Abstract
The rev6-1 allele was isolated in a screen for mutants deficient for UV-induced reversion of the frameshift mutation his4-38. Preliminary testing showed that the rev6-1 mutant was substantially deficient for UV-induced reversion of arg4-17 and ilv1-92 and markedly UV sensitive. Unlike other REV genes, which encode DNA polymerases and an associated subunit, REV6 has been found to be identical to POL30, which encodes proliferating cell nuclear antigen (PCNA), the subunit of the homotrimeric sliding clamp, in which the rev6-1 mutation produces a G178S substitution. This substitution appears to abolish all DNA damage-tolerance activities normally carried out by the RAD6/RAD18 pathway, including translesion replication by DNA polymerase ζ/Rev1 and DNA polymerase η, and the error-free, recombination-dependent component of this pathway, but has little effect on the growth rate, suggesting that G178S may prevent ubiquitination of lysine 164 in PCNA. We also find that rev6-1 mutation can be fully complemented by a centromere-containing, low copy-number plasmid carrying POL30, despite the presumed occurrence in the mutant of sliding clamp assemblies that contain between one and three G178S PCNA monomers as well as the fully wild-type species.
THE RAD6/RAD18 DNA damage-tolerance pathway of budding yeast, Saccharomyces cerevisiae, promotes the completion of genome replication when nascent strand elongation by the replicase is stalled at sites of template damage. At least two mechanisms may be employed for this purpose: translesion replication by specialized DNA polymerases, a process that is often inaccurate and thus generates mutations, and an accurate process based on recombination between partially replicated sister strands (Zhang and Lawrence 2005). The particular process used is regulated by modification of K164 in PCNA (Hoege et al. 2002), the subunit of the homotrimeric sliding clamp that enhances DNA polymerase processivity. K164 is modified by SUMO during normal replication, whereas monoubiquitination of this residue by the Rad6 E2 ubiquitin conjugase present in a Rad6–Rad18 heterodimer promotes translesion replication (Stelter and Ulrich 2003). Polyubiquitination of K164, in which ubiquitin is conjugated to K36 of ubiquitin itself by an Mms2–Ubc13–Rad5 complex, promotes the accurate recombination-dependent process (Ulrich and Jentsch 2000).
Investigation of the translesion replication component of the DNA damage-tolerance pathway has been greatly advanced by the identification of mutants isolated specifically for a reduction in the frequency of induced mutagenesis. These were first isolated in yeast—and in fact in any organism—by Lemontt (1971), producing alleles of genes designated REV1, REV2 (now renamed RAD5), and REV3. Following this lead, we isolated alleles of REV4 through REV7 and NGM2 (Lawrence et al. 1985a,b; Nisson and Lawrence 1986). Of these, REV3 encodes the catalytic subunit of DNA polymerase ζ (Morrison et al. 1989; Nelson et al. 1996a), and REV7 (Torpey et al. 1994) encodes an additional subunit of this enzyme. REV1 encodes a deoxycytidyl transferase activity, but also possesses a more general function in translesion replication (Larimer et al. 1989; Nelson et al. 1996b, 2000). Although the rev4 and rev5 mutations isolated were found on outcrossing to exhibit deficiencies in mutagenesis that were too small for further analysis, rev5-1 has been found recently (M. Villasmil and P. E. M.Gibbs, unpublished data) to be an allele of POL32, which encodes a subunit of DNA polymerase δ, a subunit shown to influence induced mutagenesis (Huang et al. 1997, 2000). However, the functions of the two remaining loci, NGM2 and REV6, have not been previously identified.
The REV6 was a particularly interesting gene for further study because the phenotype of rev6-1 strains was in several respects similar to that of rev1, rev3, or rev7 mutants, suggesting that the REV6 gene product might constitute a hitherto unidentified role in translesion replication. The rev6-1 allele was isolated in a screen of mutagen-treated cells for strains that exhibited much-reduced frequencies of mutagenesis as measured by the UV-induced reversion of the frameshift allele his4-38 (Lawrence et al. 1985a). Further testing showed that it also reduced the UV-induced reversion frequency of arg4-17 (ochre) and ilv1-92 (missense), suggesting that its deficiency for induced mutagenesis, like that of rev1, rev3, or rev7 mutants, was likely to be global. Similarly, the rev6-1 mutant was also found to be sensitive to UV. However, although these phenotypes were similar to the other rev mutants, no direct comparison was made. To investigate the function of the REV6 gene, we cloned and sequenced the rev6-1 allele and, surprisingly, found that it is an allele of POL30, encoding the sliding clamp protein, proliferating cell nuclear antigen (PCNA), in which it produces a G178S substitution. This substitution not only causes a deficiency in translesion replication but also abolishes the error-free, recombination-dependent component of the RAD6/RAD18 pathway. We suggest that the rev6-1 allele be renamed pol30-61.
MATERIALS AND METHODS
Yeast strains:
HSZ1-1C (MATα rev6-1 arg4-17 his3-Δ1 ilv1-92 leu2-3,112 ura3-52) was used in the screen for a REV6 clone. Deletions of REV1, REV3, RAD30, and POL32 were created in CL1265-7C (MATα arg4-17 his3Δ-1 leu2-3,112 trp1 ura3-52) by the gene-blaster method (Alani et al. 1987), and the rev6-1 allele inserted by gene replacement (Rothstein 1991), to provide strains for assessing the comparative influence of these mutants on UV-induced mutagenesis and survival. The rev6-1 strain was also used to examine the influence of this mutation on the bypass of specific lesions by translesion replication. HSZY12, a rad1Δ derivative of this strain, was used to investigate the effect of the rev6-1 mutation on lesion bypass employing the error-free recombination process.
Cloning the REV6 locus and its rev6-1 allele:
A clone carrying the REV6 locus was isolated by transforming the rev6-1 strain HSZ1-1C with a yeast genomic library constructed in YCplac 33 (Briggs and Butler 1996), supplied by Scott Butler (University of Rochester School of Medicine and Dentistry), followed by a screen for clones carrying plasmids that complemented the UV sensitivity of the rev6-1 mutant. To this end, master plates of transformants, together with REV6 and rev6-1 controls, on synthetic complete medium lacking uracil were replicated using a rod-type replicator onto a series of plates containing yeast extract peptone dextrose medium that were exposed to 0, 20, 40, 60, 80, and 100 J/m2 of 254 nm UV and incubated in the dark for 1–3 days. UV-resistant transformants were retested in the same way and levels of arg4-17 reversion examined by spreading ∼4 × 107 cells on each of two plates of synthetic complete medium lacking uracil and arginine, one irradiated with 15 J/m2 254 nm UV and the other unirradiated, to determine whether they exhibited frequencies of ARG4 revertants characteristic of REV6 strains rather than the much lower frequencies found in rev6-1 mutants. Composition of the media used is as described (Sherman 2002). Sequence analysis of the ends of the genomic insert in a complementing plasmid of this kind indicated that it contained a 12.6-kb segment of chromosome II carrying eight intact open reading frames (ORFs). The subcloning of portions of this 12.6-kb segment identified a 1.83-kb sequence that was fully complementing and that encoded the POL30 locus on one strand and the YBR089W overlapping ORF on the other strand. All other segments failed to complement. To establish that complementation of the rev6-1 phenotype depended on POL30 rather than on YBR089W, the POL30 cys22 TGT codon was converted to a TGA stop codon, a change that generated a synonymous alteration in the YBR089W reading frame. This substitution was introduced using a QuickChange (Stratagene, La Jolla, CA) site-directed mutagenesis kit and the following PCR primers: 5′-GGAAATTGACCAACTGGACTCAATCTTTGAAACCATC-3′ and 5′-GATGGTTTCAAAGATTGAGTCCAGTTGGTCAATTTCC-3′. The identity of the rev6-1 mutation was determined by recovering the genomic allele using plasmid gap repair (Rothstein 1991) and sequencing the entire gene using Big Dye cycle sequencing and an ABI Genetic Analyzer model 3100. The gapped plasmid was generated by removing the 2.2-kb BamHI/HpaI sequence encompassing the whole POL30 gene, from a 4-kb HindIII genomic fragment derived from the original 12.6-kb genomic insert, cloned into YCplac33.
UV mutagenesis, survival, and growth rate:
Frequencies of arg4-17 revertants were estimated by growing each strain at 30° with vigorous shaking for 4 days to late stationary phase in liquid YPD medium and plating ∼4 × 107 washed cells on synthetic complete medium without arginine for revertants and suitable dilutions on synthetic complete medium with arginine for estimates of survival. Survival curves were determined by plating washed cells on YPD medium. Plates were irradiated with 2.5, 5.0, 7.5, 10.0, and 12.5 J/m2 254 nm UV or left unirradiated. All plates were handled under illumination from gold fluorescent lights and incubated at 30° in the dark. Growth rates were examined by inoculating 100 ml of synthetic complete medium, or for transformed strains such medium lacking uracil, in side-arm flasks with 1 × 105 cells of an overnight culture grown in the same medium, followed by incubation at 30° with vigorous shaking. Turbidity of the culture was measured with a Klett-Summerson colorimeter.
Construction of plasmids carrying site specific DNA lesions:
Duplex plasmids carrying a T-T cis–syn cyclobutane dimer or abasic site uniquely located within a 28-nt single-stranded region, used to estimate frequencies of translesion replication, were constructed as described (Gibbs and Lawrence 1995; Gibbs et al. 2005). To produce the dimer-containing species, 36-mers were constructed by ligating an 11-mer carrying this lesion, produced and purified as described (Banerjee et al. 1988), to a flanking 12-mer and 13-mer, followed by the annealing of the gel-purified product to an unphosphorylated complementary 28-mer and ligation of this duplex insert into EcoRI–PstI-digested pYPOG1, using a two-step process that promoted high ligation efficiency. Following denaturation of the 28-mer, carried out in the presence of a molar excess of its complementary sequence, the gapped circular material was purified by electrophoresis and quantitated by picogreen fluorometry (Molecular Probes, Eugene, OR) using a Turner Quantech fluorometer. Gapped circular constructs containing an abasic site were produced similarly, except that 36-mers carrying a dUMP nucleotide were synthesized and the construct treated after purification with uracil N-glycosylase (New England Biolabs, Beverly, MA).
Duplex plasmids carrying specifically located T-T pyrimidinone pyrimidine (6-4) adducts in each strand at staggered positions 28 bp apart, used to investigate the effect of the rev6-1 mutation on lesion bypass by the error-free recombination process, were constructed as described (Ozgenc et al. 2005; Zhang and Lawrence 2005). To this end, a 72-mer and complementary 80-mer were annealed and ligated into EcoRI–PstI-digested pYPOG1 by a method that promoted high yields of the desired product. The complementary 80- and 72-nt strands that form the duplex sequence inserted into either pYPOG1 or pYPOG2 were assembled individually by ligating together oligonucleotides annealed to complementary scaffold oligomers as described (Ozgenc et al. 2005), followed by purification of the required single-stranded species by electrophoresis through a 12% denaturing polyacrylamide gel. The 72-mer strand was assembled from the following oligonucleotides in the order given: 5′-TCCACGGTACCTTAG-3′, 5′-GCAAGTTGGAG-3′, 5′-GGTTACCAGTAGCTCGTACCC TCCCCCTTGCGAGTCCAGCAAGACC-3′. For the 80-mer they were 5′-AATTGGTCTTGCGGACTC-3′, 5′-GCAAGTTGGAG-3′, 5′-GGTACGAGCT ACTGGTAACCCTCCCCCTTGCTAAGGTACCGTGGATGCA-3′. A T-T (6-4) photoadduct was placed at the unique T-T site in the 11-mer 5′-GCAAGTTGGAG-3′ as described (Ozgenc et al. 2005) and incorporated into both the 72-mer and the 80-mer. A complete set of strands was also assembled using lesion-free 11-mer to act as controls. Following ligation of the 72-mer/80-mer insert into pYPOG1, covalently closed product was purified by electrophoresis through a 0.7% agarose gel and quantitated with picogreen fluorometry (Molecular Probes) using a Turner Quantech fluorometer.
RESULTS
Isolation of a REV6 clone:
To investigate the molecular role performed by the REV6 gene, we isolated a clone carrying a wild-type copy of this locus by transforming the rev6-1 strain HSZ1-1C with a yeast genomic library in the low copy-number plasmid YCplac33 (Briggs and Butler 1996) followed by the screening of transformants for plasmids that complemented the UV sensitivity of the rev6-1 mutant. One such UV-resistant transformant, found among 4732 screened, contained a yeast genomic sequence from chromosome II spanning nucleotides 414638–427283, which contains eight intact ORFs. Subcloning various parts of this sequence into YCplac33 indicated that a 1.83-kb BamHI/BglII fragment, but no other segment, fully complemented rev6-1 UV sensitivity. This region contains the POL30 gene on one strand and the substantially overlapping YBR089W ORF on the other. Two pieces of evidence show that complementation of the rev6-1 mutant phenotype depends on a functional POL30 and is not dependent on YBR089W. First, sequence analysis of the REV6/POL30 allele segregating in the same pedigree as HSZ1-1C revealed that it carried three single-nucleotide polymorphisms compared to the GenBank sequence, two producing synonymous changes in the POL30 ORF (Phe57, TTC to TTT; Thr89, ACA to ACG) and one outside this ORF. Of these, the Phe57 polymorphism converted the Trp138 TGG of the YBR089W reading frame into a TGA stop codon, which, since the fragment complements, suggests that complementation cannot depend on YBR089W. Second, complementation was abolished when a site-directed change was made in the POL30 Cys22 codon, converting it from TGT to a TGA stop codon, a change that is synonymous in the YBR089W reading frame. The rev6-1 mutation therefore encodes a mutant form of PCNA that substantially reduces UV-induced mutagenesis but, since rev6-1 strains appear to grow at the usual rate, does not seem to materially impair the role of the sliding clamp in normal replication.
To identify sequence alteration present in the rev6-1 allele, we recovered the POL30 locus from the rev6-1 mutant HSZ1-1C by gapped plasmid repair (Rothstein 1991), transforming this strain with pYHS7 from which the POL30 ORF had been deleted by digestion with BamHI and HpaI. Sequence analysis of the whole of the POL30 locus in the repaired plasmid showed that it contained a single G:C-to-A:T substitution that produced a glycine178-to-serine substitution. That this single alteration in POL30 is indeed responsible for the rev6-1 mutant phenotype was confirmed by the observation that strain HSZY1, created by replacing POL30 in strain CL1265-7C with the mutant allele (Rothstein 1991), possesses both the UV sensitivity and UV-induced mutation deficiency characteristic of the original rev6-1 mutant. We conclude that in addition to a K164R substitution, which is known to impair all RAD6/RAD18-dependent DNA damage-tolerance processes (Hoege et al. 2002; Stelter and Ulrich 2003), a single substitution of at least one other residue can impair UV mutagenesis and therefore presumably translesion replication. This finding also raises the questions of whether the rev6-1 mutation impairs translesion replication by all bypass enzymes in yeast and also whether it decreases the efficiency of the error-free component of the RAD6/RAD18 pathway.
Characterization of the rev6-1 phenotype:
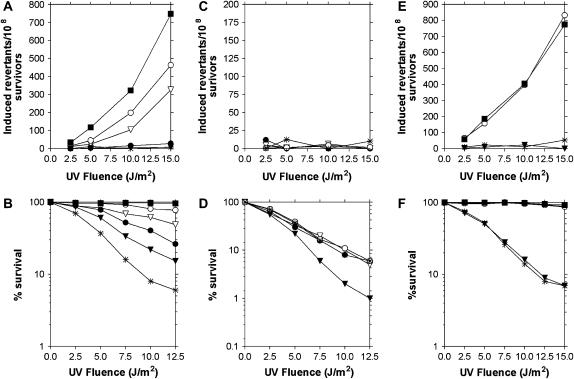
We compared the phenotype of the rev6-1 mutant to that of isogenic REV6 strains carrying a rev3, rev1, rad30, or pol32 deletion by measuring the frequency of ARG4 revertants induced by irradiation with 254 nm UV and UV sensitivity in each of these strains. Double mutants carrying rev6-1 and one of these deletions were also examined (Figure 1). Although the frequency of induced ARG4 revertants was decreased in each of the single mutant strains, and sensitivity to cell killing increased, the extent of these differences varied widely. As shown in Figure 1A, the frequency of ARG4 revertants in the rev6-1 strain is much lower than that in the REV6 strain and is essentially no different from the similarly low frequencies seen in the rev1Δ and rev3Δ strains. In contrast, the frequency of ARG4 revertants is decreased to a much smaller extent in the rad30Δ and pol32Δ strains. The rev6-1 mutant is also least like the rad30Δ and pol32Δ strains with respect to survival following exposure to 254 nm UV. As can be seen in Figure 1B, the rad30Δ and pol32Δ strains, even though slightly more sensitive than the wild-type, are the least sensitive of the mutant strains tested. The rev6-1 mutant, on the other hand, exhibits much greater sensitivity and is in fact appreciably more sensitive than either of the rev1Δ or rev3Δ strains, perhaps the consequence of impairment of the error-free process. Each of the double mutants exhibits the same very low frequencies of UV-induced ARG4 revertants as the rev6-1 single mutant, and the rev1Δ, rad30Δ, and pol32Δ mutants are also each epistatic to the rev6-1 allele with respect to sensitivity to killing by UV. Interestingly, however, the rev3Δ rev6-1 strain is more UV sensitive than either of the component single mutants, perhaps reflecting the PCNA-independent activities of DNA polymerase ζ.
Figure 1.—
Frequencies of UV-induced reversion of arg4-17 (A, C, and E) and percentage of survival following UV irradiation (B, D, and F) in REV6, rev6-1, rev1Δ, rev3Δ, rad30Δ, and pol32Δ strains and in rev6-1 rev1Δ, rev6-1 rev3Δ, rev6-1 rad30Δ, and rev6-1 pol32Δ double mutants. A–F: ▪, REV6+; *, rev6-1. Additionally, in A and B: ○, rad30Δ; ▿, pol32Δ; •, rev1Δ; ▾, rev3Δ. In C and D: ○, rev6-1 rev1Δ; ▿, rev6-1 pol32Δ ; •, rev6-1 rad30Δ; ▾, rev6-1 rev3Δ. In E and F: ○, rev6-1 + YCplac33 POL30; ▾, rev6-1 + YCplac33. Data are the average of two or three replicate experiments.
To investigate the impact of the rev6-1 mutation on the translesion replication and error-free recombination-dependent components of the RAD6/RAD18 DNA damage-tolerance pathway more directly, we have transformed a rev6-1 strain with plasmids carrying specifically located DNA lesions of two types, each designed to examine one of these two processes (Gibbs et al. 2005; Zhang and Lawrence 2005). To examine translesion replication, the plasmids carried either an abasic site or T-T cys–syn cyclobutane dimer placed centrally within a 28-nt single-stranded region. Plasmid replication, and hence the production of transformants, requires the filling of this gap, which entails translesion replication. The frequency of this event can therefore be estimated by normalizing the number of transformants to the number obtained with an equal amount of lesion-free control plasmid. Because the bypass frequencies of the two lesions are twofold different, we present the data in Table 1 as a percentage of the frequency in the wild type strain, a procedure that makes it easier to compare the contributions of the various gene functions to the bypass of each lesion. As shown in Table 1, the G178S substitution in rev6-1 essentially abolishes the ability to replicate past both an abasic site and a T-T cyclobutane dimer, indeed to a level lower than that in strains lacking DNA polymerase ζ (rev3Δ) or Rev1 (rev1Δ) and matched only, at least for the T-T dimer, in strains simultaneously deficient for pol ζ and pol η (rad30Δ) or for Rev1 and pol η. Because, as can be seen in Table 1, pol ζ/Rev1 are largely responsible for replication past abasic sites whereas pol η is responsible for replication past the T-T dimer, these results show that the G178S substitution impairs the activity of each of these three enzymes.
TABLE 1.
Bypass frequencies, expressed as percentage of the wild-type frequency, for abasic sites and T-T cyclobutane dimers in rad30Δ, rev3Δ, rev1Δ, pol32Δ, and rev6-1 strains
| Lesion bypass, % of wild-type frequency
|
|||
|---|---|---|---|
| Abasic site (O)
| |||
| Strain | T-O | O-T | T-T cis–syn dimer |
| Wild-typea | 100.0 | 100.0 | 100.0 |
| rev6-1 | 1.3 | 0.5 | 1.3 |
| rad30Δa | 80.7 | 87.1 | 15.3 |
| rev3Δa | 4.3 | 5.5 | 93.6 |
| rev1Δa | 3.8 | 2.9 | 98.5 |
| rad30Δ rev3Δa | — | — | 0.0 |
| rad30Δ rev1Δa | — | — | 1.0 |
| pol32Δa | 4.7 | 3.6 | 112.1 |
Each lesion is located at the T-T site in the sequence 5′-GCAAGTTGGAG-3′. Actual bypass frequencies and the number of replicate experiments (N) for the different lesions in the wild-type strain were: T-O, 32.3% ± 4.2% (N = 5); O-T, 24.1% ± 2.8% (N = 3); T-T dimer, 60.1% ± 5.8% (N = 16).
Data from Gibbs et al. (2005).
In addition to investigating the impact of the rev6-1 mutation on translesion replication, we also examined its influence on the error-free, recombination-dependent component of the RAD6/RAD18 pathway. To do this, we transformed an excision-defective (rad1Δ) strain and its isogenic rev6-1 derivative with plasmids that carried a T-T pyrimidinone (6-4) pyrimidine photoadduct [T-T (6-4) photoadduct] in each strand of the duplex, at staggered positions 28 bp apart. C-C mismatches placed opposite the T-T (6-4) photoadducts allow the unambiguous identification of the events leading to the completion of replication: recombination between partially replicated sister strands within the interlesion region or translesion replication on one or the other strand. Each event has a unique signature sequence, which in the case of recombination is the presence in replicated plasmids of G:C,G:C and C:G,C:G doublets at the sites previous occupied by the T-T lesions. Although these photoadducts present a severe impediment to translesion replication, an astonishing 54.7% of the plasmids are replicated in the rad1Δ strain, with 50.7% of them achieving this by a recombination-dependent process (Zhang and Lawrence 2005) (Table 2). The rev6-1 mutation reduces this to 19.7% (Table 2), a frequency similar to that found in the strain deleted for RAD5 (21.9%), a gene whose function is required for the RAD6/RAD18 error-free process. As discussed below and in Zhang and Lawrence (2005), the remainder of recombination occurs by a different process for which the function of Rad52 is required. We conclude that the G178S substitution interferes with two, and perhaps all, of the RAD6/RAD18-dependent DNA damage-tolerance processes in yeast; it not only abolishes the PCNA-dependent activities of pol ζ, pol η, and Rev1 but also impairs the error-free recombination process.
TABLE 2.
Percentage of total plasmids replicated, percentage replicated by a recombination-dependent process, and percentage replicated using translesion replication in an excision repair-defective (rad1Δ) strain and its isogenic rev6-1, rad5Δ, rad18Δ, rad52Δ, and rad18Δ rad52Δ deletion derivatives
| Strain | Relevant genotype | % plasmids replicated | % by recombination | % by translesion replication | No. plasmids sequenced | No. replicate experiments |
|---|---|---|---|---|---|---|
| POGY9a | rad1Δ | 54.7 ± 3.2 | 50.7 | 4.0 | 182 | 5 |
| HSZY12 | rad1Δ rev6-1 | 22.1 ± 4.6 | 19.7 | 2.4 | 165 | 5 |
| POGY13a | rad1Δ rad5Δ | 23.5 ± 1.9 | 21.9 | 1.6 | 165 | 5 |
| POGY12a | rad1Δ rad18Δ | 15.5 ± 1.6 | 15.5 | 0.0 | 84 | 6 |
| HSZY9a | rad1Δ rad52Δ | 28.6 ± 3.6 | 24.2 | 4.4 | 169 | 5 |
| HSZY7a | rad1Δ rad18Δ rad52Δ | 2.3 ± 0.8 | 2.3 | 0.0 | 22 | 11 |
Data from Zhang and Lawrence (2005).
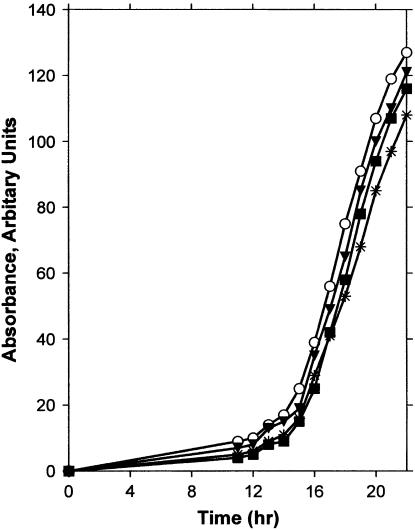
Although it is clear that processes promoting the replication of lesion-containing DNA templates are defective in rev6-1 strains, we observed no obvious deficiency in the growth rate of unirradiated cells carrying this mutation, suggesting that G178S PCNA functions reasonably well in the replication of undamaged DNA. To specifically examine this issue, we measured change in absorbance over time in logarithmically growing cultures of isogenic REV6 and rev6-1 strains transformed with either YCplac33 POL30 or YCplac33 (Figure 2). These results show that growth rate in the rev6-1 mutant is at best only slightly slower than that in the wild-type strain, suggesting that G178S PCNA functions almost normally during replication on undamaged DNA templates.
Figure 2.—
Growth rate monitored by turbidity (arbitrary absorbance units) and viable cell titer in REV6+ and rev6-1 strains transformed with either YCplac33 POL30 or YCplac33. ▪, REV6; *, rev6-1; ○, rev6-1 with YCplac33 POL30; ▾, rev6-1 with YCplac33. Data are the average of three experiments.
DISCUSSION
We find that rev6-1, a mutation isolated by virtue of its deficiency in UV-induced mutagenesis, is an allele of POL30 that encodes a G178S substitution in PCNA. This substitution appears to abolish the effective mobilization by the homotrimeric sliding clamp—of which PCNA is the subunit—of pol ζ, pol η, and Rev1 for translesion replication. A comparison of UV-induced reversion frequencies of arg4-17 and survival in an isogenic series of rev6-1, rev1Δ, rev3Δ, rad30Δ, and pol32Δ mutant strains (Figure 1) shows that induced reversion is almost completely absent in the rev6-1 mutant, as it also is in the rev1Δ and rev3Δ strains, in contrast to the rad30Δ and pol32Δ strains, which reduce reversion frequencies only modestly. These observations suggest that the G178S substitution in PCNA inhibits translesion replication by pol ζ and Rev1, but do not exclude a similar effect with pol η. In keeping with this conclusion, UV-induced reversion of arg4-17 is essentially absent in double mutants carrying rev6-1 and one of the deleted genes. The rev6-1 mutant is also more sensitive to killing by UV than the other strains and in fact is more sensitive than the rev3Δ strain, consistent with the G178S alteration impairing a DNA damage-tolerance process other than translesion replication, as discussed below. All but one of the double mutants is epistatic to rev6-1 with regard to UV sensitivity. The rev6-1 rev3Δ strain, however, is more sensitive than either single mutant, perhaps because pol ζ promotes survival in processes other than those concerned with DNA damage tolerance or those dependent on PCNA.
The conclusion that the G178S abolishes virtually all capacity for translesion replication was also supported by transforming a rev6-1 strain with plasmids that carried either an abasic site or T-T dimer within a 28-nt single-stranded region of an otherwise duplex vector, which provides a direct estimate of lesion bypass frequencies. We chose a T-T dimer because it is bypassed in vivo almost exclusively by pol η; although, as shown in Table 1, dimer bypass can be performed by pol ζ/Rev1 when pol η is absent, in its presence they do so only in ≤0.6% of bypass events (Gibbs et al. 2005). An abasic site, conversely, is bypassed predominantly by pol ζ/Rev1. Bypass frequencies for both of these lesions were no more than ∼1% of the wild type frequency in the rev6-1 mutant, suggesting that the G178S substitution abolishes the activity of both polymerases in translesion replication. Regarding the dimer, such a result is similar to what is seen in the rad30Δ rev3Δ and rad30Δ rev1Δ strains but is unlike the bypass frequency in the pol32Δ mutant, which is at wild-type level. This difference between rev6-1 and pol32Δ strains suggests that unlike Pol32, which appears to influence mutagenesis indirectly via interactions with Srs2 or checkpoint proteins (Gibbs et al. 2005), the G178S substitution influences lesion bypass DNA polymerases more directly.
In addition to abolishing translesion replication, the PCNA G178S substitution in the rev6-1 strain also inhibits the error-free, recombination-dependent component of the RAD6/RAD18 pathway, presumably the reason for the greater sensitivity to UV of the rev6-1 strain compared to the rev3Δ mutant. Although 50.7% of the plasmids carrying T-T (6-4) photoadducts completed replication by a recombination-dependent mechanism in the rad1Δ control strain (Table 2), only 19.7% did so in the rad1Δ rev6-1 strain. The latter value is similar to the 21.9% observed in the rad1Δ rad5Δ strain, suggesting that the G178S substitution essentially abolishes the recombination-dependent component of the RAD6/RAD18 pathway for which RAD5 function is essential. The remaining fraction of plasmids using a recombination mechanism to complete replication in the rad1Δ rev6-1 and rad1Δrad5Δ strains was generated by a different process, which is not part of the RAD6/RAD18 pathway, and instead uses RAD52. As the very small fraction (2.3%) of plasmids replicated in the rad1Δ rad18Δ rad52Δ strain shows (Table 2), the RAD18- and RAD52-dependent processes between them account for nearly all of the recombination observed in the rad1Δ control strain. Approximately 60–70% can be ascribed to the RAD6/RAD18-dependent process, with the remaining 30–40% resulting from the RAD52-dependent mechanism (Zhang and Lawrence 2005). Although all of the strains used in these experiments carry a RAD1 deletion, and this gene is known to be involved in certain types of recombination (Rattray and Symington 1995), there is no obvious reason why its absence should influence template strand switching, although it may well influence the RAD52-dependent process.
The rev6-1 mutant therefore appears to be deficient in both the translesion replication and the error-free, recombination-dependent components of the RAD6/RAD18 DNA damage-tolerance pathway. At the same time, unirradiated cells of this mutant grow at a rate that is little different from that of the wild type (Figure 2), resulting in a phenotype that is remarkably similar to that of the PCNA K164R mutation. The G178 residue is located in a β-sheet at the junction between the monomers composing the sliding clamp, and the side chain of a substituted serine at this site appears to show an unfavorable steric interaction with Y114 located in the adjacent monomer. The PCNA G178S substitution may therefore inhibit modification of K164, perhaps by changing the structure of the sliding clamp sufficiently to prevent binding of the Rad6/Rad18 heterodimer that monoubiquitinates this residue. Alternatively, the altered structure may prevent the association of the sliding clamp with other proteins required for the activities carried out by the RAD6/RAD18 pathway.
Surprisingly, the rev6-1 mutation is fully recessive to the POL30 wild-type allele, with respect to both survival and UV-induced reversion of arg4-17 (Figure1, E and F), and can be fully complemented by a centromere-containing, low copy-number plasmid carrying the POL30 gene, although not by the vector alone, despite the presumed occurrence in the mutant of sliding clamp assemblies that contain anywhere between one and three G178S PCNA monomers as well as the fully wild-type species. Such complementation is in marked contrast to the dominant negative phenotype observed in heterozygous diploids carrying alleles encoding either the ED104,105AA or the DE256,257AA alterations together with the wild type (Torres-Ramos et al. 1996). The observation that the growth rate of the rev6-1 mutant is not appreciably different from that of the wild type (Figure 2) suggests that its complementation by the POL30 gene is not the consequence of the instability—and thus a higher turnover rate—of clamp assemblies that contain G178S PCNA, resulting in a largely wild-type clamp being used for almost all replication. Perhaps the presence of even a single wild-type PCNA monomer may be sufficient to allow normal function of the RAD6/RAD18 pathway, either because clamps containing mutant and wild type PCNA assume a fully normal structure or because association is intrinsically confined to a single monomer.
Acknowledgments
We thank J. Scott Butler for the yeast genomic library; Meng-Er Huang, Michael Resnick, and Jim Westmoreland for plasmids; and Ravi Basavappa for advice and assistance. This work was supported by U.S. Public Health Service grant GM-60652 from the National Institutes of Health.
References
- Alani, E., L. Cao and N. Kleckner, 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S. K., R. B. Christensen, C. W. Lawrence and J. E. Leclerc, 1988. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc. Natl. Acad. Sci. USA 85: 8141–8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, M. W., and J. S. Butler, 1996. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics 143: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, P. E. M., and C. W. Lawrence, 1995. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol. 251: 229–236. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. E. M., J. MacDonald, R. Woodgate and C. W. Lawrence, 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol 32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct, and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G.-L. Moldovan, G. Pyrowolakis and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Huang, M.-E., E. Cadieu, J. L. Souciet and F. Galibert, 1997. Disruption of six novel yeast genes reveals three genes essential for vegetative growth and one required for growth at low temperature. Yeast 13: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Huang, M.-E., A. De Calignon, A. Nicolas and F. Galibert, 2000. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38: 178–187. [DOI] [PubMed] [Google Scholar]
- Larimer, F. W., J. R. Perry and A. A. Hardigree, 1989. The REV1 gene of Saccharomyces cerevisiae; isolation, sequence, and functional analysis. J. Bacteriol. 171: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. W., B. R. Krauss and R. B. Christensen, 1985. a New mutations affecting induced mutagenesis in yeast. Mutat. Res. 150: 211–216. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. W., G. Das and R. B. Christensen, 1985. b REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 200: 80–85. [DOI] [PubMed] [Google Scholar]
- Lemontt, J. F., 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, A., R. B. Christensen, J. Alley, A. K. Beck, E. G. Bernstine, et al., 1989. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 171: 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. R., C. W. Lawrence and D. C. Hinkle, 1996. a Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649. [DOI] [PubMed] [Google Scholar]
- Nelson, J. R., C. W. Lawrence and D. C. Hinkle, 1996. b Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson, J. R., P. E. M. Gibbs, A. M. Nowicka, D. C. Hinkle and C. W. Lawrence, 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37: 549–554. [DOI] [PubMed] [Google Scholar]
- Nisson, P. E., and C. W. Lawrence, 1986. The isolation and characterization of ngm2, a mutation that affects nitrosoguanidine mutagenesis in yeast. Mol. Gen. Genet. 204: 90–97. [DOI] [PubMed] [Google Scholar]
- Ozgenc, A. I., E. S. Szekeres and C. W. Lawrence, 2005. In vivo evidence for a recA- independent recombination process in Escherichia coli that permits the completion of replication in DNA containing UV damage in both strands. J. Bacteriol. 187: 1974–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., and L. S. Symington, 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R., 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast, pp. 281–301 in Guide to Yeast Genetics and Molecular Biology, Part B (Methods in Enzymology, Vol. 194), edited by C. Guthrie and G. R. Fink. Academic Press, San Diego. [DOI] [PubMed]
- Sherman, F., 2002. Getting started with yeast, pp. 3–41 in Guide to Yeast Genetics and Molecular and Cell Biology, Part B (Methods in Enzymology, Vol. 350), edited by C. Guthrie and G. R. Fink. Academic Press, San Diego. [DOI] [PubMed]
- Stelter, P., and H. D. Ulrich, 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Torpey, L. E., P. E. M. Gibbs, J. Nelson and C. W. Lawrence, 1994. Cloning and sequencing of REV7, a gene whose function is required for DNA damage-induced mutagenesis in Saccharomyces cerevisiae. Yeast 10: 1503–1509. [DOI] [PubMed] [Google Scholar]
- Torres-Ramos, C. A., B. L. Yoder, P. M. J. Burgers, S. Prakash and L. Prakash, 1996. Proc. Natl. Acad. Sci. USA. 93: 9676–9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, H. D., and S. Jentsch, 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19: 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and C. W. Lawrence, 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA 102: 15954–15959. [DOI] [PMC free article] [PubMed] [Google Scholar]