Abstract
The AMT4 locus of the green alga Chlamydomonas reinhardtii, which we mapped to the long arm of chromosome 8, provides a good experimental system for the study of transposition. Most mutations that confer resistance to the toxic ammonium analog methylammonium are in AMT4 and a high proportion of spontaneous mutations are caused by transposon-related events. Among the 15 such events that we have characterized at the molecular level, 9 were associated with insertions of the retrotransposon TOC1, 2 with a small Gulliver-related transposon, and 1 with the Tcr1 transposon. We found that Tcr1 is apparently a foldback transposon with terminal inverted repeats that are much longer and more complex than previously realized. A duplication of Tcr1 yielded a configuration thought to be important for chromosomal evolution. Other mutations in AMT4 were caused by two mobile elements that have not been described before. The sequence of one, which we propose to call the Bill element, indicates that it probably transposes by way of a DNA intermediate and requires functions that it does not encode. The sequence of the other and bioinformatic analysis indicates that it derives from a miniature retrotransposon or TRIM, which we propose to call MRC1 (miniature retrotransposon of Chlamydomonas).
THE AMT4 gene of Chlamydomonas reinhardtii codes for an NH3 gas channel that also transports the toxic analog methylamine (CH3NH2) (Kim et al. 2005). Like other AMT genes, AMT4 is highly expressed under nitrogen-limiting conditions (González-Ballester et al. 2004) and appears to be the major methylamine transporter. We have shown previously that most spontaneous mutations to methylamine (methylammonium) resistance occur in AMT4 and have briefly described several transposon-induced lesions (Kim et al. 2005).
Transposable elements can be classified by their replicative intermediates: class I transposons, the retrotransposons, replicate by means of an RNA intermediate, whereas class II transposons replicate through a DNA intermediate (Sabot et al. 2004). Members of both these major families have been described in C. reinhardtii. TOC1 (transposon of Chlamydomonas) and REM1 are retrotransposons (Day et al. 1988; Pérez-Alegre et al. 2005) and Gulliver, Pioneer, and Tcr3 (transposon of C. reinhardtii) are class II elements (Ferris 1989; Graham et al. 1995; Wang et al. 1998). Transposable elements may be further classified according to their transposition mechanism (Curcio and Derbyshire 2003). TOC1 has recently been shown to belong to the DIRS-1 subclass of retrotransposons (Goodwin and Poulter 2004), along with DIRS-1 from Dictyostelium discoideum (Cappello et al. 1985), PAT from Panagrellus redivivus (de Chastonay et al. 1992), and Kangaroo from Volvox carterei (Duncan et al. 2002). The replication of these retrotransposons involves a circular intermediate, which integrates by means of a tyrosine transposase. REM1 is related to the Ty3-gypsy long terminal repeat (LTR) retrotransposon class (Pérez-Alegre et al. 2005), which integrates by means of a DDE transposase (named for the acidic amino acid residues aspartate-aspartate-glutamate, which coordinate the metal ions required for catalysis) (Curcio and Derbyshire 2003). The Gulliver and Pioneer transposons have been included in the Activator (Ac) and Dissociation (Ds) family, first described by McClintock (1947, 1951). These, too, utilize a DDE transposase. The Tcr1 element, one of three named TCR transposons, was first encountered by Schnell and Lefebvre (1993) and later used by Ferris et al. (1996) to clone the FUS1 gene. It has not been assigned with respect to class or transposition mechanism.
Previous studies have indicated that C. reinhardtii carries a wide variety of transposable elements (Lefebvre and Silflow 1999). Online access to the nearly complete genome (v3.0) of this organism at the Joint Genome Institute (JGI) has brought increased awareness of the diversity and abundance of these elements. The nuclear genome of C. reinhardtii is a patchwork of repetitive elements, some of which are intact functional transposons, but most of which are at least partially degraded. We have used molecular and bioinformatic analysis of transposons recovered at the AMT4 locus to characterize a wide variety of naturally occurring transposition events. These provide additional information on the types of mutations that can be caused by previously known transposons and indicate that additional elements in the genome are mobile.
MATERIALS AND METHODS
Strains, culture conditions, and isolation of spontaneous mutants:
Parental Chlamydomonas strains were 4A+ and CC-125 (Kim et al. 2005). Additional strains used for mapping the AMT4 locus were obtained from the Chlamydomonas Culture Collection. They were CC-641 (pf12 nic1 mt+), CC-602 (pf1 mt−), CC-604 (pf3 mt−), CC-3321 (pf27 mt−), CC-1112 (ac206 mt−), CC-399 (ani1 mt+), and CC-899 (pf10 mt+). All strains were maintained on solid TAP medium (Harris 1989). The nitrogen source was 10 mm NH4Cl or 2.5 mm arginine as appropriate. Cells were grown at 25° under continuous light (30 μE).
One set of spontaneous mutants was isolated from a population of 4A+ cells that had been transformed to zeocin resistance with the pSP124S vector (Lumbreras et al. 1998; Soupene et al. 2004) with the intention of obtaining insertional mutants at an AMT locus. None of the 400 transformant lines tested was resistant to 0.1 mm methylammonium (Kim et al. 2005). However, single methylammonium-resistant colonies arose from them. These were purified twice and retested for methylammonium resistance. The 48 resistant lines chosen for further study were then maintained on TAP ammonium plates (a condition under which three of the four AMT genes are not expressed).
Another set of mutants was derived from strains CR46 and CR50 (Kim et al. 2005). These strains are resistant to 0.1 but not 1.0 mm methylammonium. Each strain was streaked onto TAP arginine plates with 1 mm methylammonium. Resistant colonies were picked and purified as described above.
RNA isolation and RT–PCR:
Cultures were grown on an orbital shaker in 30-ml scintillation vials (7-ml culture) or in 125-ml flasks (20-ml culture) in TAP/arginine or TAP/ammonium medium under continuous light to a chlorophyll a + b content of ∼8 μg/ml. Chlorophyll a + b was determined according to the method of Wintermans and de Mots (1965). RNA was isolated as described previously (Kim et al. 2005). Expression of the AMT4 gene was analyzed by RT–PCR using the one-step RT–PCR kit (Roche) initially following instructions of the manufacturer. The 5′-end of the AMT4 transcript was amplified by RT–PCR using forward primer ATGF and reverse primer TILL2R and the 3′-end was amplified using forward primer short2F and reverse primer AMT4-ENDR (Figure 1; Table 2 of Kim et al. 2005).
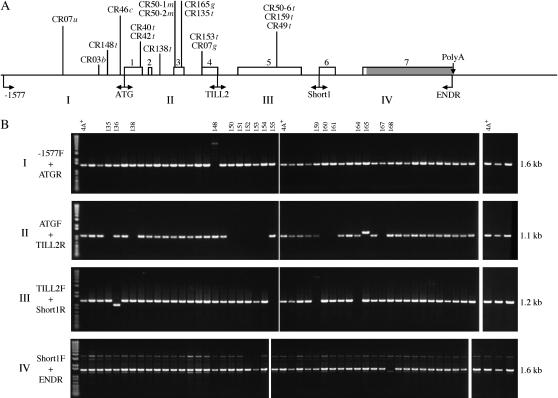
Figure 1.—
Location of transposon-related mutations in AMT4. (A) Diagram of the AMT4 gene showing the relative locations of macrolesions in various strains above the line and locations of primers used to amplify the four segments of the gene (see text) below the line. Each strain name is followed by a lowercase italic letter to indicate the transposon responsible for the lesion: t for TOC1, c for Tcr1, b for Bill, g for Gulliver-like, and m for MRC1. The unknown upstream lesion of CR07 is indicated as CR07u. The seven exons are shown as numbered blocks with the 3′ noncoding tail of exon 7 shaded. (B) Use of PCR to identify macrolesions in the 48 strains from the second selection for methylammonium resistance (CR132–CR180; see text). The four primer sets were those indicated in A, which yielded products of 1.6, 1.1, 1.2, and 1.6 kb, respectively, for regions I–IV. In spite of the short PCR extension time, the TOC1 insert in CR148 is visible. CR136 has a deletion of 276 bp in exon 5 of the AMT4 gene.
TABLE 2.
Summary of macrolesions in AMT4
| Selection | Total methylammonium- resistant lesions | Total lesions in AMT4 | Macrolesions in AMT4 | Types of macrolesions |
|---|---|---|---|---|
| Study 1 (Kim et al. 2005) | 16 | 12 | 6 | 3 TOC1 |
| 1 Gulliver-related | ||||
| 1 Tcr1 | ||||
| 1 “Bill” | ||||
| Selection 2a (this work) | 48 | NDb | 17 (7 amplified) | 5 TOC1 |
| 1 Gulliver-related | ||||
| 1 deletion | ||||
| Selection 3c (this work) | 18d | ≥15 | 15 | 12 Tcr1e |
| 2 MRC1f | ||||
| 1 TOC1f |
Selection for resistance to 0.1 mm methylammonium.
Not determined.
Selection for resistance to 1 mm methylammonium in CR46 (Tcr1 in 5′ UTR of AMT4) and CR50 (base-pair substitution at a splice site in AMT4).
Not necessarily independent.
From CR46.
From CR50.
DNA isolation and PCR:
Cultures were grown in scintillation vials in 7 ml TAP/ammonium medium with constant agitation and continuous light. They were harvested at stationary phase and DNA was isolated using a DNA extraction kit (Stratagene, La Jolla, CA). PCR was performed with the Long Template PCR system (Roche) as described previously (Kim et al. 2005). Primers for the various regions of the AMT4 gene are indicated in Table 2 of Kim et al. (2005). In many instances the structure of the DNA and the size of the transposon insert required modification of the PCR conditions. For amplification of Tcr1, the following additional primers were used: Tcr1F (TGGCAGCACGCTTTTGAGGC); Tcr1LR (AGCTCCGCGGTGTAGCTTCC); Tcr15820R (TACTCCGCATCCCTTCAACCATGT); Tcr1LRF (GGAAGCTACACCGCGGAGCT); Tcr17365F (AATCGGTACCGCTGCGATGTAGAA); −785F (ACTGTCGCCGCTGCGCAAGGC); −110F (ATAGATGTCATGCTCATGAGC); and 291R (ACGCAAGGAAAGCCGTGGAA).
Cloning of RT–PCR products from CR07:
Products from RT–PCR reactions with the AMT4 primers ATGF and TILL2R and “old mRNA” from CR07 were cleaned up using the QIAGEN (Valencia, CA) PCR purification kit. PCR fragments were cloned using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). The resulting mixture was used to transform DH5α cells and cells were plated on LB medium supplemented with 25 μg/ml kanamycin. Twenty-two colonies were picked and plasmid was purified with the QIAprep Spin miniprep kit (QIAGEN). Plasmids were sequenced with T7 and M13 reverse primers.
DNA sequencing:
Sequencing was performed by the University of California, Berkeley, Sequencing Facility using the ABI PRIZM Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Primers for sequencing were the same as those described above for PCR and RT–PCR.
RESULTS
Selection of spontaneous mutations in AMT4 and identification of transposon-induced lesions:
Several selections for spontaneous methylammonium resistance were used to isolate transposon-induced lesions in AMT4. Methylammonium greatly retards growth of C. reinhardtii but is not lethal and analog resistant lines are easily identified as green colonies over a white (bleached) background. Six strains (CR03, CR07, CR40, CR42, CR46, and CR49) were selected and identified as described previously (Kim et al. 2005). Forty-eight additional strains were selected for resistance to 0.1 mm methylammonium and happen to have been isolated from zeocin-resistant transformants (see materials and methods). Transposon-induced lesions in these derivatives were identified as described below. As expected, methylammonium resistance in the two lines tested segregated from zeocin resistance in genetic crosses (data not shown). Finally, derivatives of two parental strains resistant to 0.1 but not 1 mm methylammonium, CR46 and CR50 (Kim et al. 2005), were selected for resistance at the higher level of analog. CR46, which was found to contain the transposon Tcr1 in the 5′ UTR of AMT4 (see below), yielded ∼10 times more colonies resistant to 1 mm methylammonium than CR50, which has a single base change at the intron 4/exon 5 boundary (Kim et al. 2005). We screened 12 derivatives of CR46 and 6 derivatives of CR50 as described below. In each case, derivatives came from a single petri plate and were not necessarily independent.
For purposes of molecular analysis of changes in AMT4, the gene and its promoter were divided into four regions of ∼1 kb each and PCR primers were designed to amplify them (Figure 1A). Region I encompassed the promoter and regions II–IV each encompassed a third of the gene. Macrolesions were identified by screening for fragments that had altered mobility on a 0.8% agarose gel or putatively by failure to amplify a fragment (Figure 1B). A size alteration of 50 bp was easily detected visually, whereas smaller changes may have been overlooked. Among strains isolated in the second selection, only one (CR148) had a macrolesion in region I and, likewise, a single strain (CR168) had a putative macrolesion in region IV. Eleven strains carried macrolesions or putative macrolesions in region II and 4 of these (CR135, CR138, CR153, and CR165) are described below. Four strains carried macrolesions in region III of AMT4 and 1 (CR159) is described in detail. Even with long extension times, we failed to amplify fragments from CR150–CR154, CR160, CR161, and CR167 (lesions in region II); CR155 and CR164 (lesions in region III); and CR168 (lesion in region IV). These strains may carry large transposons and/or structurally complex sequences problematic for PCR (see below for Tcr1) or chromosomal rearrangements. They have not been pursued further. Assuming that 75% of the lesions to methylammonium resistance obtained in the second selection were in AMT4, as was the case in our first study (Kim et al. 2005), about half the lesions in AMT4 were macrolesions in both cases.
Among strains isolated in the third selection, all 12 derivatives of CR46 carried macrolesions related to the original lesion and are described below. Three of 6 derivatives of CR50, which was used as a control, carried macrolesions, the smallest of which (a 29-bp insertion) was detected by sequencing regions at which the other two macrolesions occurred. These are described below. The remaining derivatives of CR50 did not have sequence changes near the original lesion or the macrolesions in derivatives and were not studied further.
Molecular characterization of TOC1 inserts:
Eight strains with inserts derived from the TOC1 transposon (Day and Rochaix 1991a) were obtained from the first two selections and seven of these were resistant to 1 mm methylammonium, despite the fact that they were selected at lower concentrations. An additional strain was obtained as a derivative of strain CR50 in the third selection. The nine inserts were at seven separate positions: one was upstream of the translational start, two were in exon 1, one was in exon 3, one in exon 4, one in exon 5, and one in intron 2 (Figure 1 and Table 1). Only the strain carrying TOC1 in intron 2 (CR138) remained sensitive to methylammonium at 1 mm and showed higher residual methylammonium uptake than strains carrying other inserts (data not shown). The seven insert sites had several similarities (Figure 2): (1) the region within 20 nucleotides to either side was less guanine-cytosine (GC) rich than the Chlamydomonas genome generally (62% GC overall); (2) GC content dropped to ≤50% within four nucleotides to either side; (3) at least two CA doublets were found within eight nucleotides to either side; and (4) an A/T base pair was present one nucleotide removed from the insert site. Orientation of TOC1 relative to the direction of transcription of the AMT4 gene is indicated in Figure 2 for each of the inserts. Transcription of TOC1 is in the same direction as that of AMT4 in three cases and in the opposite direction in six cases. The TOC1 insert sites are spaced at regular intervals along the AMT4 gene (see Figure 1A): in CR148 at −104; CR40 at +176; CR138 at +419; CR135 at +685; CR153 at +952; and CR49, CR159, and CR50-6 (all independent) at +2074. The distance between them varies between 243 and 280 nucleotides; the distance between the last two insertion sites is 1122 nucleotides (= 4 × 280). Only the insert in CR42, which lies at +210, is an exception. It is separated from that in CR40 by only 34 nucleotides.
TABLE 1.
Genetic lesions in AMT4 from second and third selections
| Strain | Allele | Type of mutation | Site in AMT4 |
|---|---|---|---|
| CR135 | amt4-13 | TOC1 insert | Exon 3, after nt 685 |
| CR138 | amt4-14 | TOC1 insert | Intron 2, after nt 419 |
| CR148 | amt4-15 | TOC1 insert | 5′ untranslated region |
| CR153 | amt4-16 | TOC1 insert | Exon 4, after nt 952 |
| CR159 | amt4-17 | TOC1 insert | Exon 5, after nt 2074 |
| CR165 | amt4-18 | Gulliver-related insert | Exon 3, after nt 680 |
| CR50-1 | amt4-12m1 | Novel MRC1 insert | Exon 3, after nt 620 |
| CR50-2 | amt4-12m2 | Novel MRC1 insert | Exon 3, after nt 620 |
| CR50-6 | amt4-12m3 | TOC1 insert | Exon 5, after nt 2074 |
| CR46-1 | amt4-8m1 | Tcr1 modified | 5′ untranslated |
| CR46-2 | a | Tcr1 modified | 5′ untranslated |
| CR46-3 | a | Tcr1 modified | 5′ untranslated |
| CR46-4 | a | Tcr1 modified | 5′ untranslated |
| CR46-5 | amt4-8m2 | Mini Tcr1 | 5′ untranslated |
| CR46-6 | amt4-8m3 | Two Tcr1 | 5′ untranslated and 5′ coding |
| CR46-7 | b | Mini Tcr1 | 5′ untranslated |
| CR46-8 | b | Mini Tcr1 | 5′ untranslated |
| CR46-9 | a | Tcr1 modified | 5′ untranslated |
| CR46-10 | c | Two Tcr1 | 5′ untranslated and 5′ coding |
| CR46-11 | c | Two Tcr1 | 5′ untranslated and 5′ coding |
| CR46-12 | a | Tcr1 modified | 5′ untranslated |
May be duplicates of amt4-8m1.
May be duplicates of amt4-8m2.
May be duplicates of amt4-8m3.
Figure 2.—
Sequence surrounding the seven separate sites of the TOC1 insertion in AMT4. The TOC1 inserts, which occurred at the position of the open space, are omitted. They were aligned left to right and their orientation with respect to AMT4 (F, forward; R, reverse) is given after the strain numbers. The sites of TOC1 insertion in CR159 and CR50-6 are the same as in CR49 and each transposon is oriented in the same direction. Conserved adenine nucleotides 2 bp from the insert site are in boldface type and the CA and TG dinucleotides near the insert sites are underlined (see text).
The TOC1 inserts in six strains (CR40, CR135, CR138, CR148, CR153, and CR50-6) appeared to be intact retrotransposons (5.66 kb; Day and Rochaix 1991a): the size of the PCR fragment amplified from the genome was ∼6 kb larger than expected (Figure 1B and not shown) and sequences of 700 bp from each end of the insert were identical to the GenBank sequences for the termini of TOC1 (X56231). The locations of these inserts are shown in Figure 1A and Table 1. One is diagrammed in Figure 3.
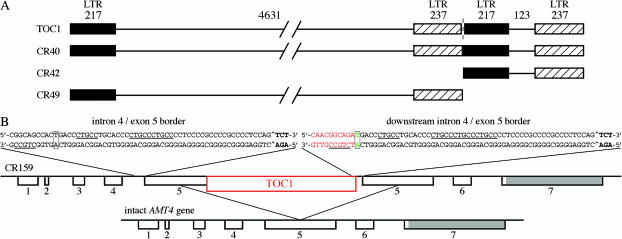
Figure 3.—
Diagram of TOC1 and TOC1-related inserts in AMT4. (A) Diagram of TOC1 and the inserts in CR40 (intact TOC1; 5662 bp), CR 42 (right half; 577 bp), and CR49 (left half; 5085 bp). LTRs are indicated in black and with cross-hatching. The direction of transcription of TOC1 is from left to right, as is the numbering of nucleotides, which corresponds to the standard numbering. The numbers of base pairs in each region are given above the line for TOC1. Intact TOC1 was also found in strains CR135, CR138, CR148, CR153, and CR50-6. (B) Diagram of modifications to AMT4 in CR159 and sequence near the left boundaries of the intron 4/exon 5 duplications that bracket the TOC1 insert (red). The diagram is oriented with respect to the AMT4 gene and therefore the TOC1 insert has the opposite orientation to that in A and the numbering is reversed. The start of exon 5 is indicated by boldface nucleotides and the normal splice site is indicated with a “^” in both upstream and downstream copies of the duplication. The boxed green “T/A” in the downstream copy is the common nt 161 of TOC1 and 1340 of the AMT4 gene. The boxed “T/A” in the upstream copy is nt 1340 of AMT4. Red sequence is from TOC1. Repeated CTGCC sequences (see text) are underlined. Exon numbers are indicated below the line for the AMT4 gene.
The TOC1-related inserts in two strains (CR42 and CR49) were only fragments of the retrotransposon (Figure 3). The 577-bp insert in strain CR42 is the “right” side of the transposon, consisting of the 217- and 237-bp segments of the LTR and the 123-bp unique region between them that is thought to contain the initiation site for replication. The 5-kb insert in CR49 appears to be the “left” side of the TOC1 transposon, consisting of the 217- and 237-bp segments of the LTR and the 4631-bp intervening region (Figure 3A). It is the precise complement of the insert in strain CR42.
The TOC1-related insert in CR159 was much more complex than others. The insertion site was the same as that in CR49, which carries the left side of TOC1, and CR50-6, which carries an intact element. In all three cases the insertion site was in the middle of the large exon 5 (Figure 3) and all three elements inserted with the same orientation—antiparallel to the direction of AMT4 transcription. However, the insert in CR159 lacks nucleotides (nt) 1–160 or 1–161 of TOC1 and carries in its place an almost exact duplication of the 734 or 735 nucleotides of AMT4 preceding the insert site. The duplication appears to lie just downstream of the insert site and to differ from the corresponding upstream sequence only by the presence of an additional repeat of the five nucleotides CTGCC in intron 4 (all copies of the repeat are underlined in Figure 3). The nucleotide in the downstream duplication in green in Figure 3 is common between AMT4 (nt 1340) and TOC1 (nt 161). The experimentally determined start of TOC1 transcription is the “G” at 165 (Day and Rochaix 1991b), which lies at the middle of the underlined sequence in red in Figure 3B. Although the AMT4 transcript could theoretically be correctly spliced in CR159 if intron 4, the upstream duplication of exon 5, and TOC1 were treated as a single intron of ∼6.5 kb, this apparently did not occur because CR159 behaved like an amt4 null strain with respect to its level of methylammonium resistance and its methylammonium uptake (data not shown). Oddly, the splice junction involved, which is indicated by a “^,” is the same one altered in CR50 (Kim et al. 2005).
Gulliver-related inserts:
One strain with a Gulliver-related insert (CR165) was obtained from the second selection and another with a more complex history (CR07) in the first study (Kim et al. 2005). Lesions in the two strains were very similar (Figure 4). The insertion of 238 bp in CR165 is located between 8-bp target-site duplications of GCTACGGC in exon 3 of AMT4 and is near the site of the TOC1 insertion in CR135. The 15-bp imperfect inverted repeats at the ends of the CR165 insert are identical to those of Gulliver and the similarity to the Gulliver sequence extends for another 18 nucleotides on the left and 45 nucleotides on the right. On the basis of the termini of the Gulliver sequence (AF019750 and AF019751), 14 Gulliver transposons are present in the JGI Chlamydomonas genome, a number consistent with an earlier estimate based on Southern analysis (Ferris 1989). Each of these has at least one sequencing gap. No other section of the Gulliver sequence is similar to the sequence of the insert in CR165. A Blastn search indicated that the 238-bp insertion in CR165 is closely related to >200 sequences in the Chlamydomonas genome.
Figure 4.—
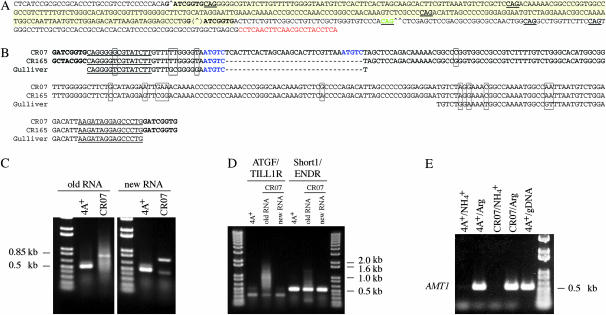
Gulliver-related elements in AMT4 and effects of the CR07 insert on splicing (RT–PCR). (A) Sequence of the AMT4 gene of CR07 from the end of intron 3 to the end of exon 4. Target-site duplications are in boldface type and the insert sequence is highlighted in yellow. The splice site leading to the large RT–PCR product (C) is indicated with a “^” and the splice site leading to the small RT–PCR product is indicated with a “^^.” The position of the splice site used in parental strain 4A+, which was destroyed by the insertion, is indicated with a “(^).” Potential 3′ splice junction signals are underlined and the splice signal leading to the small RT–PCR product (C) is in green. The complement of the TILL2R primer (see text) is in red. (B) Comparison of the inserts in strains CR165 and CR07 with each other and with Gulliver termini (AF019750 and AF019751). Only Gulliver sequence similar to that of inserts in CR165 or CR07 is shown. The target-site duplications are in boldface type and the terminal inverted repeats are underlined. In addition, CR07 contains an insert of 32 bp with respect to CR165 (indicated by dashes for CR165). It is flanked by a 5-bp target-site duplication in purple. Nucleotides that differ between the three sequences are boxed. (C) RT–PCR products for the beginning of AMT4 from strains 4A+ and CR07. Products were amplified from RNA prepared in October 2002 (old RNA) and February 2004 (new RNA) with the ATGF and TILL2R primers (Figure 1). Strains were grown on arginine as nitrogen source. (D) RT–PCR products for 5′ and 3′ regions of AMT4 that bracket the region of the CR07 insertion. Primers were ATGF and TILL1R or Short1 and ENDR, respectively. The TILL1R primer is at the 3′-end of exon 3 and its sequence is given in Kim et al. (2005). (E) RT–PCR of the AMT1 gene from strains 4A+ and CR07 grown on arginine or ammonium. “Old RNA” was used for CR07. The PCR product from 4A+ genomic DNA (gDNA) is shown for comparison. Primers were AM2 and AM3 (Kim et al. 2005).
The insertion of 270 bp in CR07 (highlighted in yellow in Figure 4A) is located between 8-bp target-site duplications of GATCGGTG that disrupt the splice junction between intron 3 and exon 4 of AMT4. The 15-bp imperfect inverted repeats at the ends of the insertion differ from those of Gulliver by only a single nucleotide. Similarities to Gulliver extend for an additional 18 nucleotides on the left and 45 nucleotides on the right, as they do for the insert in CR165. Between these regions of similar sequence, the CR07 insert carries an additional 32 nucleotides not present in the CR165 insert (Figure 4B). The 32 nucleotides include a 5-bp target-site duplication of Gulliver-related sequence, ATGTC, and a 27-nucleotide fragment of the MRC1 transposon discussed below. The entire 270-bp insert of CR07 is present in only one copy in the Chlamydomonas genome (JGI v3.0, Scaffold_2 at 2791714–2791983) and that copy is identical to it. Nine other copies of the 238-bp insert in CR165 are also located on Scaffold_2.
To ascertain the effects of the CR07 insert on splicing, we amplified AMT4 mRNA by RT–PCR and determined the sequence of the two RT–PCR products obtained (Figure 4C “new RNA”). Both RNA and DNA were prepared ∼21/2 years after the strain was isolated. The larger product was due to splicing at the original splice site between intron 3 and exon 4 to add 278 nucleotides to the transcript and the smaller product was due to splicing at the first CAG within exon 4 to reduce the mature transcript by 44 nucleotides at the exon 4 start (Figure 4A). Although there are three potential splice sites within the insert (Figure 4A), none was used.
We noted with interest that RT–PCR products for AMT4 from CR07 were different from those that we had observed from RNA prepared earlier (Figure 4C “new RNA” vs. “old RNA”). The earlier RNA preparation, which had been made about a year after CR07 was isolated, yielded one prominent band corresponding to the larger product described above and a very visible smear around it (see Figure 5A of Kim et al. 2005). Obtaining the smear from the earlier RNA preparation was reproducible (Figure 4C “old RNA”) and unique to mRNA from CR07 (Kim et al. 2005 and additional data not shown for CR40 and CR42, which gave no RT–PCR product for AMT4 under identical conditions). Smearing also occurred with the “old” mRNA preparation in regions of AMT4 that did not carry the insertion (Figure 4D), but was unique to AMT4; i.e., amplification of mRNA for AMT1-3 was normal in CR07 (Figure 4E and data not shown). These results indicated that CR07 had changed during the 11/2 years between the times the RNA preparations were made, during which CR07 had been maintained nonselectively on TAP medium with NH4Cl as nitrogen source. Sequence of the RT–PCR smear from “old RNA” (Figure 4C) was the same as that for the larger product obtained from “new RNA” and showed minimal background contamination. To attempt to learn more about background products, we cloned 22 of them (see materials and methods) and determined their sequences. Two clones had sequence corresponding to the larger product described above; 17 had sequence for TILL2R, one of the two amplification primers, at both ends; 1 had the TILL2R primer at one end and no primer at the other; and 2 were unreadable. Each of 14 clones that had TILL2R at both ends contained a different DNA fragment that was nearly identical to a unique sequence at the JGI database. No Gulliver-related sequence was nearby, but in five cases a putative retrotransposon-like polyprotein (POL) gene was close. In all instances, the border sequences had some resemblance to the TILL2R primer, but at only one end. Unfortunately, we had not isolated DNA earlier and so could not explore directly whether the nature of the insert in CR07 had originally been different from the one that we characterized. However, on the basis of a comparison of sequences for the “old” and “new” RNA preparations by RT–PCR and the unique copy of the CR07 insert on Scaffold_2, we suspect that the insert itself did not change over time.
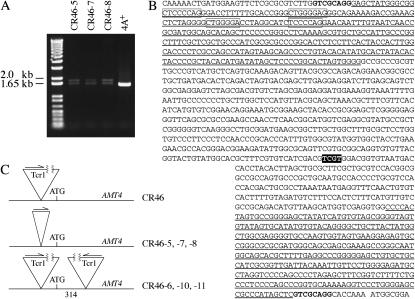
Figure 5.—
Tcr1 elements in AMT4. (A) RT–PCR products obtained from derivatives of CR46 carrying small Tcr1-related inserts that remain at the original insertion site (CR46-5, CR46-7, and CR46-8). The primers were −1577F and ATGR (Figure 1). (B) Complete DNA sequence of the small inserts (mini Tcr1) in CR46 derivatives CR46-5, CR46-7, and CR46-8. Terminal inverted repeats of 314 bp characterized in this work are underlined. Direct target-site repeats are in boldface type. The boxed sequences are examples of those that could pair with one another to yield stem-loop structures (1 with 2 or 3, or 2 with 4). The TCTG sequence present in one copy in mini Tcr1 but two copies in Tcr1 (see results) is indicated by letters on a solid background. The “ATG” translational start of AMT4 is indicated by a space just preceding it (bottom line). (C) Diagram of the Tcr1 insert in CR46, smaller inserts in the derivatives characterized in B, and derivatives that carry two Tcr1 inserts (CR46-6, CR46-10, and CR46-11). Arrows indicate the orientation of Tcr1.
In addition to the insertion between intron 3 and exon 4, CR07 also carries a macrolesion upstream of AMT4 (Kim et al. 2005), which we were unable to characterize. We know this macrolesion lies upstream of −785, because we could amplify the entire region from −785 to the 3′-end of AMT4 by PCR. However, even with long extension times, we could not amplify the segment from −1577 to −785. We did not attempt to localize the lesion further within this 800-bp region. Attempts to amplify portions of the lesion with nested primers upstream of AMT4 and random primer sets were unsuccessful, as were attempts using upstream AMT4 primers and primers internal to the Gulliver-like insertion within the AMT4 gene in CR07. We know that the Gulliver-like primers were properly designed, because we could amplify sequences adjacent to Gulliver-like elements elsewhere in the genome.
Tcr1-related inserts:
One strain with a Tcr1 insertion (CR46) was obtained from the first selection. We were able to partially characterize the insertion in this strain only after characterizing derivatives of it (mini Tcr1) and it is discussed more fully below. The CR46 insert was upstream of the ATG start for AMT4 and the strain was resistant to only 0.1 mm methylammonium. All 12 derivatives of the strain selected at 1 mm methylammonium also had Tcr1 inserts that apparently derived from the original one. These were of three different types (Figure 5C). The derivatives that were most tractable molecularly (CR46-5, CR46-7, and CR46-8), which may not have occurred independently (Table 1), had lesions of reduced size at the same position as the lesion in CR46. The target-site duplication of GTCGCAGG between −15 and −8 relative to the ATG start of AMT4 remained. Even these smaller inserts were difficult to amplify by PCR and difficult to sequence, apparently because they had complex internal structure due to multiple long inverted repeats. With a variety of primers external to the inserts (−1577 and ATGR, −1577 and +500R, −89F and +500R, and −785F and +500R; materials and methods and Kim et al. 2005), we obtained two PCR products that appeared to differ in size by ∼200 bp and whose sizes suggested an insert in the range of a few base pairs to ∼200 bp (Figure 5A). We thought initially that the larger of these PCR products carried the entire insert and that the smaller arose because DNA polymerase jumped across the insert. However, when we tried to clone the DNA from these bands, we did not find any clones with >∼20 bp of insert. Thus, both products were apparently small and were artifacts of the PCR reaction. We also attempted to sequence these two PCR products directly using the AMT4 primers listed above but could read only the target-site duplication and 10 bp into the inverted repeats. Although the sequence beyond that was very difficult to read, we guessed at additional sequence until we had enough to search the JGI database. By that means, we found sequences that looked like the ends of the sequence originally reported for Tcr1 (Ferris et al. 1996) and designed primers internal to Tcr1 (see materials and methods). Use of one external primer with one internal to Tcr1 allowed us to obtain good PCR products and sequence, which revealed that the insert size, 1.35 kb, was much larger than we had expected (Figure 5B). Moreover, the perfect terminal inverted repeats were 314 bp rather than the 140 bp reported previously (Ferris et al. 1996). Not only were the inverted repeats unusually long, but also within them were contained smaller direct and inverted repeats (indicated in Figure 5B) that could give rise to a wide variety of secondary structures. The four inverted repeats that are boxed are examples of ones that could give rise to alternative stem-loop structures (see Figure 5 legend). The same inverted repeats could also form structures with sequences in the long inverted repeat at the other end of the insert. A complex DNA secondary structure or structures may account for the difficulties of amplifying the small Tcr1-derived inserts, which we call mini Tcr1, by PCR and sequencing them. The complete 1.35-kb sequence for mini Tcr1 was identical to the “Tcr1” sequences on JGI Scaffold_2 (1,463,060–1,486,080; 3,005,034–3,018,530) and Scaffold_17 (503,592–513,076).
Using the same Tcr1 and AMT4 primers described above, we verified that CR46 itself carried Tcr1. We were able to amplify a 5.8-kb fragment from the left end of the transposon and a 1.2-kb fragment from the right end (using primer pairs −110F and 5820R and +500R and 7365F, respectively). From the 5.8-kb fragment we obtained 850 bp of sequence adjacent to AMT4 and 1 kb of sequence at the opposite end. We obtained the sequence of the entire 1.2-kb fragment. We were unable to amplify the Tcr1 sequence between the endpoints of the 5.8- and 1.2-kb fragments. Our inability to amplify internal sequences might have been due to structural complexity problematic for PCR or to a gross underestimate of transposon size (Ferris et al. 1996). Indeed, “giant” transposons of >100 kb with multiple foldback elements (see discussion) have been described in Drosophila (Lovering et al. 1991).
Three derivatives of CR46 (CR46-6, CR46-10, and CR46-11), again not necessarily independent, appear to retain the original lesion present in CR46 and to have acquired a second Tcr1 lesion in exon 2 of AMT4 (Figure 5C). The second copy is flanked by the 8-bp target duplication GTGCCGGT and is separated from the first by 314 bp, the precise length of the Tcr1 terminal inverted repeat. On the basis of the sizes of the 5.8- and 1.2-kb fragments, the two inserts appear to be the same but opposite in orientation. This was confirmed by 500 bp of sequence from the end of the 5.8-kb fragment adjacent to AMT4 and the sequence of the entire 1.2-kb fragment.
Tcr1 lesions in the remaining derivatives of CR46 (CR46-1, CR46-2, CR46-3, CR46-4, CR46-9, and CR46-12) appeared to remain at the original site, and we were able to verify that they have the same termini as the insertion in CR46 (the same sequence for 500 bp and 1.2 kb at the left and right ends, respectively, and a fragment of 5.8 kb amplified with the −110F and 5820R primers). Like the insertion in CR46, insertions in these derivatives have been resistant to further analysis. We have been unable to account for the increased methylammonium resistance of these derivatives with respect to CR46.
The newly described Bill element:
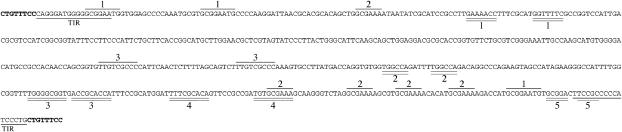
One strain carrying a newly described element (CR03) was obtained in the first study (Kim et al. 2005). The element is located at −295 relative to the translational start of AMT4 and confers resistance to 0.1 mm methylammonium but not 1 mm. It is 558 bp long (Figure 6) and flanked by an 8-bp target-site duplication. The element ends in perfect 17-bp inverted repeats and contains other direct and inverted repeats internally that may contribute to a complex structure. Major portions of the element appear at 15 locations on 14 scaffolds around the JGI assembly of the C. reinhardtii genome. At 7 of these locations the entire 558 bp are present with ≥98% identity to the element in CR03. In 6 of these 7 cases, the element is flanked by an 8-bp direct repeat, presumably of the target sequence. Each direct repeat is different and none is the same as the one in CR03. In one of the 7 cases, the insert is within the putative reverse transcriptase gene of a homolog of the Chlorella Zepp retrotransposon (Higashiyama et al. 1997). In no instance was the insert sequence in CR03 found to be part of a longer sequence in the JGI assembly, suggesting that it is an intact transposable element, not a fragment of a larger transposon. In keeping with literary tradition for naming transposable elements, we propose that the insert in CR03 be called “Bill” after our former president. The resemblance to the name of one of the authors and a 16th-century playwright is coincidental.
Figure 6.—
Sequence of the newly described “Bill” element. The sequence of the insert in CR03 is given with the 8-bp target-site duplication in boldface type and the terminal inverted repeats (TIR) underlined. Internal inverted repeats are double underlined and numbered and internal direct repeats are overlined and numbered. They give a sense of the complexity of the internal structure of the element.
Elements related to the newly described MRC1 element:
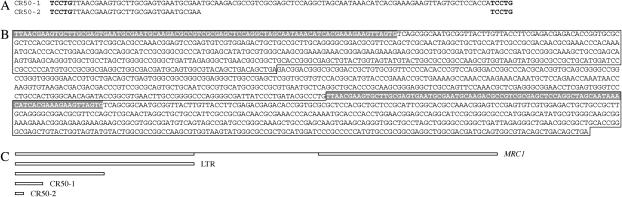
The two strains with lesions related to the newly described MRC1 element (see below and discussion) were isolated from CR50 in the third selection. CR50 carries a single base change in intron 4 of AMT4 (Kim et al. 2005), and derivatives CR50-1 and CR50-2 carry insertions after nucleotide 620 in exon 3. In each case, there is a 5-bp duplication of the target sequence TCCTG. Between this duplication CR50-2 carries an additional 29 bp and CR50-1 carries an additional 94 bp, the first 29 of which are identical to those in CR50-2 (Figure 7A). A Blastn search of the C. reinhardtii genome using the 94-bp sequence found in CR50-1 yields >100 related sequences. Examination of these indicated that the 94-bp sequence was at the terminus of a 600-bp sequence that was also abundant in the Chlamydomonas genome. Moreover, in several JGI scaffolds, the 600-bp sequence brackets a sequence of 420 bp, the same in all cases, and thus appears to serve as an LTR for a 1.6-kb element (Figure 7B). We propose to call this putative retrotransposon MRC1 (miniature retrotransposon of Chlamydomonas) for reasons given below and in the discussion. One of the complete MRC1 elements is in the last intron of the gene for the catalytic subunit of the DNA primase associated with the DNA polymerase α. This MRC1 element is only 4 kb away from an element that was annotated by Patrick Ferris as a 6-kb “Osser”-like retrotransposon of Volvox, complete with 217-bp LTRs (Lindauer et al. 1993), apparently the only such element in the C. reinhardtii genome.
Figure 7.—
Sequences related to the newly described MRC1 element. (A) Sequence comparison for the CR50-1 (94 bp) and CR50-2 (29 bp) inserts. Each insert has the same 5-bp target-site duplication (boldface type) and the first 29 bp of CR50-1 are identical to those of CR50-2. (B) Sequence of the putative retrotransposon MRC1. The LTRs are boxed and the sequence identical to that of CR50-1 is indicated by letters on a shaded background. (C) Diagram of intact MRC1, a single LTR, and portions of the LTR in CR50-1 and CR50-2. Unique sequence in the intact element is indicated with a single line.
In addition to the large number of copies of the putative LTR for MRC1 in the Chlamydomonas genome, there are a large number of fragments of it, all of which begin with the 29-bp sequence present in CR50-2 and extend for various lengths (Figure 7C). Many of these fragments are flanked by 5-bp duplications, which are presumably target-site duplications. We postulate that the inserts in CR50-1 and CR50-2, like the fragments in other places in the genome, are residues of insertion and decay of the MRC1 element. If they occurred as different decay events from the same original insertion, the original insertion probably occurred in the CR50 culture before it was subjected to selection for a higher level of methylammonium resistance. The decay events may have occurred either before or after exposure to 1 mm methylammonium.
Summary of macrolesions:
In our first study, which included selections for methylammonium resistance at both low and high concentrations (Kim et al. 2005), we obtained 12/16 strains with lesions in AMT4. Of these, six had transposon-induced macrolesions (three TOC1 related, one Gulliver related, one Tcr1 related, and one carrying the Bill element) (Table 2). In a second selection for methylammonium resistance, we obtained 48 strains, of which 17 carried macrolesions or putative macrolesions in AMT4. Of the seven macrolesions that we were able to amplify by PCR and therefore characterize, five were TOC1 related, one was Gulliver-related, and one was a deletion. With the exception of the TOC1 insert in an intron (CR138), the macrolesions that we characterized resulted in complete loss of function of AMT4, as assessed by loss of [14C]methylammonium uptake in strains carrying them (data not shown) and by the high level resistance of these strains to the analog. In the third selection, we studied 18 derivative strains resistant to higher levels of methylammonium than their parental strains. Fifteen of these carried transposon-related lesions, although they did not necessarily arise independently. Strikingly, all derivatives of a strain initially carrying Tcr1 in the promoter region for AMT4 carried Tcr1-related secondary lesions. By contrast, derivatives of a strain carrying a base-pair substitution at a splice site within AMT4 occurred less frequently and no secondary lesion was related to the primary lesion. Although two derivatives carried MRC1-related lesions, these presumably occurred de novo.
Mapping of AMT4:
To better define the AMT4 system, we located the gene. Strain CR02, which carries a 2-bp deletion in exon 2 (Kim et al. 2005), was crossed to a number of mapping strains. Early crosses to strains with centromere-linked markers ac17 [linkage group (LG) III], pf2 (LG XI), y-1 (LG XVI/XVII), pf27 (LG XII/XIII), and pf10 (LG XIX) yielded a distance of 30–35 cM from the centromere. A cross between CR02 and CC-604 (pf3) yielded a ratio of parental ditype:nonparental ditype:tetratype tetrads of 9:1:24, which indicates that AMT4 is ∼45 cM from the PF3 locus. Together, the results place AMT4 on the right arm of linkage group VIII at a greater distance from the centromere than any genetic marker previously mapped to chromosome 8. The current version of the JGI Chlamydomonas genome (v3.0) places AMT4 on Scaffold_2 (1,198,141–1,202,241). Two of three Tcr1 sequences, nine Gulliver-related sequences of the type found in CR165, and the unique Gulliver-related sequence of CR07 are also assigned to Scaffold_2. In conflict with our findings, two physical markers for LG XIV, GP221 and GP324, have been assigned to Scaffold_2, but the assignment appears tentative.
DISCUSSION
Spontaneous mutations to methylammonium resistance at the AMT4 locus of C. reinhardtii carry signatures of five different transposable elements, including examples in both major classes. Two of the elements and a number of the events that occurred are novel. The variety and frequency of transposon-related events at AMT4 suggest that the Chlamydomonas genome is dynamic with respect to both the mobilization and rapid disintegration of transposons. Below we discuss new findings about previously described transposons and the novel transposable elements that we encountered. Then we return briefly to the usefulness of the AMT4 system and the evolution of the Chlamydomonas genome.
TOC1:
The previously described retrotransposon TOC1 or fragments of it have been found associated with a number of mutated loci in Chlamydomonas (Moseley et al. 2002), but movement of the transposon into a specific locus has not been encountered during the course of a mutant selection since the initial description of its movement into the oxygen-evolving enhancer gene OEE1 (Day et al. 1988). We observed nine independent events due to TOC1, three of which we had briefly described earlier (Kim et al. 2005). From nine inserts, we were able to deduce common features of the target site for TOC1 (Figure 2 and results). The periodicity of TOC1 insertion sites along the AMT4 gene (243–280 nucleotides) suggests a relationship between the choice of target site and chromatin organization, particularly nucleosome placement. Nucleosome structure appears to be an important factor in target-site selection for retroviral integrases generally (Pryciak and Varmus 1992; Müller and Varmus 1994; Pruss et al. 1994), as do particular features of DNA structure (bending) and sequence associated with nucleosome formation (Craig 1997). The fact that three of nine independent TOC1 insertions (those in CR49, CR159, and CR50-6) occurred at exactly the same site and with the same orientation in AMT4 suggests that more specific local features of DNA sequence or structure may also contribute to target-site selection for this retrotransposon.
We can infer from the mechanism of transposition of TOC1 that the mutations in CR42 and CR49, which carry complementary portions of the transposon, probably occurred as a consequence of two events. Transposition of intact TOC1 was probably followed by excision of portions of the transposon by homologous recombination between the 217- or 237-bp regions, respectively, of its unusual long terminal repeats. The type of fragmentation of TOC1 seen in CR42 was observed previously in a partial revertant obtained from a TOC1 insertion into the OEE1 gene (Day et al. 1988). In that case, the original mutation was in intron 2 of the gene so the reversion event did not require precise excision. As others have noted (Schnell and Lefebvre 1993), screens to detect mobile elements by virtue of unstable mutations have not generally worked for retrotransposons: their mode of replication and gradual incomplete disintegration over time do not lend themselves to reconstruction of the parental genotype.
The sort of TOC1-related insertion in strain CR159 has not been seen previously. CR159 carries a nearly complete copy of TOC1 between almost identical 734-bp direct repeats of AMT4 sequence from exon 5 (Figure 3). The repeats bear analogy to the long terminal repeats of the transposon and, together with information on the portion of TOC1 that is deleted, suggest a mechanism by which the insertion could have arisen. The size of the duplication, 739 bp (734 + 5 bp repeat), minus the size of the TOC1 deletion, 160 bp, is 579 bp, which is very close to the size of the “right side” of TOC1 (i.e., the portion present in CR42). The portion of TOC1 deleted is that just prior to the start of TOC1 message (Day and Rochaix 1991b). These features suggest that TOC1 originally inserted correctly and was then replicated abortively. In the secondary event, the primer for TOC1 replication would presumably have been the AMT4 transcript rather than a tRNA and reverse transcriptase would have copied the AMT4 sequence upstream of the insertion. The resulting DNA would have circularized with TOC1 nt 161 juxtaposed to AMT4 nt 1340. The insert in CR159 would then have come about by completely replacing the original TOC1 insert with this modified form. Replacement could not have come about by simple homologous recombination and may have required gene conversion or a complex interaction between the original insert and the modified TOC1. The secondary event postulated, which was possible because AMT4 was actively transcribed, offers a remedy to the problem of permanent gene inactivation by retrotransposons. The remainder of TOC1 in CR159 is bracketed by a duplication of AMT4 sequence and could be precisely excised by homologous recombination to restore parental AMT4 sequence. Unfortunately, we do not yet have a selection for reversion of mutations in AMT4 (see below) and hence cannot monitor for such an event. If the type of replication error that produced the lesion in CR159 occurs generally when TOC1 inserts into actively transcribed genes, it would allow true reversion of lesions caused by TOC1 and allow TOC1 to maintain a congenial relationship with the Chlamydomonas genome. It would also make TOC1 useful in transposon tagging of genes, as proposed for the Tos retrotransposons of rice (Hirochika et al. 1996).
Since the discovery of TOC1 (Day et al. 1988), its termini have been used as a measure of repetitive element frequency in genomic studies of C. reinhardtii (Li et al. 2003), and TOC1 mobilization, as assessed globally by Southern analysis, has been used to assess loci involved in transgenic transcriptional silencing (Wu-Scharf et al. 2000; Jeong et al. 2002). Silencing and suppression of transposon movement appear to share common regulatory elements. We think the AMT4 system will provide a useful adjunct to global monitoring tools by allowing direct assessment and quantitation of transposition per se, as opposed to transposon disintegration or rearrangement, and by allowing explicit molecular characterization of the events involved.
Gulliver:
We obtained two strains, CR165 and CR07, carrying small inserts with imperfect terminal inverted repeats similar to those of Gulliver (Ferris 1989). These inserts occur in >200 copies in the C. reinhardtii genome and may be members of a family of small elements (Ds) capable of transposition only with the assistance of other functions (McClintock 1947, 1951). Alternatively, these elements may belong to the transposon family known as miniature inverted repeat transposable elements, distinguished as nonautonomous elements of high copy number in plant genomes (Feschotte et al. 2002). In either case, on the basis of the similarities of the terminal inverted repeats, Gulliver may have served as the corresponding Ac to mobilize these elements into AMT4.
The Gulliver-like element in CR07 differs from that in CR165 by virtue of a 32-bp internal sequence, which appears to be derived from a mini-retrotransposon (see MRC1 below). There is a single identical copy of this element in the Chlamydomonas genome and hence it is this element that probably moved into AMT4 and excised from its original location. Subsequent movement of the element out of AMT4 could also be followed by virtue of the tag.
We considered several possible explanations for the difference between the “old” and “new” RNA samples from CR07 (see results) that could have led to the differences in RT–PCR products shown in Figure 4. First, the insert itself might have changed; second, variable splicing might have occurred initially; third, AMT4 mRNA might have been degraded due to an RNA interference (RNAi) effect caused by the presence of the insertion in the message; fourth, the Gulliver-like insertion in CR07 might have been mobilized at high frequency when the strain was first isolated. Unfortunately, as sketched below, there are problems with each explanation. It is unlikely that the insert in CR07 changed because it carries a unique tag that distinguishes it from >200 similar inserts in the Chlamydomonas genome. Variable splicing to yield the smear of products obtained by RT–PCR from “old RNA” is difficult to reconcile with the clean sequence obtained from the smear and the nature of the sequences cloned from it (see results). Although degradation of AMT4 mRNA by RNAi accounts for obtaining smears for RT–PCR products in all regions of AMT4, even those that did not contain the insertion, it is not easily reconciled with sequences of the clones mentioned above and it is not obvious why similar effects did not occur with other insertions. Finally, mobilization of the 270-bp insert along with a portion of AMT4 provides a fairly straightforward explanation for sequences of the clones but does not easily account for obtaining RT–PCR smears from regions of AMT4 that did not carry the insertion.
The CR07 strain contains an uncharacterized macrolesion between −785 and −1577 upstream of AMT4. We could obtain no evidence that this lesion was Gulliver-related and the behavior of CR07 in genetic crosses—reduced zygote survival and germination rates without skewing of the ratio of methylammonium resistant to sensitive segregants (Kim et al. 2005)—could be easily explained if the upstream lesion were a chromosomal rearrangement. Although we do not have a satisfactory explanation for it, evidence that CR07 has changed during the time we studied it is reasonably strong. If the changes are related to Gulliver, they may occur again in strains with less complex histories, which would allow them to be better characterized.
Tcr1:
Tcr1 (present in CR46) is a foldback (FB) transposon (Potter et al. 1980): at 314 bp, its long inverted repeats are much longer than previously believed (140 bp; Ferris et al. 1996), are internally complex, and probably give rise to cruciform (foldback) structures in both DNA and RNA (Figure 5; see results). Tcr1 is >7 kb in length and longer and/or more complex in sequence and/or structure than previously thought.
A small derivative of Tcr1, mini Tcr1 (present in strains CR46-5, CR46-7, and CR46-8), is 1.35 kb in size and is present at three locations in the Chlamydomonas genome (scaffolds 2 and 17). It contains 830 bp from the left side and 511 bp from the right side of Tcr1 (original insert in CR46; Figure 5B). The segment of Tcr1 that was deleted is bounded by two copies of the 4-bp sequence TCGT, which occurs only once in mini Tcr1. Given the secondary structure that can be assumed by Tcr1, the deletion may have arisen by slipped mispairing during DNA replication (Streisinger et al. 1966).
The Tcr1 insert in CR46 appears to be present in two copies in its derivatives CR46-6, CR46-10, and CR46-11. The duplicated copies of Tcr1 are facing one another (Figure 5C). The secondary event producing these derivatives may have been a “continuation” of the transposition process. Tcr1 appears to have measured a precise distance, 314 bp, which is the length of its long inverted repeats, and inserted again in the opposite orientation. The second event was accompanied by a new target-site duplication.
The Tcr1 element in CR46 was located at −7, just prior to the ATG start of AMT4. AMT4 was transcribed in CR46 not only under the appropriate nitrogen-limiting growth conditions (González-Ballester et al. 2004; Kim et al. 2005), but also when cells were grown with NH4Cl as the nitrogen source (data not shown). Due to the size and location of the transposon, it is likely that transcripts originated within it, although this was not demonstrated. CR46 was resistant to only low levels of methylammonium. Derivatives resistant to higher levels of the analog were readily obtained and all contained lesions related to Tcr1. Resistance of derivatives in which there was a second copy of the transposon within AMT4, described above, was easily understood. More interesting were the derivatives in which a portion of the transposon excised to yield mini Tcr1. Whereas mini Tcr1 may lack a promoter or completely disrupt function of the AMT4 promoter, it is also possible that transcripts are made and that the long inverted repeats of Tcr1 form double-stranded RNA that results in their destruction (RNAi effect). These issues remain to be studied.
FB elements, first described by Potter et al. (1980), have since been seen in many organisms, including plants (Rebatchouk and Narita 1997), and have been shown to play a role in gene silencing (Puig et al. 2004). Although their mode of transposition remains unknown, they are considered to be class II elements. They are known to be associated with chromosomal rearrangements (Caceres et al. 1999; Kidwell and Holyoake 2001) and the relationship is probably causal: pairs of oppositely oriented FB elements lead to inversions and translocations and thus to exon shuffling and genetic variation (Kidwell and Lisch 2001; Moschetti et al. 2004). Studies of FB elements in various organisms have offered compelling hypotheses as to how such rearrangements occur but have not addressed the origin of the FB pairs required. As discussed above, we were able to monitor directly the events that produced such a pair of Tcr1 elements in CR46 and derivatives like CR46-6. Homologous recombination between the two Tcr1 elements in the derivatives would generate an inversion within AMT4 and homologous recombination between one of the two Tcr1 elements and other similar elements in the Chlamydomonas genome could lead to translocations.
The “Bill” element:
The newly described Bill element is a 558-bp element carrying terminal inverted repeats different from those of Gulliver and apparently causing 8-bp target-site duplications (Figure 6 and results). Thus, it appears to be a class II element of the Ds type (McClintock 1947, 1951). Our finding it at the AMT4 locus demonstrates that the Bill element is mobile. Although there are repetitive elements of a size similar to Bill throughout the Chlamydomonas genome (v3.0), to our knowledge none has been confirmed as mobile. Whereas the TOC1 and Gulliver-related elements or identifiable fragments of them appear frequently in the Chlamydomonas genome (∼40 and ∼200 occurrences, respectively), the Bill element occurs only 15 times and with little sequence variation. Given that Ds elements have been useful for gene disruption and transposon tagging (Bancroft and Dean 1993; Takken et al. 1998; Kuromori et al. 2004), the Bill element might be useful for these purposes in Chlamydomonas (Kempken and Windhofer 2001). Clearly its value would be increased if the corresponding Ac element were known and could be controlled. The Ds element Jordan has been used extensively for gene inactivation and tagging in Volvox (Miller et al. 1993; Kirk and Nishii 2001).
The Bill element in CR03, the only strain carrying such an element, was located at −295 with respect to the ATG start of AMT4. As for CR46, transcription of AMT4 occurred under nitrogen excess as well as under nitrogen-limiting conditions (data not shown). Whether transcripts came from within the Bill element and hence were not regulated or regulation of AMT4 transcription was disrupted was not assessed. CR03 was resistant to only low levels of methylammonium, but derivatives resistant to higher levels of the analog did not occur frequently. Thus we obtained no evidence that the Bill element could readily rearrange or transpose to nearby locations.
MRC1:
The small 94- and 29-bp insertions in two of our strains (CR50-1 and CR50-2) appear to be left terminal fragments of a putative miniature retrotransposon of 1.6 kb (Figure 7). This newly described element, which we propose to call MRC1 for miniature retrotransposon of Chlamydomonas, consists of an ∼420-bp unique sequence bracketed by ∼600-bp LTRs. It is the first transposon in miniature (TRIM) to be described in Chlamydomonas. We postulate that the inserts in CR50-1 and CR50-2, like fragments in other places in the genome, are residues of insertion and decay of the MRC1 element. Like the insertions in CR50-1 and CR50-2, 6 of 12 related fragments in the Chlamydomonas genome that we checked appear to be flanked by 5-bp target-site repeats.
TRIMs have been described in a number of plants including Arabidopsis (the Katydid elements), maize, rice, legumes, and tobacco (Witte et al. 2001; Havecker et al. 2004). By virtue of their movement and rapid disintegration, they are implicated in the restructuring of plant genomes. MRC1 appears to have moved into AMT4 and left behind small pieces in CR50-1 and CR50-2. Fragments of TRIMs accompanied by target-site duplications are found throughout plant genomes, often as solo LTRs. This is also true for MRC1. Plant TRIM-related sequences are reported to be associated with nested retrotransposons. On Scaffold_1 of the Chlamydomonas genome, a sequence identical to the MRC1-related insert in CR50-1 appears to be nested within a miniature transposable element that has LTRs of 464 bp. As noted above, it is also nested in the Gulliver-related element of CR07.
The AMT4 system:
The AMT4 system offers several advantages for the study of transposition. First, even at low levels of analog (50–100 μm), most lesions to methylammonium resistance (80%) lie within this one gene. Second, because methylammonium resistance does not kill Chlamydomonas, but slows its growth, mutational lesions yielding resistance can occur in the presence of the analog (i.e., Chlamydomonas can evolve under selective pressure) and, indeed, dark-green colonies appear over the white background of parental cells for many weeks. The system has proved useful for monitoring not only spontaneous transposon-induced lesions but also UV-induced mutations (J. Hsu, K.-S. Kim, W. Inwood and S. Kustu, unpublished results). After UV mutagenesis, dark-green colonies grew up rapidly (within a week) and the 14 strains that we characterized at the molecular level had microlesions that were easily rationalized in terms of the mode of action of UV. Selection was at high methylammonium (1 mm) and the lesions appeared to completely inactivate AMT4. Hence, inactivation of the gene does not require the transposon-related events that we have observed so frequently among spontaneous mutants. Rather, transposon-mediated events occur as a result of the behavior of the Chlamydomonas genome. A third advantage of the AMT4 system is that selecting for spontaneous resistance to low levels of methylammonium (50 or 100 μm) can be used to isolate and study transposable elements in the promoter region and 5′ UTR for the gene. We obtained two such events (a Tcr1 insertion in CR46 in the 5′ UTR and the Bill insertion in CR03 in the promoter region) by doing so. CR46 readily gave rise to derivatives resistant to higher levels of methylammonium (1 mm), all of which carried lesions due to rearrangement or further transposition of Tcr1. CR03 did not give rise to derivatives readily and therefore we have no reason to believe that the presence of a Bill element at a particular location facilitates its subsequent rearrangement or movement nearby. In one instance (CR148), insertion of the TOC1 transposon into the AMT4 promoter region yielded resistance to 1 mm methylammonium initially. We have not explored in depth the effect of transposon insertions in the AMT4 promoter region on transcription of the gene or its regulation. In light of current evidence that single regulatory changes can account for major morphogenetic differences in natural populations of plants and animals (Shapiro et al. 2004; Wang et al. 2005), this is a fertile area for future study. Finally, selecting for resistance to low levels of methylammonium can yield transposons in introns of AMT4 (e.g., TOC1 in CR138). CR138 readily yielded derivatives resistant to higher levels of methylammonium, perhaps by means of an event similar to that which occurred in CR159 or an additional nearby transposition event. Thus the system is useful for studying the behavior of particular transposons at particular locations upon increased selective pressure. The worst failing of the AMT4 system at present is the lack of a selection for reversion. Although preliminary efforts to demonstrate a growth defect of amt4 null strains at low concentrations of ammonium have been unsuccessful, we continue to look for a phenotype(s) that will allow us to select for revertants. Even at present it is clear that the system can be used to learn a great deal more about the natural history of transposition in C. reinhardtii. Our first few selections provided evidence for new transposable elements and new types of events associated with known elements without showing any signs of a limitation in their variety.
Evolution of the C. reinhardtii genome:
The role of transposons in sculpting genomes in response to environmental changes is increasingly being accepted (Kidwell and Lisch 2001). Application of nonlethal selection pressure of the kind imposed by growth of C. reinhardtii on methylammonium allows one to see the types of changes brought about by transposition and has allowed us to look at insertions into the promoter region and 5′ UTR, a splice site, and an intron of AMT4. Changes such as these are expected to have important roles in the responses of an organism to its environment and thus in its evolution (McClintock 1984; Grandbastien 1998; Kumar and Bennetzen 1999; Sabot et al. 2004).
Acknowledgments
We thank Damon Lisch and Michael Freeling for their interest and encouragement and an anonymous reviewer for leading us to further explore the TOC1 insert sites. This work was supported by National Institutes of Health grant GM38361 and a Miller Research Professorship (to S.K.).
References
- Bancroft, I., and C. Dean, 1993. Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, M., J. M. Ranz, A. Barbadilla, M. Long and A. Ruiz, 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418. [DOI] [PubMed] [Google Scholar]
- Cappello, J., K. Handelsman and H. F. Lodish, 1985. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell 43: 105–115. [DOI] [PubMed] [Google Scholar]
- Craig, N. L., 1997. Target site selection in transposition. Annu. Rev. Biochem. 66: 437–474. [DOI] [PubMed] [Google Scholar]
- Curcio, M. J., and K. M. Derbyshire, 2003. The outs and ins of transposition: from Mu to Kangaroo. Nat. Rev. Mol. Cell Biol. 4: 865–877. [DOI] [PubMed] [Google Scholar]
- Day, A., and J. D. Rochaix, 1991. a A transposon with an unusual LTR arrangement from Chlamydomonas reinhardtii contains an internal tandem array of 76 bp repeats. Nucleic Acids Res. 19: 1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, A., and J. D. Rochaix, 1991. b Structure and inheritance of sense and anti-sense transcripts from a transposon in the green alga Chlamydomonas reinhardtii. J. Mol. Biol. 218: 273–291. [DOI] [PubMed] [Google Scholar]
- Day, A., M. Schirmer-Rahire, M. R. Kuchka, S. P. Mayfield and J. D. Rochaix, 1988. A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J. 7: 1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chastonay, Y., H. Felder, C. Link, P. Aeby, H. Tobler et al., 1992. Unusual features of the retroid element PAT from the nematode Panagrellus redivivus. Nucleic Acids Res. 20: 1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, L., K. Bouckaert, F. Yeh and D. L. Kirk, 2002. Kangaroo, a mobile element from Volvox carteri, is a member of a newly recognized third class of retrotransposons. Genetics 162: 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P. J., 1989. Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher plants. Genetics 122: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P. J., J. P. Woessner and U. W. Goodenough, 1996. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7: 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., N. Jiang and S. R. Wessler, 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341. [DOI] [PubMed] [Google Scholar]
- González-Ballester, D., A. Camargo and E. Fernández, 2004. Ammonium transporter genes in Chlamydomonas: the nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol. Biol. 56: 863–878. [DOI] [PubMed] [Google Scholar]
- Goodwin, T. J., and R. T. Poulter, 2004. A new group of tyrosine recombinase-encoding retrotransposons. Mol. Biol. Evol. 21: 746–759. [DOI] [PubMed] [Google Scholar]
- Graham, J. E., J. G. Spanier and J. W. Jarvik, 1995. Isolation and characterization of Pioneer1, a novel Chlamydomonas transposable element. Curr. Genet. 28: 429–436. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M.-A., 1998. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3: 181–187. [Google Scholar]
- Harris, E., 1989. The Chlamydomonas Sourcebook. Academic Press, New York.
- Havecker, E. R., X. Gao and D. F. Voytas, 2004. The diversity of LTR retrotransposons. Genome Biol. 5: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T., Y. Noutoshi, M. Fujie and T. Yamada, 1997. Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J. 16: 3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., K. Sugimoto, Y. Otsuki, H. Tsugawa and M. Kanda, 1996. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93: 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, B.-R., D. Wu-Scharf, C. Zhang and H. Cerutti, 2002. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. USA 99: 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempken, F., and F. Windhofer, 2001. The hAT family: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma 110: 1–9. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., and A. J. Holyoake, 2001. Transposon-induced hotspots for genomic instability. Genome Res. 11: 1321–1322. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., and D. R. Lisch, 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- Kim, K. S., E. Feild, N. King, T. Yaoi, S. Kustu et al., 2005. Spontaneous mutations in the ammonium transport gene AMT4 of Chlamydomonas reinhardtii. Genetics 170: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, D. L., and I. Nishii, 2001. Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Dev. Growth Differ. 43: 621–631. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Kuromori, T., T. Hirayama, Y. Kiyosue, H. Takabe, S. Mizukado et al., 2004. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37: 897–905. [DOI] [PubMed] [Google Scholar]
- Lefebvre, P. A., and C. D. Silflow, 1999. Chlamydomonas: the cell and its genomes. Genetics 151: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. B., S. Lin, H. Jia, H. Wu, B. A. Roe et al., 2003. Analysis of Chlamydomonas reinhardtii genome structure using large-scale sequencing of regions on linkage groups I and III. J. Eukaryot. Microbiol. 50: 145–155. [DOI] [PubMed] [Google Scholar]
- Lindauer, A., D. Fraser, M. Brüderlein and R. Schmitt, 1993. Reverse transcriptase families and a copia-like retrotransposon, Osser, in the green alga Volvox carteri. FEBS Lett. 319: 261–266. [DOI] [PubMed] [Google Scholar]
- Lovering, R., N. Harden and M. Ashburner, 1991. The molecular structure of TE146 and its derivatives in Drosophila melanogaster. Genetics 128: 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras, V., D. R. Stevens and S. Purton, 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14: 441–447. [Google Scholar]
- McClintock, B., 1947. Cytogenetic studies of maize and Neurospora. Carnegie Inst. Washington Year Book 46: 146–152. [Google Scholar]
- McClintock, B., 1951. Chromosome organization and genic expression. Cold Spring Harbor Symp. Quant. Biol. 16: 13–47. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1984. The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- Miller, S. M., R. Schmitt and D. L. Kirk, 1993. Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetti, R., R. M. Marsano, P. Barsanti, C. Caggese and R. Caizzi, 2004. FB elements can promote exon shuffling: a promoter-less white allele can be reactivated by FB mediated transposition in Drosophila melanogaster. Mol. Genet. Genomics 271: 394–401. [DOI] [PubMed] [Google Scholar]
- Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn et al., 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H. P., and H. E. Varmus, 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13: 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Alegre, M., A. Dubus and E. Fernández, 2005. REM1, a new type of long terminal repeat retrotransposon in Chlamydomonas reinhardtii. Mol. Cell. Biol. 25: 10628–10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, S., M. Truett, M. Phillips and A. Maher, 1980. Eucaryotic transposable genetic elements with inverted terminal repeats. Cell 20: 639–647. [DOI] [PubMed] [Google Scholar]
- Pruss, D., R. Reeves, F. D. Bushman and A. P. Wolffe, 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269: 25031–25041. [PubMed] [Google Scholar]
- Pryciak, P. M., and H. E. Varmus, 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69: 769–780. [DOI] [PubMed] [Google Scholar]
- Puig, M., M. Caceres and A. Ruiz, 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl. Acad. Sci. USA 101: 9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebatchouk, D., and J. O. Narita, 1997. Foldback transposable elements in plants. Plant. Mol. Biol. 34: 831–835. [DOI] [PubMed] [Google Scholar]
- Sabot, F., D. Simon and M. Bernard, 2004. Plant transposable elements, with an emphasis on grass species. Euphytica 139: 227–247. [Google Scholar]
- Schnell, R. A., and P. A. Lefebvre, 1993. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M. D., M. E. Marks, C. L. Peichel, B. K. Blackman, K. S. Nereng et al., 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428: 717–723. [DOI] [PubMed] [Google Scholar]
- Soupene, E., W. Inwood and S. Kustu, 2004. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. USA 101: 7787–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger, G., Y. Okada, J. Emrich, J. Newton, A. Tsugita et al., 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31: 77–84. [DOI] [PubMed] [Google Scholar]
- Takken, F. L., D. Schipper, H. J. Nijkamp and J. Hille, 1998. Identification and Ds-tagged isolation of a new gene at the Cf-4 locus of tomato involved in disease resistance to Cladosporium fulvum race 5. Plant J. 14: 401–411. [DOI] [PubMed] [Google Scholar]
- Wang, H., T. Nussbaum-Wagler, B. Li, Q. Zhao, Y. Vigouroux et al., 2005. The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. C., R. A. Schnell and P. A. Lefebvre, 1998. Isolation and characterization of a new transposable element in Chlamydomonas reinhardtii. Plant Mol. Biol. 38: 681–687. [DOI] [PubMed] [Google Scholar]
- Wintermans, J. F., and A. de Mots, 1965. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta 109: 448–453. [DOI] [PubMed] [Google Scholar]
- Witte, C. P., Q. H. Le, T. Bureau and A. Kumar, 2001. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 98: 13778–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf, D., B. Jeong, C. Zhang and H. Cerutti, 2000. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290: 1159–1162. [DOI] [PubMed] [Google Scholar]