment. Similarly, a 7-bp SRE core element also appears in many of the same ergosterol biosynthetic genes in C. albicans (Silver et al. 2004).
Upc2p has further been proposed to regulate the transcription of a number of anaerobically expressed genes (Kwast et al. 1998). For example, Upc2p induces the expression of the anaerobic cell wall-related genes in the PAU family (Abramova et al. 2001; Wilcox et al. 2002). Combined with the induction of sterol influx, a largely anaerobic process, these results suggest that Upc2p and Ecm22p play a role in coordinating the transcriptional response to hypoxia. The role of Ecm22p under anaerobic conditions has not been documented. Furthermore, the transcriptional targets of Upc2p and Ecm22p under oxygen-limited conditions are largely unknown.
In this article we demonstrate synthetic lethality between genes encoding transcription factors that decrease sterol biosynthesis (UPC2, ECM22, and HAP1) and mutations in ergosterol biosynthetic genes that by themselves are viable (ERG2, ERG6, and ERG28). In addition, we identify mutations in GDA1 and YND1 as suppressors of this synthetic lethality. Gda1p was shown previously to play a role in mannosylation of sphingolipids (Abeijon et al. 1993). Ynd1p is related to Gda1p and we demonstrate that it also plays a role in sphingolipid synthesis. These results demonstrate that inviability caused by changes in sterols can be suppressed by compensatory changes in sphingolipids.
MATERIALS AND METHODS
General:
Yeast complete (YEPD), synthetic complete (CSM), and bacterial media were prepared as described (Ausubel et al. 1998). Yeast extract, yeast nitrogen base, Bacto-peptone, and Bacto-agar were from Difco. Molecular biology and genetic procedures were performed according to conventional protocols (Ausubel et al. 1998). Gene-specific oligonucleotides were synthesized by Invitrogen.
Yeast strains and molecular techniques:
Yeast strains used in this study are isogenic with the strain W303-1A (MATa ade2-1, can1-1, trp1-1, ura3-1, his3-11, 15, leu2-3, 112) (Thomas and Rothstein 1989). Deletion mutant strains were generated by homologous recombination with PCR products (Erdeniz et al. 1997) generated using the S. cerevisiae pRS303, -304, -305, and -306 vectors to generate HIS3, TRP1, LEU2, and URA3 selectable markers (Sikorski and Hieter 1989) and oligonucleotides that contained 45–50 bp of gene-specific sequence. Disruptions containing the kanamycin (G418) resistance marker were obtained using the plasmid pFA6 as template. Oligonucleotides used to create various deletions are listed in Table 1. All disruptions were verified by PCR. The Ty transposon insertion at the 3′ end of the HAP1 gene was identified by PCR using oligonucleotides specific to HAP1 and the Ty1 transposon (CTTCCTTTTATCAAAGCAT CTTG and CGAGGATTTAGGAATCCATAAA, respectively). The hap1Δ mutation was created as follows: An integrating plasmid PdAG1-HS (kindly provided by A. Gower and J. L. Pinkham) containing a 1347 deletion within HAP1 spanning the HindIII site to SalI site was digested with EcoRI for targeted integration into the HAP1 locus of SCY325. URA3 transformants were selected and colonies placed on 5-fluoroorotic acid (FOA) medium to select for a strain (SCY2127) containing the hap1Δ allele. PCR screening primers for the characterization of the HAP1 locus are indicated in Table 1. Plasmid pU6ΔsacI containing the normal UPC2 gene was used as the rescue plasmid in all suppressor screens (A. Tinklenberg and S. L. Sturley, unpublished data). Standard protocols for matings, sporulation, tetrad analyses, and transformation are as described (Burke et al. 2000).
TABLE 1.
Primers used in this study
| Primer name | Primer sequence | Purpose |
|---|---|---|
| frwUPC2 | CGGTAAACGTAAATTCCATAACAAATCAA | UPC2 disruption |
| AGAATGGGTGCGATAACTGTAAAAGAAG | ||
| AAGTggcgggtgtcggggctggc | ||
| revUPC2 | CTATCAGGTTCAGATTGCCTTTGGTAGAA | |
| AGATCTAAAAGCTTAGCGATGTTACTGGT | ||
| ACttgccgatttcggcctattg | ||
| frwECM22 | CCGATGATGGGAATGCTGGACAAGAAAG | ECM22 disruption |
| AGAGAAGGATGCTGAACTGATTGAGGTTG | ||
| Gggcgtacgctgcaggtcgac | ||
| revECM22 | CGCGATGCAGTTTGTCCAAATATGCTAAA | |
| GTTATCAAGTACGGTGAATCAATTTCTAC | ||
| Ggatcgatgaattcgagctcg | ||
| frwERG2 | CCACTCCTTTTGTTGATTGGTGTTGTAGGC | ERG2 disruption |
| TACATTATGAACGTATTGTTCACTACCTG | ||
| GTggcgggtgtcggggctggc | ||
| revERG2 | CAAGTTCTTACCCATGTCCCTGGCAGTCA | |
| GGTAGACAGTTCTATATAGAGTGTATAAA | ||
| TCttgccgatttcggcctattg | ||
| frwERG6 | GGCCCAATTCACTAGGGAGTTACATGGTG | ERG6 disruption |
| ATGATATTGGTAAAAAGACAGGTTTGAGT | ||
| GCTGGCGGGTGTCGGGGCTGGC | ||
| revERG6 | CGTTTTCTGGCTTCCTAGCGACGAAAAGC | |
| ATCATTGGAGTGAATAACTTGGACTTACC | ||
| ACTTGCCGATTTCGGCCTATTG | ||
| frwERG28 | CTTTACGGTATTACCGATAGGAAACTTCT | ERG28 disruption |
| ATTTTATGATTTTTTCGTTCGGGGACGGAA | ||
| CTGGCGGGTGTCGGGGCTGGC | ||
| revERG28 | CGTTAGAGGAGAGAGGTAGGGATACTTA | |
| CAGAAATGAAGCCAAATTGGCAGCTTTTT | ||
| GAGTTGCCGATTTCGGCCTATTG | ||
| frwHDA3 | GATTTACTACGCATTTTAGACACGAAACC | HDA3 disruption |
| AATACCTACAATTGTTGATGCTACTACTCT | ||
| Gtggcgggtgtcggggctggc | ||
| revHDA3 | CGTGGAATTGTTAAGTTTTCCAAGTTGTCC | |
| ATCGTCTTCGATAACTGTCCCACTAACTCC | ||
| ttgccgatttcggcctattg | ||
| frwTUP1 | GGGAAGAAAGAAATCAGCTTTCCATCCAA | TUP1 disruption |
| ACCAATATGACTGCCAGCGTTTCGAATAC | ||
| GCtggcgggtgtcggggctggc | ||
| revTUP1 | GTGTGGTGGCCACGGACAATACAAAGTCT | |
| TTATGCCCGATATACGTAACTTCACAAGT | ||
| GCttgccgatttcggcctattg | ||
| frwYND1 | ATTTCCCCGTCTGCCCTCTTATGCTCATAG | YND1 disruption |
| AAAACACTAATGATCGGTTTGGTATCGTC | ||
| Atggcgggtgtcggggctggc | ||
| revYND1 | CAGGTCGTAAGTTTGACGGAGCGAGATTA | |
| GCGGACTGACTTGCTGTCCTTTGTGGTTCG | ||
| Tttgccgatttcggcctattg | ||
| frwIES1 | GTATTGGAAACAGTTCATGATTAACTAAG | IES1 disruption |
| TAAAATCGAAAGCTAAATAAAGGCGACA | ||
| CTCtggcgggtgtcggggctggc | ||
| revIES1 | CTTAAACATGGCGGTTGCTTCTTCGTCATC | |
| AATTTCCGCGCGTTCGATTTTATTTAAAAG | ||
| ttgccgatttcggcctattg | ||
| MNN11fwd | GAAGAAAGCCAATTTTCAACTTGGAATCA | MNN11 disruption |
| TCCTGCCGGTTTGTTGTTGGTGTTAGCTGT | ||
| Gtggcgggtgtcggggctggc | ||
| MNN11rev | GTAAAGAAAGATCGAGATTCATCAATAA | |
| AGCATCTTGATCAACAAAATGAATATACT | ||
| TGGttgccgatttcggcctattg | ||
| UME6fwd | CAGCGCACAGGAACTAGGACACTACCGC | UME6 disruption |
| ACTCAAACCATTTGCATGGACCTTAACTC | ||
| ACGtggcgggtgtcggggctggc | ||
| UME6rev | GTCTTTCCTCGGTACACTTCTTTTTCCTTA | |
| ATCTACAAATCCAGCAACCAGTACGGGAC | ||
| Cttgccgatttcggcctattg | ||
| MNN9fwd | AATTTTAGTATTTTTCAAACAGAAGAATT | MNN9 disruption |
| TAAAAGAGCTAGAATAAAAAGTTAGGAA | ||
| ACAtggcgggtgtcggggctggc | ||
| MNN9rev | ATTAATGGCCTATAAGTGGCAATTTCTGC | |
| ATAACCCTCGACAATAATCTCGTCATCAC | ||
| CCttgccgatttcggcctattg | ||
| GDA1fwd | CAATCTTTATTGGCGAACAGTTAAGGGTC | GDA1 disruption |
| CTCTCGAGAAGAAACATTAAGACATCATC | ||
| GCtggcgggtgtcggggctggc | ||
| GDA1rev | AACAATTTGTGTAGACCCTCCGCCTAAGT | |
| CAAAAACGGCAGCAGTAGGTAACTTGGG | ||
| GCCttgccgatttcggcctattg | ||
| GDA1-5′GAL | CGGGATCCCACAAAAAACATGGCGCCCA | pYES2-GDA1 plasmid |
| GDA1-3′GAL | CGGAATTCGAAAAGCGGTGTCCATGTTC | |
| inPCR1 | 5′-TAAGTTGGGTAACGCCAGGGTTTTC-3′ | Nested/inverse PCR for RsaI, TaqI |
| inPCR2 | 5′-TTCCATGTTGCCACTCGCTTTAATG-3′ | |
| inPCR7 | CCTCAGGAAGATCGCACTCC | Inverse PCR for TaqI |
| inPCR8 | GTGATGGTGCTGCGTTGGAG | |
| MTn3-SEQ1 | CCCCCTTAACGTGAGTTTTCGTTCCACT | Sequencing InPCR |
| HAP1-frw | ACTTCCTTTTATCAAAGCATCTTG | hap1Δ check in SCY2127 |
| HAP1-2rev | GACGTATCCCATTCTGAAACGCAAC | |
| LW1 | ATTCCAACGGTTCCATCGTTAACC | Hap1 wild-type check |
| LW2 | TATCCCATTCTGAAACGCAACG | |
| LW1 | ATTCCAACGGTTCCATCGTTAACC | Hap1Ty check |
| LW3 | CGAGGATTTAGGAATCCATAAA |
In the disruption primers, lowercase letters correspond to conserved regions bordering the TRP1, KANMX, HIS3, LEU2, and NATR genes in the pRS304, pFA6, pRS303, pRS315, and pAG36 vectors, respectively. All primers are designed so that left to right corresponds to 5′ → 3′.
Strains used in this study were derived from W303. SCY325 and SCY328 are common Sturley laboratory strains. All other strains listed in Table 2 were derived for this study and are deletions, with the exception of hap1Ty.
TABLE 2.
Strains used in this study
| Strain | Genotype |
|---|---|
| W303 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1, ura3-1 can1 |
| SCY325 | MATα ade2-1 his3-11,15 leu2-3,112,trp1-1, ura3-1 can1 |
| SCY328 | MATaade2-1 his3-11,15 leu2-3,112,trp1-1ura3-1 can1 |
| SCY1832 | MATα upc2Δ∷URA3 hap1Ty |
| SCY1987 | MATα upc2Δ∷URA3 |
| SCY1996 | MATα ecm22Δ∷LEU2 |
| SCY2127 | MATα hap1Δ |
| MVS36 | MATaerg28Δ∷HIS3 |
| MVS40 | MATaerg2Δ∷HIS3 |
| MVS41 | MATα erg6Δ∷HIS3 |
| MH1-3B | MATα hap1Δ upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacIa |
| M28-2D | MATaerg28Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| M6-3B | MATaerg6Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| M2-7A | MATα erg2Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| MVS139 | MATα ynd1Δ∷LEU2 |
| M5B-3 | MATaupc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| MVS3 | MATaerg3Δ∷TRP1 upc2Δ∷URA3 ecm22Δ∷LEU2 |
| MVS14 | MATaerg5 Δ∷TRP1 upc2Δ∷URA3 ecm22Δ∷LEU2 |
| MVS18 | MATaerg4Δ∷TRP1 upc2Δ∷URA3 ecm22Δ∷LEU2 |
| BBY102 | MATaynd1Δ∷LEU2 erg2 Δ∷HIS3 upc2 Δ∷TRP1 ecm22 Δ∷KANMX4/pU6ΔSacI |
| BBY103 | MATaynd1Δ∷LEU2 hap1Δ upc2 Δ∷TRP1 ecm22 Δ∷KANMX4/pU6ΔSacI |
| BBY105 | MATα ynd1 Δ∷LEU2 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY107 | MATaynd1Δ∷LEU2 erg2Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY112 | MATaynd1Δ∷LEU2 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY113 | MATaynd1Δ∷LEU2 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY114 | MATagda1Δ∷HIS3 |
| BBY125 | MATα gda1Δ∷HIS3 hap1Δ upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY131 | MATα ynd1Δ∷LEU2 erg28Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| BBY137 | MATagda1Δ∷HIS3 erg28Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY29 | MATα gda1Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY31 | MATagda1Δ∷HIS3 erg6Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY34 | MATagda1Δ∷HIS3 erg6Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY38 | MATα gda1Δ∷HIS3 erg2Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY47 | MATα ynd1Δ∷LEU2 erg6Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
| SBY56 | MATα ynd1Δ∷LEU2 hap1Δ∷HIS3 upc2Δ∷TRP1 ecm22Δ∷KANMX4/pU6ΔSacI |
All strains are derived from W303. SCY325 and SCY328 are common laboratory strains. All others were generated in this study.
pU6ΔSacI is a complementing plasmid containing UPC2.
Transposon mutagenesis:
Fifteen pools of the mTn-lacZ/LEU2 transposon library were received as a generous gift from Michael Snyder's lab (Burns et al. 1994). UltraMAX DH5α-FT Competent Cells were transformed with the pools of DNA according to standard bacterial transformation protocols. Transformants were selected on LB medium containing 60 μg/ml ampicillin. On average 250,000 colonies could be harvested from each pool. DNA purification was performed according to standard DNA midi-prep protocols (Ausubel et al. 1998). S. cerevisiae DNA from amplified pools containing the 6.6-kb mTN-lacZ/LEU2 cassette was restricted with NotI and transformed into the appropriate strains containing the UPC2 rescue plasmid pU6ΔSacI. For hap1Δ upc2Δ ecm22Δ /pUΔSacI, 33,754 LEU2 transformants were obtained. For the erg2, erg6, and erg28 mutants in the upc2Δ ecm22Δ background, 10,804, 39,771, and 1095 LEU2 transformants were obtained, respectively. Transformants that were able to lose the rescue plasmid on FOA medium became candidates for containing suppressor mutations. To locate the site of genomic insertion inverse PCR was used as described on the Gottschling web site (http://www.fhcrc.org/science/labs/gottschling/misc/ipcr.html) and in Ochman et al. (1988). Genomic DNA from S. cerevisiae from putative suppressor strains was extracted according to standard protocols (Ausubel et al. 1998) and digested with AciI, AluI, HaeIII, HpaII, RsaI, or TaqI that corresponded to known restriction sites in the mTn-LacZ/LEU2 transposon cassette. The restriction site defined the sequence for which both the inverse PCR and sequencing primers would be used. Inverse PCR was performed using primers InPCR1, -2, -7, and -8. A PCR product using primers InPCR7 and InPCR 8 was used as a template for a second round of PCR with the primers InPCR1 and InPCR2. Using this method, PCR products were produced that were amenable to DNA sequencing. DNA purification for nucleotide sequencing was performed using the QIAquick Method (QIAGEN). Sequencing was performed at the Indiana University Biochemistry and Biotechnology Facility using mTn3-SEQ1, which anneals at position 15–42 of the mTn-lacZ/LEU2 transposon cassette.
Sterol analyses:
Sterols were extracted following published protocols (Gachotte et al. 1999). Gas chromatography analysis of sterols was conducted on an HP5890 series II GC, using a DB-5 capillary column (15 m × 0.25 mm i.d., 0.2 μm film thickness) with nitrogen as carrier gas (30 cm/sec) and was programmed from 195° to 300° (195° for 3 min, 5.5°/min to 300° then held for 10 min). Gas-chromatography–mass spectrometry analyses were performed with an HP5890 GC coupled to an HP5972 mass selective detector. Electron impact MS (70 eV, scanning from 40 to 700 or 650 atomic mass units, at 1-sec intervals) was performed using the following conditions: DB-5MS column (20 m × 0.18 mm i.d., 0.18 μm film thickness), helium as carrier gas (30 cm/sec), detector temperature 180°, column temperature 100°–300° (100° for 1 min, 10°/min to 300° then held for 15 min); and DB-5 (10 m × 0.25 mm i.d., 0.25 μm film thickness), helium as carrier gas (50 cm/sec), column temperature 40°–300° (40° for 1 min, 30°/min to 300° then held for 4 min). All injections were run in splitless mode.
Sphingolipid and membrane microdomain analyses:
Lipids were extracted from cells radiolabeled with [3H]myo-inositol, separated by chromatography on Whatman HP-K plates and detected by using a BioScan apparatus (Dickson et al. 1997). The chromatography solvent was 55:45:10 (chloroform:methanol:0.25% KCl) (Puoti et al. 1991). Detergent-insoluble fractionations were carried out according to Bagnat et al. (2000) with a few minor alterations. A total of 10 OD600 units of exponential phase cells were lysed in TNE (50 mm Tris–HCl, pH 7.4/150 mm NaCl/5 mm EDTA) buffer by bead-beating mechanical disruption at 4° in the presence of protease inhibitors. The supernatant (450 μl) was incubated for 30 min on ice with pre-chilled Triton X-100 at final concentration of 1% v/v. A total of 400 μl of this mixture with 2 volumes of 60% OPTIPREP were then overlaid with 2.7 ml of 30% OPTIPREP and finally with 0.5 ml TNE protease buffer and centrifuged for 3 hr at 45,000 rpm in a SW50.1 rotor at 4°. A total of 500 μl fractions 1–8 were removed from the top of the gradient. The detergent-insoluble (raft-associated) proteins reside predominately in fraction 2 and sometimes in fraction 1. Fractions were stored at −80°. All immunoblots were performed on 8% SDS–PAGE gels with a 5% stacking gel. α-Gas1p or α-Pma1p antibody was used at a 1:2500 dilution followed by α-goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:4000 dilution). Amersham ECL assays were used to detect the proteins.
RESULTS
The upc2Δ ecm22Δ double mutant exhibits synthetic lethality in combination with various mutations affecting sterol synthesis:
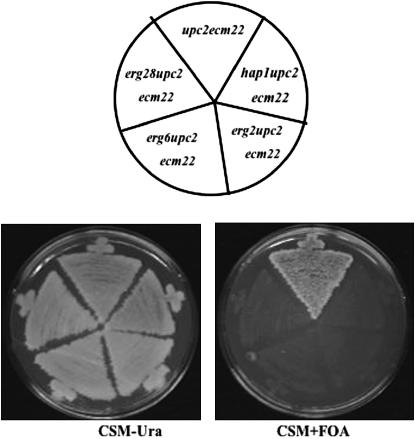
In contrast to other findings, Parks' group (Shianna et al. 2001) found that the double mutant upc2Δ ecm22Δ was inviable in their genetic background. The original upc2-1 allele (Lewis et al. 1988) was derived from strain S288C, in which a Ty1 element disrupts the 3′ region of the HAP1 ORF (Gaisne et al. 1999). The HAP1 gene encodes a heme-responsive transcription factor that regulates sterol biosynthesis (Turi and Loper 1992; Kennedy et al. 1999). In a upc2-1 gain-of-function mutant, HAP1 is upregulated and furthermore acts as a modifier of sterol uptake (Wilcox et al. 2002). Given the roles of Upc2p and Ecm22p in the regulation of sterol biosynthesis, we further investigated the cross-talk between the HAP1 and UPC2/ECM22 pathways by attempting to generate a triple mutant by crossing an ecm22Δ mutant to a upc2Δ hap1Ty strain. Whereas every possible double mutant segregant from the cross was recovered, all triple mutant segregants (hap1Ty upc2Δ ecm22Δ) died (50 tetrads dissected). However, triple hap1Ty upc2Δ ecm22Δ mutants containing a URA3-marked plasmid with a wild-type copy of UPC2 (pU6ΔSacI) were viable. upc2Δ ecm22Δ strains containing a deletion allele of HAP1 were also inviable; dissection of ∼39 tetrads also failed to yield a viable hap1Δ upc2Δ ecm22Δ strain. Thus both the hap1TY and hap1Δ alleles in combination with upc2Δ ecm22Δ were inviable. The inability of hap1Ty (or hap1Δ) upc2Δ ecm22Δ/pU6ΔSacI strains to grow on medium containing 5-FOA, which selects against the URA3 containing plasmid, further supports the inviability of the triple mutant combination (Figure 1).
Figure 1.—
The triple mutants hap1Δ upc2Δ ecm22Δ, erg2Δ upc2Δ ecm22Δ, erg6Δ upc2Δ ecm22Δ, and erg28Δ upc2Δ ecm22Δ contain a UPC2 rescue plasmid (pUΔSacI) and are able to grow on CSM-Ura medium but cannot grow on CSM + FOA, which negatively selects against plasmid retention. The upc2Δ ecm22Δ control strain grows on both media.
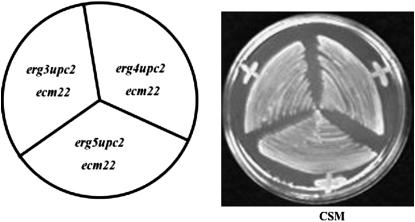
In previous studies, we and others have demonstrated that the combined deletion of UPC2, ECM22, and ERG2 was synthetically lethal (Vik and Rine 2001; Valachovic et al. 2004). However, this lethality could be suppressed by a mutation in ELO3, which is necessary for synthesis of the C26 fatty acids found in sphingolipids (Valachovic et al. 2004). In the current study, we investigated whether upc2Δ ecm22Δ was synthetically lethal in combination with other viable ergosterol deletion mutants, such as erg3Δ, erg4Δ, erg5Δ, erg6Δ, and erg28Δ. Our results demonstrate that ERG6 and ERG28 but not ERG3, ERG4, or ERG5 are essential genes in the absence of UPC2 and ECM22 (Figures 1 and 2). These results are summarized in Table 3, in which we demonstrate that viable triple mutants of hap1Δ (40 tetrads dissected), erg6Δ (33 tetrads dissected), and erg28Δ (38 tetrads dissected) with upc2Δ ecm22Δ were not observed. Triple mutants containing hap1Δ upc2Δ ecm22Δ, erg6Δ upc2Δ ecm22Δ, and erg28Δ upc2Δ ecm22Δ were, however, recovered from crosses containing the rescue plasmid (pU6ΔSacI) with a normal UPC2 allele. These strains grew on CSM-Ura but failed to grow on FOA medium as expected if the triple mutants are inviable (Figure 1). Finally, attempts to disrupt HAP1, ERG6, or ERG28 wild-type alleles in a upc2Δ ecm22Δ strain were unsuccessful, supporting the hypothesis that mutating these genes in the double deletion background mutant is a lethal event.
Figure 2.—
The triple mutants erg3Δ upc2Δ ecm22Δ, erg4Δ upc2Δ ecm22Δ, and erg5Δ upc2Δ ecm22Δ are all able to grow on CSM medium without a rescue plasmid.
TABLE 3.
Genetic analyses to determine viability of hap1Δ, erg2Δ, erg6Δ, and erg28Δ in a upc2Δ ecm22Δ mutant background
| Mating | No. of tetrads dissected | Viable triple mutantsa |
|---|---|---|
| erg3Δ upc2Δ × upc2Δ ecm22Δ | 23 | 19 |
| erg4Δ upc2Δ × upc2 Δ ecm22Δ | 24 | 24 |
| erg5Δ upc2Δ × upc2Δ ecm22Δ | 18 | 10 |
| erg6 Δ upc2Δ × upc2 Δ ecm22Δ | 33 | 0 |
| erg28Δ upc2Δ × upc2Δ ecm22Δ | 38 | 0 |
| hap1Δ × upc2Δ ecm22Δ | 40 | 0 |
Number of viable triple mutant isolates in a upc2ecm22 background.
Deletion of upc2 and ecm22 alters sterol profile:
An 11-bp SRE responsible for UPC2-mediated sterol regulation of ERG2 was found in eight other sterol biosynthetic genes in S. cerevisiae and a similar element was found in several ergosterol biosynthetic genes in C. albicans (Vik and Rine 2001; Silver et al. 2004). The upc2Δ ecm22Δ double mutant accumulates large amounts of two sterol intermediates, episterol and ergosta-5,7-dien-ol, at the expense of end-product sterol (Valachovic et al. 2004). The single deletion mutants upc2Δ and ecm22Δ had sterol profiles essentially like that of the wild type. Initially, we looked at erg3Δ, erg4Δ, and erg5Δ alone and in a upc2Δ ecm22Δ genetic background to determine exactly how the sterol profiles were altered. The principal sterol that accumulates in erg3Δ is ergosta-7,22,-dienol (70% of total sterol), but in a erg3Δ upc2Δ ecm22Δ mutant the principal sterol is episterol (41%) followed by fecosterol (13.9%) and ergosta-7-enol(13.3%), not a dramatic difference in sterol composition. Similar results were observed in an erg4Δ strain compared to the erg4Δ upc2Δ ecm22Δ triple mutant. The former accumulates almost exclusively ergosata-5,7,22,24(28) sterols but the triple mutant accumulates mostly episterol, two closely related erg4Δ-type sterols, ergosta-5,7,24(28), ergosta-5,7,22, 24(28), as well as zymosterol. Finally, the sterol profiles of erg5Δ and the erg5Δ upc2Δ ecm22Δ were essentially the same, accumulating principally ergosta-5,7 diene (74.6% and 67.6%), respectively. These results suggest that these triple mutants are likely viable because the sterols produced are not significantly different from those that occur in the erg3Δ, erg4Δ, and erg5Δ single mutants.
Isolation of suppressors of inviable upc2Δ ecm22Δ triple mutants:
Previously we isolated a UV-induced suppressor mutation of an erg2Δ upc2Δ ecm22Δ triple mutant. Genetic analysis of the suppressor showed that it was resistant to the morpholine tridemorph (Baloch et al. 1984), suggesting that the suppressor may be elo2 or elo3 (Valachovic et al. 2004). This suppressor was identified as elo3, which has been demonstrated to suppress lethal mutations of erg2 (Silve et al. 1996), although in the majority of genetic backgrounds erg2Δ mutants are not lethal (Ashman et al. 1991).
Similar attempts to identify viable suppressors of triple mutants of hap1, erg6, erg2, or erg28 in the upc2Δ ecm22Δ background were unsuccessful. Therefore transposon mutagenesis using hap1Δ upc2Δ ecm22Δ, erg2Δ upc2Δ ecm22Δ, erg6Δ upc2Δ ecm22Δ, and erg28Δ upc2Δ ecm22Δ strains containing the UPC2 rescue plasmid pU6ΔSacI were transformed with a mutagenized transposon library-mTn-lacZ/LEU2. Transformants were subsequently plated onto synthetic complete medium containing FOA to select loss of the pU6ΔSacI plasmid. Colonies purified on CSM-leu media that were able to grow on FOA medium were then considered as candidates for carrying a suppressor mutation. These colonies were then subjected to PCR analysis to ensure that a wild-type version of the UPC2 was not present. Approximately 40,000 transformants were obtained for triple mutants of erg6, 34,000 for hap1, 11,000 for erg2, and 1100 for erg28. Transposon screening by PCR allowed us to identify 116 different candidate suppressor genes. The largest category (14.7%) involved genes encoding transcription factors and the next largest class (10.3%) involved genes required for chromatin remodeling and chromatin structure. In total, 15 categories of genes were identified and the ones that were isolated multiple times are listed in Table 4.
TABLE 4.
Genes disrupted by mTn3 in multiple backgrounds
| M2-7A: erg2upc2ecm22 | M6-3B: erg6upc2ecm22 | MH1-3B: hap1upc2ecm22 | M28-2D: erg28upc2ecm22 | |
|---|---|---|---|---|
| YND1 | 1 | 1 | 1 | — |
| YER049w | 1 | 2 | 1 | — |
| HDA3 | — | 1 | 1 | — |
| RPN9 | — | 1 | 1 | — |
| IES1 | 1 | 2 | — | — |
| SSN2 | 1 | — | 1 | — |
| BCK2 | — | 2 | — | — |
| RDN25-1 | — | 3 | — | 3 |
| RDN37-1 | — | 3 | — | 3 |
| TAR1 | — | 3 | — | 1 |
| YNL011C | 1 | 4 | — | — |
| YNL010W | 1 | 4 | — | — |
M2-7A [erg2Δupc2Δecm22Δp(U,U)], M6-3B [ergΔupc2Δecm22Δp(U,U)], MH1-3B [hap1Δupc2Δecm22Δ(U,U)], and M28-2D [erg28Δupc2Δecm22Δp(U,U)] were all transformed with the mTn3 library resulting in several genes disruped in multiple backgrounds.
Putative transposon suppressors were confirmed by independently creating gene deletions:
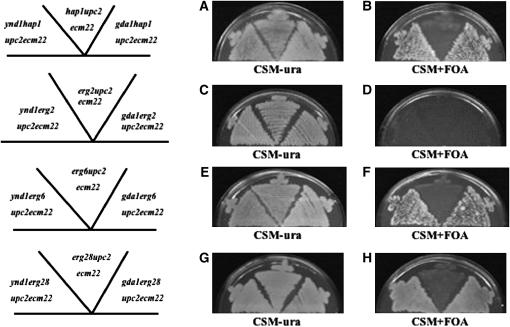
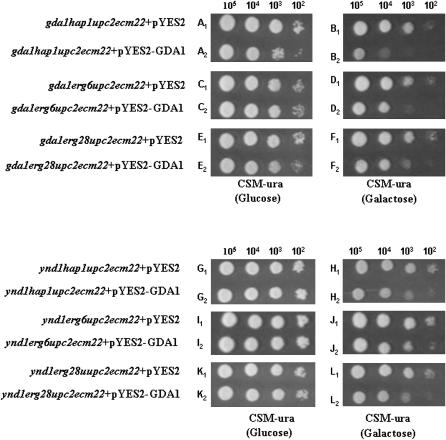
While the transposon mutagenesis screen indicated many putative suppressors, it was necessary to independently confirm these mutations by creating knockouts of putative genes and crossing these into our strains to generate viable quadruple mutants (e.g., mutxΔ hap1Δ upc2Δ ecm22Δ). We chose several categories of genes to independently disrupt: TUP1 and HDA3 are two genes involved in chromatin repression; transposon mutagenesis indicated that in the upc2Δ ecm22Δ background, tup1 rescued hap1 and hda3 suppressed both hap1 and erg6. A mutation in OSH1 encoding an oxysterol-binding protein suppressed erg6Δ and a mutation in UME6 (a transcriptional regulator) suppressed erg2Δ in upc2Δ ecm22Δ strains. Various crosses of these independently derived disrupted genes to the triple mutants containing a rescue plasmid indicated that tup1Δ was not a suppressor of hap1Δ; hda3Δ was not a suppressor of hap1Δ, erg2Δ, or erg6Δ; osh1Δ was not a suppressor of erg6Δ; and ume6Δ was not a suppressor of erg2Δ. However, ynd1Δ that was initially identified as a suppressor of hap1Δ, erg2Δ, and erg6Δ mutants by transposon mutagenesis was confirmed by independently derived disruptions to suppress hap1Δ, erg6Δ, and erg28Δ mutants but unexpectedly not the erg2Δ triple mutant (summarized in Table 5). In various independent crosses involving the generation of triple or quadruple mutants, segregants containing the pU6ΔSacI rescue plasmid were plated onto FOA medium. For hap1Δ, erg6Δ, and erg28Δ mutants that also contained the ynd1Δ allele in the upc2Δ ecm22Δ background, viability (suppression) was indicated as the ability to lose the rescue plasmid. However, in two distinct matings in which ynd1Δ erg2Δ upc2Δ ecm22Δ quadruple mutants were generated, no quadruple mutant strain was able to lose the rescue plasmid. YND1 encodes an apyrase (nucleoside diphosphatase and nucleoside triphosphatase activity) and has 20% amino acid identity to GDA1, which also encodes a nucleoside diphosphatase. GDA1 is required for transporting GDP-mannose into the Golgi lumen (Gao et al. 1999). GDA1 mutants are defective in mannosylation of both proteins and sphingolipids (Yanagisawa et al. 1990; Ashman et al. 1991; Abeijon et al. 1993). The ynd1Δ gda1Δ double mutant grows slowly and demonstrates defects in cell morphology and protein glycosylation. An independently derived gda1Δ mutant strain was similarly mated to all four triple mutants and similar results were obtained. The gda1Δ mutation suppressed hap1Δ, erg6Δ, and erg28Δ triple mutants but not the erg2Δ upc2Δ ecm22Δ triple mutant (Table 6). Figure 3 demonstrates that both ynd1 and gda1 deletions suppress lethality of hap1Δ, erg6Δ, and erg28Δ triple mutants but not the erg2Δ triple mutant. To determine whether GDA1 expression would reverse the suppression of the gda1Δ-suppressed quadruple mutant, the GDA1 open reading frame was introduced into the pYES2 expression vector that contains the galactose-inducible promoter GAL1. On media containing glucose both empty vector control (pYES2) and pYES2-GDA1 did not adversely affect growth of the hap1Δ, erg6Δ, and erg28Δ mutants in gda1Δ upc2Δ ecm22Δ strains. However, on galactose-containing media in which expression of the GDA1 is induced, the quadruple mutants grew worse than when GDA1 was not expressed (Figure 4, compare B1 and B2, D1 and D2, and F1 and F2) indicating that GDA1 expression did reverse suppression. Minimal GDA1 expression under noninducible conditions (glucose medium) showed similar growth in the absence or presence of wild-type GDA1 (Figure 4, compare A1 and A2, C1 and C2, and E1 and E2). Unexpectedly, the inducible expression of GDA1 was also able to prevent growth of ynd1 hap1Δ (or erg6Δ or erg28Δ) upc2Δ ecm22Δ strains. These results suggest that overexpression of GDA1 can reverse the suppression due to a mutation of the related gene YND1, suggesting lesions in both GDA1 and YND1 genes are suppressing through a similar mechanism.
TABLE 5.
Synthetically lethal backgrounds in which ynd1Δ was tested as a suppressor
| Mating | Tetrads dissecteda | Viable quadruplesb | Inviable quadruplesc | Viable triplesd |
|---|---|---|---|---|
| hap1upc2ecm22/p × ynd1upc2ecm22/pe | 41 | 44 | 0 | 43 |
| erg2upc2ecm22/p × ynd1erg2upc2ecm22/p | 18 | 0 | 9 | 16 |
| ynd1erg2upc2ecm22/p × ynd1upc2ecm22/p | 46 | 0 | 65 | 20 |
| erg6upc2ecm22/p × ynd1upc2ecm22/p | 54 | 28 | 0 | 15 |
| erg28upc2ecm22/p × ynd1upc2ecm22/p | 50 | 30 | 0 | 54 |
Matings were made to produce quadruple mutants composed of a synthetically lethal triple mutant background plus ynd1. All mutations are deletions.
Number of tetrads dissected to produce the quadruple mutants.
Number of quadruple mutants where ynd1 suppressed lethality as indicated by the ability to lose the accompanying rescue containing UPC2 plasmid on FOA medium.
Number of quadruple mutants in which ynd1 did not suppress lethality as indicated by the inability to grow on FOA medium.
Number of synthetically lethal triple mutants produced by the cross as a control and not plated on FOA medium.
URA3-containing rescue plasmid carrying UPC2 allele.
TABLE 6.
Synthetically lethal backgrounds in which gda1Δ was tested as a suppressor
| Mating | Tetrads dissecteda | Viable quadruplesb | Inviable quadruplesc | Viable triplesd |
|---|---|---|---|---|
| hap1upc2ecm22/p × gda1upc2ecm22/pe | 35 | 30 | 0 | 17 |
| erg2upc2ecm22/p × gda1upc2ecm22/p | 45 | 0 | 24 | 20 |
| erg6upc2ecm22/p × gda1upc2ecm22/p | 37 | 30 | 0 | 16 |
| erg28upc2ecm22/p × gda1upc2ecm22/p | 39 | 34 | 0 | 30 |
Matings were made to produce quadruple mutants composed of a synthetically lethal triple mutant background plus gda1. All mutations are deletions.
Number of tetrads dissected to produce the quadruple mutants.
Number of quadruple mutants where gda1 suppressed lethality as indicated by the ability to lose the accompanying rescue containing UPC2 plasmid on FOA medium.
Number of quadruple mutants in which gda1 did not suppress lethality as indicated by the inability to grow on FOA medium.
Number of synthetically lethal triple mutants produced by the cross as a control and not plated on FOA medium.
URA3-containing rescue plasmid carrying UPC2 allele.
Figure 3.—
(A and B) Suppression of hap1Δ upc2Δ ecm22Δ by ynd1 and gda1 mutations. The quadruple mutants ynd1hapΔ upc2Δ ecm22Δ and gda1hapΔ upc2Δ ecm22Δ will grow on CSM-Ura or CSM + FOA medium, which selects against plasmid retention of the rescue plasmid, pUΔSacI. Only the triple mutant hap1Δ upc2Δ ecm22Δ cannot grow without the rescue plasmid on CSM + FOA medium. (C and D) The erg2Δ upc2Δ ecm22Δ triple mutant is not suppressed by ynd1 and gda1 as loss of plasmid results in inviability. (E and F) Suppression of erg6Δ upc2Δ ecm22Δ by ynd1 and gda1 mutations. The quadruple mutants ynd1erg6Δ upc2Δ ecm22Δ and gda1erg6Δ upc2Δ ecm22Δ will grow on CSM-Ura or CSM + FOA medium, which selects against plasmid retention of a rescue plasmid. erg6Δ upc2Δ ecm22Δ cannot grow without the rescue plasmid on CSM + FOA medium. (G and H) Suppression of erg28Δ upc2Δ ecm22Δ by ynd1 and gda1 mutations. The quadruple mutants ynd1erg28Δ upc2Δ ecm22Δ and gda1erg28Δ upc2Δ ecm22Δ will grow on CSM-Ura or CSM + FOA medium. erg28Δ upc2Δ ecm22Δ cannot grow without the rescue plasmid on CSM + FOA medium.
Figure 4.—
Overexpression of a plasmid containing the GDA1 ORF under the control of a galactose-inducible GAL1 promoter. Expression of GDA1 reverses suppression due to gda1 and ynd1 mutations. Expression of GDA1 in a gal1 hap1Δ upc2Δ ecm22Δ background reduces viability on galactose medium more than on glucose medium (B2 vs. A2) as expected. Similar growth was observed with vector controls (A1 and B1). However, GDA1 expression on galactose medium also reduced suppression of the ynd1hap1Δ upc2Δ ecm22Δ quadruple mutant (H2 vs. H1 but not on glucose medium (G1 and G2). Similar results were observed for suppression of the gda1erg6Δ upc2Δ ecm22Δ, ynd1erg6Δ upc2Δ ecm22Δ, gda1erg28Δ upc2Δ ecm22Δ, and ynd1erg2Δ upc2Δ ecm22Δ strains.
Sterol analyses of the ynd1- and gda1-suppressed strains:
We were interested in determining what the sterol profiles of the suppressed quadruple mutants might look like since these strains might be regarded as having a sterol composition that was minimally adequate for viability. The sterol profiles of the hap1Δ, erg6Δ, erg28Δ single mutants, the upc2Δ ecm22Δ double mutant, and the ynd1Δ- and gda1Δ-suppressed quadruple mutants are given in Tables 7–9. The hap1 sterol profile is essentially that of wild type (as are the single ynd1 and gda1 mutants). The upc2Δ ecm22Δ sterol profile indicated a decrease in ergosterol and increase in sterol intermediates. ynd1Δ hap1 upc2Δ ecm22Δ and gda1Δ hap1upc2Δ ecm22Δ are capable of synthesizing ergosterol at very low levels (3.5 and 3.1%, respectively) and also have nearly identical sterol profiles in which the predominant sterol is zymosterol, similar to that of the erg6Δ single mutant. gda1Δ erg6Δ upc2Δ ecm22Δ and ynd1Δ erg6Δ upc2Δ ecm22Δ are also nearly identical (Table 8). The differences between erg28Δ strains in the suppressed state are also substantial, with the quadruple mutants synthesizing much less ergosterol than an erg28 strain alone (Table 9). Quadruple mutants lacking either YND1 or GDA1 synthesize more ergosta-5,7,24(28) sterols and substantially more 4-methylzymosterol and 4,4-dimethylzymosterol than the erg28 single mutant. It is unlikely that these latter two would function as membrane sterols due to the C-4 methyl groups.
TABLE 7.
Sterol analysis of ynd1hap1upc2ecm22 and gda1hap1upc2ecm22
| Sterols | hap1 | upc2ecm22 | ynd1hap1upc2ecm22 | gda1hap1upc2ecm22 |
|---|---|---|---|---|
| Squalene | 1.4 | 5.3 | 21.4 | 16.4 |
| Zymosterol | 5.6 | 11.5 | 6.8 | 1.7 |
| Ergosterol | 69.4 | 10.8 | 3.5 | 3.1 |
| Ergosta-7,22-dienol | 0 | 1.5 | 0.6 | 2.9 |
| 4-methyl zymosterol and fecosterol | 4.2 | 10.7 | 12.3 | 11.0 |
| Ergosta-5,7,24(28) trien-3ol and ergosta-5,7-dienol | 0 | 0 | 6.7 | 6.9 |
| Episterol | 9.7 | 21.9 | 18.7 | 31.5 |
| Ergosta 5,7-dienol | 5.5 | 24.6 | 0 | 0 |
| Ergosta 7-enol | 0 | 0.8 | 7.8 | 5.8 |
| 4-methyl fecosterol | 0.5 | 0 | 3.0 | 2.4 |
| Lanosterol | 1.0 | 3.9 | 1.2 | 2.8 |
| 4,4-dimethyl zymosterol | 2.6 | 9.1 | 17.9 | 15.4 |
All mutations are deletions. Values indicate sterol as percentage of total sterol. Sterol analyses of these strains represent the average of three independent experiments.
TABLE 9.
Sterol analysis of ynd1erg28upc2ecm22 and gda1erg28upc2ecm22
| Sterols | erg28 | ynd1erg28upc2ecm22 | gda1erg28upc2ecm22 |
|---|---|---|---|
| Squalene | 4.1 | 9.2 | 13.6 |
| Zymosterol | 3.0 | 0.9 | 0.8 |
| Ergosterol | 77.4 | 5.6 | 4.9 |
| Ergosta 7,22-dienol | 0.7 | 0.9 | 0.3 |
| 4-methyl zymosterol | 4.0 | 25.5 | 14.9 |
| Ergosta 5,7,24(28) trien-3-ol and 4-methyl cholesta-trienol | 0 | 13.9 | 22.1 |
| Episterol | 3.3 | 9.9 | 13.1 |
| 4-methyl fecosterol | 0 | 1.1 | 1.2 |
| 4-methyl cholesta-8,24-dien-3-one | 0.6 | 0 | 0 |
| 4-methyl cholestasterolsa | 5.1 | 6.2 | 8.5 |
| Lanosterol | 1.3 | 5.1 | 7.1 |
| 4,4-dimethyl zymosterol | 0.5 | 21.6 | 13.5 |
Sterol analyses of these strains represent the average of three independent experiments.
These 4-methylcholestasterols could not be unambiguously identified.
TABLE 8.
Sterol analysis of ynd1erg6upc2ecm22 and gda1erg6upc2ecm22
| Sterol | erg6 | ynd1erg6upc2ecm22 | gda1erg6upc2ecm22 |
|---|---|---|---|
| Squalene | 0.3 | 8.7 | 17.1 |
| Cholesta-5,8,24-trienol | 2.1 | 0 | 0 |
| Zymosterol | 46.7 | 53.2 | 34.9 |
| Cholesta-5,7,24-trienol | 24.9 | 16.4 | 18.2 |
| Cholesta-7,24-dienol | 3.6 | 7.9 | 10.9 |
| Cholesta-5,7,22,24-tetraenol | 20.8 | 0.4 | 0.9 |
| 4-methylzymosterol | 0.8 | 3.5 | 4.7 |
| Lanosterol | 0.2 | 3.8 | 5.3 |
| 4,4-dimethyl-zymosterol | 0.8 | 6.2 | 8.0 |
Sterol analyses of these strains represent the average of three independent experiments.
Altered sphingolipid composition in a ynd1 mutant:
Since Ynd1p appears to be related to the guanosine diphosphatase encoded by GDA1 and since a gda1 mutation alters the sphingolipid composition of cells (Abeijon et al. 1993), we examined the sphingolipid composition of a ynd1 mutant. Sphingolipids were radiolabeled to equilibrium with [3H]myo-inositol, extracted from cells, and analyzed by thin-layer chromatography. Deletion of gda1 caused a twofold reduction in the level of the major sphingolipid species, mannose-(inositol-P)2-ceramide [M(IP)2C] (Table 10). In contrast, deletion of ynd1p did not change the level of M(IP)2C (Table 10); however, it did cause the mannose-inositol-P-ceramide (MIPC) and the inositol-P-ceramides to increase ∼50%, similar to what was observed in the gda1 mutant (Table 10). We conclude from these results that Ynd1 and Gda1 play roles in sphingolipid synthesis and their loss of function leads to changes in the species of sphingolipids produced in cells.
TABLE 10.
Changes in the sphingolipid composition of gda1Δ and ynd1Δ cells
| [3H]myo-inositol phospholipids | Wild-type SPH/GPI | gda1Δ SPH/GPI | gda1Δ/wild type | ynd1Δ SPH/GPI | ynd1Δ/wild type |
|---|---|---|---|---|---|
| GPI | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| M(IP)2C | 1.89 (2.10, 1.68) | 1.01 (1.07, 0.96) | 0.53 ↓ | 1.89 (2.01, 1.77) | 1.00 |
| MIPC + IPC | 1.34 (1.25, 1.43) | 2.01 (1.88, 2.14) | 1.50 ↑ | 1.96 (1.68, 2.24) | 1.46 ↑ |
Ratios are the average of two experiments and were determined by dividing the counts per minute in each radioactive lipid peak by the counts per minute in the GPI peak. Values in parentheses represent data from two separate experiments. SPH, sphingolipid; GPI, glycerolphospho-inositol; IPC, inositolphosphorylceramide; M(IP)2C, mannosyldiphosphorylinositolceramide; MIPC, mannosylphosphorylinositolceramide.
Loss of Ynd1p and Gda1p activity normalizes cold detergent-insoluble proteins:
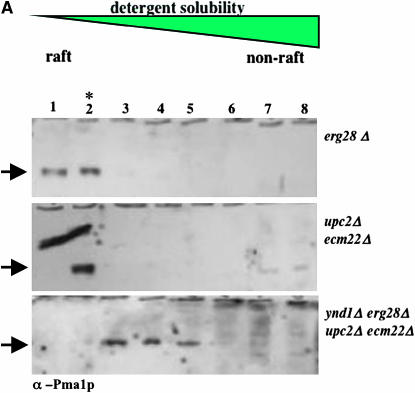
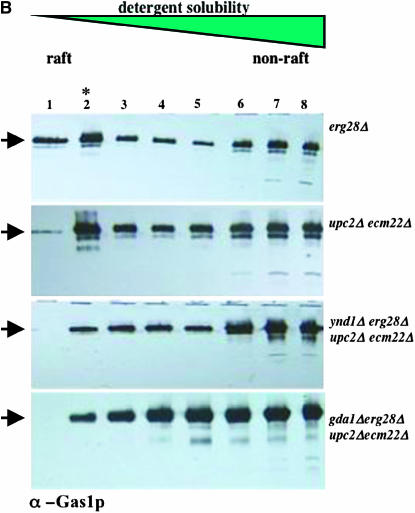
The ratio of various lipids, particularly sterols and glycosphingolipids, in the plasma membrane and other organellar membranes is directly responsible for the sequestration or complexing of certain proteins into membrane microdomains known as rafts. These are experimentally defined by their degree of solubility and thus extraction by cold detergents, such as 1% Triton X-100, and their flotation properties following density gradient fractionation. In mutants deficient in sphingolipid biosynthesis due to a temperature-sensitive allele of the LCB1 gene allele, raft formation is absent by this assay at the temperature nonpermissive for growth (Bagnat et al. 2000). We reasoned that the combined loss of the sterol biosynthetic genes and their transcription factors was a lethal event due to aberrant lipid biosynthesis and that this would produce a membrane incompatible with raft localization and viability. Deletion of YND1 or GDA1 could compensate for this situation by modulation of sphingolipid composition, and thus raft association, to produce a viable cell. We therefore assessed the localization of two known raft-associated proteins, the PMA1 and GAS1 gene products, to detergent-insoluble microdomains (rafts) using established protocols (Figure 5, A and B). Cell lysates were treated with cold Triton X-100 followed by density gradient centrifugation (Bagnat et al. 2000) and resolution by SDS–PAGE and immunoblot. In extracts from erg28Δ or upc2Δ ecm22Δ double mutants (Figure 5, A and B) and normal cells (not shown) both Pma1p and Gas1p localize to detergent-insoluble fractions 1 and 2 (peaking in fraction 2). By contrast, the same proteins partially relocalize to non-raft domains (i.e., detergent soluble), such that the proportion of these proteins in rafts is markedly diminished in the viable ynd1Δ erg28Δ upc2Δ ecm22Δ or gda1Δ erg28Δ upc2Δ ecm22Δ strains. We assume that the inviable triple mutants lack raft-associated proteins.
Figure 5.—
Cold detergent-insoluble protein localization. Protein extracts were prepared from strains of the indicted genotypes and resolved by density gradient fractionation after solubilization (or otherwise) with cold detergent. Eight fractions were collected, resolved by SDS–PAGE, and visualized following immunoblotting with antisera against the PMA1 or GAS1 proteins (A and B, respectively). Fraction 2 (indicated by the asterisk) represents the peak of detergent-insoluble protein characteristic of membrane microdomains. Uniquely in the quadruple erg28Δ upc2Δ ecm22Δ gda1Δ and erg28Δ upc2Δ ecm22Δynd1Δ strains, detergent-insoluble proteins are underrepresented relative to the other mutants and normal strains. Arrows indicate fraction localization of Pma1p (A) and Gsa1p (B).
DISCUSSION
In this article we describe the genetic interactions of two transcription factors (UPC2 and ECM22) that affect sterol biosynthesis and demonstrate that in conjunction with mutations in other ergosterol biosynthetic genes or transcription factors this can lead to inviability. We further demonstrate that such mutants can be suppressed by additional mutations in the sphingolipid biosynthetic pathway. The upc2Δ ecm22Δ double mutant strain was initially determined to be viable in one genetic background but not in another. The HAP1 gene in strain S228C has a Ty insertion at the C terminus resulting in a hap1 mutant phenotype, as indicated by loss of cytochrome c expression (Gaisne et al. 1999). However, the hap1Ty allele still allows for growth under heme-limiting conditions. In this investigation, we demonstrate that hap1Ty upc2Δ ecm22Δ or hap1Δ upc2Δ ecm22Δ represent synthetically lethal combinations and that the presence of the hap1Ty allele in the S288C background was likely responsible for the diverse finding that upc2Δ ecm22Δ was viable in one genetic background but not in another. Hap1p is also a Zn(2)–Cys(6) fungal transcription factor known to affect the sterol biosynthetic pathway, in particular the ERG11 (Turi and Loper 1992)and ERG9 genes (Kennedy et al. 1999) and the sterol esterification gene ARE2 (Jensen-Pergakes et al. 2001). In addition, levels of HMG1, encoding the major isoform of HMG-CoA reductase, are decreased more than twofold in a hap1Δ mutant as determined by microarray (Ter Linde and Steensma 2002). However, HAP1 may affect the sterol pathway indirectly as it also is required for repression by ROX1, a repressor of aerobic gene expression. A number of sterol biosynthetic genes are also known to be regulated by ROX1 (Zitomer and Lowry 1992), among them ERG11, NADPH cytochrome P450 reductase, and HMG2 (Ter Linde and Steensma 2002). Thus it is not surprising that loss of HAP1 in conjunction with deletion of UPC2 ECM22 would be lethal.
Mutations in UPC2 and ECM22 have a broad effect on sterol biosynthesis. When sterol levels are reduced due to the addition of Lovastatin, the expression of ERG2 and ERG3 substantially increase. Both gene products bind to the same regulatory elements in the ERG2 and ERG3 promoters (Vik and Rine 2001). A model has been presented in which the activation domains of Upc2p and Ecm22p are targeted by a repressor and in response to sterol depletion binding of the repressor is reversed (Davies et al. 2005). Several other ergosterol biosynthetic genes may be regulated by these transcription factors such as ERG1, ERG6, ERG8, ERG11, ERG12, ERG13, and ERG25, since all contain a consensus 11-bp sterol regulatory element in their respective promoters (Vik and Rine 2001). The UPC2 deletion in C. albicans renders cells highly susceptible to antifungals that target the sterol pathway, such as ketoconazole and fluconazole (MacPherson et al. 2005). Thus UPC2 and ECM22 appear to be global regulators of sterol biosynthesis in fungi. While not directly relevant to this investigation, a gain-of-function semidominant allele of UPC2, designated upc2-1, allows for aerobic sterol uptake (Lewis et al. 1988).
Deletion of UPC2 and ECM22 is synthetically lethal with ergosterol biosynthetic mutants:
We were curious to determine which other viable ergosterol mutations resulted in synthetic lethality in upc2Δ ecm22Δ strains. Our results suggested that erg6Δ and erg28Δ were also synthetically lethal with upc2Δ ecm22Δ but that erg3, erg4, and erg5 were not. Mutations in ERG2 and ERG6 are the most deleterious of the viable ergosterol deletions (Bard et al. 1978). erg6 mutants have altered permeability characteristics, often are unable to utilize respiratory energy sources, have reduced mating capability, and are unable to transport tryptophan (Gaber et al. 1989). Due to their inability to methylate the side chain, the sterols accumulated have a diminished mobility. Mutations in ERG2 and ERG28 also compromise growth (Ashman et al. 1991; Gachotte et al. 2001) as do mutations in ERG3. However, erg4 and erg5 mutants grow nearly as well as wild type and accumulate sterols that differ from ergosterol only in the inability to reduce double bonds in the sterol side chain (Lees et al. 1999). Both erg4 and erg5 strains contain the conjugated double bonds in the sterol B ring, as do wild-type cells. These mutants have almost wild-type sensitivities to the antifungal nystatin (Molzahn and Woods 1972). While it was expected that erg4Δ upc2Δ ecm22Δ and erg5Δ upc2Δ ecm22Δ strains would be viable, the viability of erg3Δ upc2Δ ecm22Δ was less obvious because the ERG3 lesion precludes the addition of a double bond at the C-5 position in the sterol B ring.
A clue to the effect on viability of the erg2Δ upc2Δ ecm22Δ triple mutant was obtained when a suppressor of this strain was found to contain the elo3 mutation. Virtually all erg2 mutant strains in S. cerevisiae are viable, with the single exception of an erg2 strain (Silve et al. 1996) that could be suppressed by a mutation in the sphingolipid genes ELO2 or ELO3, which encode fatty acid elongases that are necessary for synthesis of C26- fatty acyl chain found in yeast sphingolipids (Oh et al. 1997). We isolated a suppressor of erg2Δ upc2Δ ecm22Δ, which was subsequently identified as elo3 (Valachovic et al. 2004). Thus an erg2Δ upc2Δ ecm22Δ not only would have reduced sterol levels as a result of the transcription factor mutations but also would have an altered sterol profile due to loss of sterol C-8 isomerase activity. The restoration of viability in erg2 elo3 upc2Δ ecm22Δ suggests that elo3Δ suppression of erg2Δ essentially converts the quadruple mutant strain to a upc2Δ ecm22Δ double mutant strain. For reasons not yet clear the sphingolipid mutation suppressing the sterol mutation may result in an alteration of lipid raft function, thereby restoring viability. We also speculate that an inviable erg2 isolate may accumulate a lower level of sterol than is found in most erg2 genetic backgrounds. Finally, the sterol defect in a upc2Δ ecm22Δ double mutant results in an accumulation of episterol (45%). This sterol could not accumulate in an erg2 upc2Δ ecm22Δ background because of the erg2 mutation.
Transposon mutagenesis and suppression due to ynd1 and gda1 mutations:
Of 116 total transformants containing suppressors involving all four triple mutant strains (upc2Δ ecm22Δ with hap1Δ, erg2Δ, erg6Δ, or erg28Δ), 25% of the mutations implicated genes involved in transcription or in chromatin remodeling. Other categories of suppressor mutations were in genes involved in endocytosis (7%), ubiquitination (6%), and ∼5% each involved genes in phosphate metabolism, the cytoskeleton, DNA repair, metabolism, mitochondria, or RNA metabolism. Upon designing disruptions of several of these genes to independently confirm suppression we found that most did not confer suppression when mated to the triple mutants. It is likely that mutations in genes involved in repression mechanisms, such as TUP1, HDA2, HDA3, IES1, and SSN2, enhanced survival of the triple mutants long enough for other suppressors to accumulate during the transformation process and during growth on FOA medium. For this reason we confined ourselves to mutants that indicated suppression by independent disruptions and matings.
The observation that the ynd1 mutation (from the transposon library) suppressed hap1Δ, erg2Δ, and erg6Δ mutants suggested a common mechanism in the context of upc2Δ ecm22Δ. We suggest that the upc2Δ ecm22Δ double mutant reduces sterol biosynthesis to a minimal level that is still compatible with viability but introduction of ergosterol mutations into this background lowers the threshold of functional sterol below this threshold. Thus hap1Δ upc2Δ ecm22Δ strains would be inviable because HAP1 is itself a transcription factor affecting sterol biosynthesis and its loss would further diminish expression of the pathway. We subsequently demonstrated by genetic crosses that ynd1Δ suppresses the erg6Δ, erg28Δ, and hap1Δ triple mutants but not the erg2Δ triple mutant.
Not finding that ynd1 suppressed erg28Δ upc2Δ ecm22Δ was likely due to the fact that only 1095 transposon-generated transformants were screened, whereas for the other screens the total number of transformants screened was significantly higher. YND1 is a protein of 630 amino acids that functions as an apyrase, enzymes that hydrolyze both di- and triphosphate nucleotides. GDA1 is a homolog of YND1 and shares 20% amino acid identity. Both ynd1Δ and gda1Δ single mutants have decreased N-linked glycosylation and O-linked glycosylation (Gao et al. 1999). The ynd1Δ gda1Δ double mutant results in slow growth. Gda1p is involved in the import of mannose into the Golgi (Abeijon et al. 1993) and fuels the Golgi antiporter Vrg4p by cleaving GDP into GMP, thus facilitating import of GDP mannose into the Golgi as GMP is simultaneously exported (Gao and Dean 2000). Once in the Golgi, GDP-mannose becomes a substrate for various reactions involving sphingolipid synthesis and cell wall remodeling. Abeijon et al. (1993) demonstrated that elongation of O-linked carbohydrate chains is blocked at the mannobiose step, N-linked carbohydrates of carboxypeptidase are not elongated beyond the phosphatidyl inositol stage, and biosynthesis of MIPC is severely impaired in gda1 mutants. Even though we did not obtain gda1 suppressors in our transposon screening, our results indicate that a gda1-deleted strain mated to give the four different types of quadruple mutants showed the exact same profile of suppression as did ynd1. This investigation implicates for the first time a role of Ynd1p, like Gda1p, in sphingolipid biosynthesis.
Lipid rafts are mixtures of sterols and sphingolipids that form separate domains in the plasma membrane. These rafts possess specific proteins and are the targets for signaling, channels, and other functions (Bagnat et al. 2000; Brown and London 2000). In this investigation we extend what is known regarding synthetic lethal and suppressor interactions that occur between the sterol and sphingolipid pathways. Inviable isolates of erg2 and erg24 are suppressed by both elo2 and elo3 (Silve et al. 1996; Baudry et al. 2001). ARV1, which harbors defects in trafficking of sterols, accumulates novel sterol intermediates, and synthesizes higher levels of sterol esters, also shows defects in sphingolipids (Tinkelenberg et al. 2000; Swain et al. 2002). A temperature-sensitive mutant of ERG26 also had sphingolipid defects (Baudry et al. 2001), and finally elo3 is synthetically lethal with erg6 (Eisenkolb et al. 2002). One model of lipid raft organization is based on the relative sizes of the polar and nonpolar regions of sterols and sphingolipids (Fantini et al. 2002). In this model, both molecules have a cylindrical or pyramidal shape. Sterols have a small polar head region relative to their nonpolar region, and sphingolipids have a large polar region relative to their nonpolar region. In lipid rafts, these two molecules interdigitate via their complementing polar and nonpolar regions. A hypothesis for the synthetic lethality in this study is that nonviability is a result of sterol intermediates fitting poorly in lipid rafts in combination with an overall lack of bulk sterols. We hypothesize that sterol intermediates produced by the erg2, erg6, erg28, and hap1 mutations have alterations such that their polar regions, nonpolar regions, or both poorly complement the wild-type sphingolipid polar and nonpolar regions. In the case of single erg mutations there is impairment of membrane function, but not enough to prevent viability. The upc2Δ ecm22Δ double mutation causes a decrease of sterol synthesis, but this does not prevent viability. However, when these two phenomena are combined, as in ergX upc2Δ ecm22Δ, they produce synthetic lethality.
If synthetic lethality is caused in part by poor complementation of the sterol intermediates with wild-type sphingolipids, suppression of lethality by mutating YND1 or GDA1 may be due to alterations in sphingolipids that allow better interactions with sterol intermediates. Of the known suppressors, ynd1 and gda1 mutants cause an alteration in the polar head group of sphingolipids, while the elo3 mutants cause an alteration in the nonpolar region. The commonality we found in ynd1 and gda1 mutants was an increase in sphingolipids with a smaller polar head group (MIPC + IPC, Table 10) compared to M(IP2)C, which has the largest polar head group. Having a smaller polar head group would make the sphingolipid more cylindrical and less conical, which would fit better with a sterol that was less cylindrical and more conical. The quadruple mutant strains in which ynd1 and gda1suppress inviability have an increase in sterol intermediates that give a more cylindrical and less conical structure. Our results confirm a previous demonstration that gda1 mutants have reduced levels of M(IP)2C (Abeijon et al. 1993) but differ in that we also see an increase rather than a decrease in MIPC. The difference may be due to the way in which sphingolipids were radiolabeled. They labeled cells for 10 min with [3H]myo-inositol and chased with nonradioactive inositol for 90 min and thus measured the rate of sphingolipid synthesis. We labeled for >10 cell divisions and thus measured the relative abundance of sphingolipids.
In summary, strains suppressed by ynd1 and gda1 are likely to have sphingolipids that are narrower near the apex, which can complement sterols that are wider near the apex and narrower at the base. To test this hypothesis, we investigated the localization of two known raft-associated proteins, Pma1p and Gas1p, in the various viable mutant combinations. Consistent with our hypothesis, we observed a unique redistribution of these raft-associated proteins in the suppressed strains that lacked the ERG28 gene product. This redistribution was not complete; a minor proportion of the proteins remain raft associated, presumably a context that is sufficient for viability. These results confirm the altered nature of membrane microdomains due to loss of GDA1 or YND1 in the context of disturbed sterol composition due to the erg28Δ upc2Δ ecm22Δ mutations. Interestingly, we did not observe the same relocalization of Pma1p or Gas1p due to loss of ERG6, suggesting alternate modes of suppression for these pathways.
Acknowledgments
This work was supported by National Institutes of Health grants GM-62104 (M.B.), GM-41302 (R.C.D.), and DK-54320 (S.L.S). S.L.S. also acknowledges the American Heart Association (National and Heritage) and the Ara Parseghian Medical Research Foundation.
References
- Abeijon, C., K. Yanagisawa, E. C. Mandon, A. Hausler, K. Moremen et al., 1993. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J. Cell Biol. 122: 307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramova, N., O. Sertil, S. Mehta and C. V. Lowry, 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183: 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman, W. H., R. J. Barbuch, C. E. Ulbright, H. W. Jarrett and M. Bard, 1991. Cloning and disruption of the yeast C-8 sterol isomerase gene. Lipids 26: 628–632. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1998. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bagnat, M., S. Keranen, A. Shevchenko, A. Shevchenko and K. Simons, 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97: 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloch, R. I., E. I. Mercer, T. E. Wiggins and B. C. Baldwin, 1984. Inhibition of ergosterol biosynthesis in Saccharomyces cerevisiae and Ustilago maydis by tridemorph, fenpropiomorph, and fenpropidin. Phytochemistry 23: 2219–2226. [Google Scholar]
- Bard, M., N. D. Lees, L. S. Burrows and F. W. Kleinhans, 1978. Differences in crystal violet uptake and cation-induced death among yeast sterol mutants. J. Bacteriol. 135: 1146–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, K., E. Swain, A. Rahier, M. Germann, A. Batta et al., 2001. The effect of the erg26–1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 276: 12702–12711. [DOI] [PubMed] [Google Scholar]
- Brown, D. A., and E. London, 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275: 17221–17224. [DOI] [PubMed] [Google Scholar]
- Brown, M. S., and J. L. Goldstein, 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89: 331–340. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics, 2000 Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove and L. W. Parks, 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180: 4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., N. D. Lees, M. Bard and R. Dickson, 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14: 1471–1510. [DOI] [PubMed] [Google Scholar]
- Davies, B. S., H. S. Wang and J. Rine, 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25: 7375–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, R. C., E. E. Nagiec, G. B. Wells, M. M. Nagiec and R. L. Lester, 1997. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J. Biol. Chem. 272: 29620–29625. [DOI] [PubMed] [Google Scholar]
- Eisenkolb, M., C. Zenzmaier, E. Leitner and R. Schneiter, 2002. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol. Biol. Cell 13: 4414–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz, N., U. H. Mortensen and R. Rothstein, 1997. Cloning-free PCR-based allele replacement methods. Genome Res. 7: 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini, J., N. Garmy, R. Mahfoud and N. Yahi, 2002. Lipid rafts: structure, function and role in HIV, Alzheimers and prion diseases. Expert Rev. Mol. Med. 2002: 1–22. [DOI] [PubMed] [Google Scholar]
- Gaber, R. F., D. M. Copple, B. K. Kennedy, M. Vidal and M. Bard, 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9: 3447–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachotte, D., S. E. Sen, J. Eckstein, R. Barbuch, M. Krieger et al., 1999. Characterization of the Saccharomyces cerevisiae ERG27 gene encoding the 3-keto reductase involved in C-4 sterol demethylation. Proc. Natl. Acad. Sci. USA 96: 12655–12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachotte, D., J. Eckstein, R. Barbuch, T. Hughes, C. Roberts et al., 2001. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 42: 150–154. [PubMed] [Google Scholar]
- Gaisne, M., A. M. Becam, J. Verdiere and C. J. Herbert, 1999. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36: 195–200. [DOI] [PubMed] [Google Scholar]
- Gao, X. D., and N. Dean, 2000. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 275: 17718–17727. [DOI] [PubMed] [Google Scholar]
- Gao, X. D., V. Kaigorodov and Y. Jigami, 1999. YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J. Biol. Chem. 274: 21450–21456. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., B. L. Todd and P. J. Espenshade, 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120: 831–842. [DOI] [PubMed] [Google Scholar]
- Jensen-Pergakes, K., Z. Guo, M. Giattina, S. L. Sturley and M. Bard, 2001. Transcriptional regulation of the two sterol esterification genes in the yeast Saccharomyces cerevisiae. J. Bacteriol. 183: 4950–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, R. I., and G. E. Herman, 2001. Inborn errors of sterol biosynthesis. Annu. Rev. Genomics Hum. Genet. 2: 299–341. [DOI] [PubMed] [Google Scholar]
- Kennedy, M. A., R. Barbuch and M. Bard, 1999. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1445: 110–122. [DOI] [PubMed] [Google Scholar]
- Kwast, K. E., P. V. Burke and R. O. Poyton, 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201: 1177–1195. [DOI] [PubMed] [Google Scholar]
- Lees, N. D., M. Bard and D. R. Kirsch, 1999. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 34: 33–47. [PubMed] [Google Scholar]
- Lewis, T. L., G. A. Keesler, G. P. Fenner and L. W. Parks, 1988. Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast 4: 93–106. [DOI] [PubMed] [Google Scholar]
- MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond et al., 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49: 1745–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzahn, S. W., and R. A. Woods, 1972. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J. Gen. Microbiol. 72: 339–348. [DOI] [PubMed] [Google Scholar]
- Ochman, H., A. S. Gerber and D. L. Hartl, 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C. S., D. A. Toke, S. Mandala and C. E. Martin, 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272: 17376–17384. [DOI] [PubMed] [Google Scholar]
- Puoti, A., C. Desponds and A. Conzelmann, 1991. Biosynthesis of mannosylinositolphosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J. Cell Biol. 113: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shianna, K. V., W. D. Dotson, S. Tove and L. W. Parks, 2001. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 183: 830–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve, S., P. Leplatois, A. Josse, P. H. Dupuy, C. Lanau et al., 1996. The immunosuppressant SR 31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, P. M., B. G. Oliver and T. C. White, 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3: 1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, E., J. Stukey, V. McDonough, M. Germann, Y. Liu et al., 2002. Yeast cells lacking the ARV1 gene harbor defects in sphingolipid metabolism: Complementation by human ARV1. J. Biol. Chem. 277: 26177–26184. [DOI] [PubMed] [Google Scholar]
- Ter Linde, J. J., and H. Y. Steensma, 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19: 825–840. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., and R. Rothstein, 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- Tinkelenberg, A. H., Y. Liu, F. Alcantara, S. Khan, Z. Guo et al., 2000. Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem. 275: 40667–40670. [DOI] [PubMed] [Google Scholar]
- Todd, R. B., and A. Andrianopoulos, 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21: 388–405. [DOI] [PubMed] [Google Scholar]
- Turi, T. G., and J. C. Loper, 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem. 267: 2046–2056. [PubMed] [Google Scholar]
- Valachovic, M., L. I. Wilcox, S. L. Sturley and M. Bard, 2004. A mutation in sphingolipid synthesis suppresses defects in yeast ergosterol metabolism. Lipids 39: 747–752. [DOI] [PubMed] [Google Scholar]
- Vik, A., and J. Rine, 2000. Membrane biology: membrane-regulated transcription. Curr. Biol. 10: R869–R871. [DOI] [PubMed] [Google Scholar]
- Vik, A., and J. Rine, 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6395–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao et al., 2002. Transcriptional profiling identifies two members of the ATP-binding-cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277: 32466–32472. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, K., D. Resnick, C. Abeijon, P. W. Robbins and C. B. Hirschberg, 1990. A guanosine diphosphatase enriched in Golgi vesicles of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 265: 19351–19355. [PubMed] [Google Scholar]
- Zitomer, R. S., and C. V. Lowry, 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]