Abstract
Mutation is a central biological process whose rates and spectra are influenced by a variety of complex and interacting forces. Although DNA repair pathways are generally known to play key roles in maintaining genetic stability, much remains to be understood about the relative roles of different pathways in preventing the accumulation of mutations and the extent of heterogeneity in pathway-specific repair efficiencies across different genomic regions. In this study we examine mutation processes in base excision repair-deficient (nth-1) and nucleotide excision repair-deficient (xpa-1) Caenorhabditis elegans mutation-accumulation (MA) lines across 24 regions of the genome and compare our observations to previous data from mismatch repair-deficient (msh-2 and msh-6) and wild-type (N2) MA lines. Drastic variation in both average and locus-specific mutation rates, ranging two orders of magnitude for the latter, was detected among the four sets of repair-deficient MA lines. Our work provides critical insights into the relative roles of three DNA repair pathways in preventing C. elegans mutation accumulation and provides evidence for the presence of pathway-specific DNA repair territories in the C. elegans genome.
A diverse range of extrinsic and intrinsic forces contribute to the rates and patterns of spontaneous mutation experienced by various forms of life. The ability to recognize and repair DNA damage and replication errors constitutes one of the most important factors in minimizing mutation rates. Consequently, multiple DNA repair pathways evolved very early in the history of life to deal with the varied mutagenic challenges experienced by genomes. The three excision DNA repair pathways—base excision repair (BER), nucleotide excision repair (NER), and postreplicative mismatch repair (MMR)—correct DNA damage and/or replication mismatch errors through mechanisms that all involve initial binding of the damage/mismatch by a recognition factor that ultimately recruits additional proteins that coordinate the repair of the damage/error-containing strand; this is then followed by resynthesis using the other, nondigested strand as a template (Eisen and Hanawalt 1999). Heritable NER deficiencies cause the human genetic disorder xeroderma pigmentosum where those afflicted suffer from elevated and early cancer incidences (Cleaver 2005), and MMR deficiency is associated with ∼70% of human hereditary nonpolyposis colorectal cancers (Buermeyer et al. 1999). Defects in DNA repair glycosylases are also associated with some colorectal cancers (Al Tassan et al. 2002; Bai et al. 2005).
Our knowledge on the spectra of damage/errors repaired by each of the three excision repair pathways in eukaryotes has benefited from decades of intense study from a variety of biochemical and molecular genetic approaches, primarily in Saccharomyces cerevisiae and mammalian cell line systems. Studies utilizing in vitro binding assays and other similar biochemical approaches have shown that damage/error surveillance and binding are carried out by MutS homolog (MSH) heterodimers for MMR (Alani 1996; Habraken et al. 1996; Harfe and Jinks-Robertson 2000) and by a diverse array of DNA glycosylases for BER (Alseth et al. 1999; Cadet et al. 2000). For NER, two distinct dimeric protein complexes (XPA-RPA and XPC-HR23B) are evidenced to be involved in damage recognition, although their relative roles in this process remain controversial (Thoma and Vasquez 2003). BER is thought to repair diverse types of base damage and base–base mismatches whereas NER is thought to repair primarily bulky, helix-distorting lesions such as UV-induced cyclopyrimidine dimers. MMR is considered to correct mostly base–base mismatches and postreplicative loop-outs. All three pathways have been implicated in the repair of oxidative DNA damage.
Molecular genetic approaches that examine mutation processes in various excision repair-deficient backgrounds using reporter genes in DNA repair-deficient mammalian cell lines and “mutator” microbe strains have yielded important insights into the spectra of mutations prevented by each of BER, MMR, and NER (Kunz et al. 1990; Bruner et al. 1998; Earley and Crouse 1998; Reis et al. 2000; Gragg et al. 2002; Mark et al. 2002). Despite the clear successes of these reductionist approaches that rely on one or a few reporter genes to investigate repair-deficient mutation processes, many important questions about the relative roles of different repair pathways in preventing mutation accumulation remain unanswered that require more broad-based experimental approaches. Do various DNA repair pathways recognize and repair damage across the genome with equal efficiencies, or is there substantial heterogeneity? Is there extensive parity among different repair pathways in determining baseline spontaneous mutation rates and spectra, or are some pathways more important than others? In animals, how do different repair pathways contribute to germline mutation rates and spectra across generations?
The nematode Caenorhabditis elegans, most widely known for its central role in studies on animal development and neurobiology, has also recently emerged as a powerful model for investigating mutation (Denver et al. 2004; Keightley and Charlesworth 2005) and DNA repair (Boulton et al. 2002; Pothof et al. 2003) processes in eukaryotes. In particular, observations in a long-term set of N2 (common lab strain) C. elegans mutation-accumulation (MA) lines have provided uniquely direct and unbiased insights into basic mutation rates and spectra (Denver et al. 2004) in addition to their fitness consequences (Vassilieva et al. 2000; Baer et al. 2005). A subsequent analysis examining mutation processes in msh-2 and msh-6 MA lines provided a first step in extending the MA approach to DNA repair-deficient C. elegans strains to investigate the role of MMR in maintaining C. elegans genome stability (Denver et al. 2005).
This study provides a broad-based and robust analysis of germline mutation rates and patterns in BER-deficient (nth-1) and NER-deficient (xpa-1) C. elegans MA lines. Mutations were surveyed across multiple nuclear loci to enable comparisons of mutation spectra between different regions of the genome. When considered in the context of previous mutational estimates from the long-term, wild-type (WT) C. elegans MA lines and the MMR-deficient (msh-2 and msh-6) MA lines, our analyses provide critical insights into the relative roles of genes from three excision DNA repair pathways in preventing mutation accumulation across different regions of the C. elegans genome.
MATERIALS AND METHODS
Ortholog identification and generation of repair-deficient mutants:
We identified orthologs of the human NTH1 and XPA genes in C. elegans through conventional BLASTP and TBLASTN searches (Altschul et al. 1990) in Wormbase (Schwartz et al. 2004), using human and S. cerevisiae ortholog sequences as queries. Phylogenetic analyses, both neighbor-joining and maximum-parsimony approaches tested with 1000 bootstrap replications each, were then carried out in MEGA2 (Kumar et al. 2001) to ensure the ortholog status of C. elegans sequences identified by BLAST. Confirmed C. elegans open reading frames orthologous to the human NTH1 and XPA genes were then submitted to the C. elegans Gene Knockout Consortium as targets for identifying deletion mutants (Edgley et al. 2002). Deletion mutants for each of the two submitted open reading frames were isolated and provided to us as homozygotes. Deletion boundaries were determined for each of the loci by sequencing polymerase chain reaction (PCR) products generated by primers specific to each target locus (the same primers initially used by the Consortium to screen for the deletion mutants). The xpa-1 allele was already named (Park et al. 2002) and we named the deletion allele of the worm NTH1 ortholog nth-1, in concordance with standard C. elegans genetic nomenclature.
Backcrossing and mutation-accumulation procedures:
Before initiating MA experiments, each of the nth-1 and xpa-1 strains were backcrossed to N2 genomes six times (as was also done for the MMR-deficient MA lines in Denver et al. 2005) so that all DNA repair-deficient MA experiments would be carried out on highly similar genetic backgrounds. Simple deletion locus-specific PCR tests, where amplified products from deletion alleles are visibly smaller than WT alleles on agarose gels, were used to track and maintain deletion alleles throughout the backcrossing process. Fifty MA lines were then initiated for each of the two backcrossed, homozygous deletion mutants (nth-1, xpa-1). Following standard C. elegans MA procedures (Vassilieva et al. 2000), each MA line was propagated across multiple generations (an average of 39 for the nth-1 lines and 40 for the xpa-1 lines) in a benign environment (NGM plates seeded with the OP50 strain of Escherichia coli as a food source, 20°) as single, randomly selected hermaphrodites picked at the L4 larval stage. This treatment resulted in an effective population size equal to one for each MA line across generations and ensured that all but the most deleterious mutations accumulated in the germline over time in an effectively neutral fashion. Sets of generational backups were maintained at 10° for the MA lines in the event of dead or sterile worms. Individual MA lines were declared extinct if three successive attempts to transfer worms from the backup plate resulted in nonviable worms.
Mutation detection and confirmation:
Mutations were detected in each set of MA lines by PCR amplifying regions of the genome followed by direct DNA sequencing of purified PCR products. The majority of loci were randomly distributed across C. elegans chromosomes by using PCR primer pairs designed to amplify chromosomal positions selected by a random number generator. A subset of loci was designed to amplify homopolymeric nucleotide runs to specifically evaluate their mutational properties. The loci assayed here were the same as those examined in the MMR-deficient C. elegans MA line study (Denver et al. 2005). PCRs were performed using a large amount of genomic DNA (∼25,000 diploid genomes per reaction) and 2 units of Taq polymerase (Eppendorf, Madison, WI) per reaction to eliminate artifacts associated with initial amplification from small amounts of genomic DNA. PCR products were purified by solid phase reversible immobilization (Elkin et al. 2001), cycle sequenced, and analyzed on an ABI3730 DNA sequencer (Applied Biosystems, Foster City, CA) at the Indiana Molecular Biology Institute.
DNA sequence text files from the repair-deficient MA lines, backcrossed progenitor line controls, and the published N2 target sequence were subjected to multiple alignment using CLUSTALW (Higgins et al. 1994) to identify putative mutations in the MA lines. Putative MA line-specific mutations identified in the alignments were then visually scrutinized on the electropherogram data to eliminate base-caller errors and other sequencing artifacts. Putative mutations supported by clear, unambiguous electropherogram data were then evaluated on the opposite strand (sequencing reaction in the opposite direction), using internal primers where necessary. Only those mutations supported by reliable electropherogram data on both strands, and receiving Phred scores >33, were considered for this study. DNA sequences containing new mutations reported in this study were deposited in GenBank under accession nos. DQ674279–DQ674313.
Calculation of mutation rates:
Mutation rates in the MA lines were calculated using the equation μ = m/(LnT), where μ is the mutation rate (per nucleotide site per generation), L is the number of MA lines, n is the number of nucleotide sites surveyed, and T is the time in generations. The standard errors of the mean (SEM) for mutation rates were calculated using the equation SEM = (μ/(LnT))1/2. SEM values for mutation rates are shown in parentheses throughout the text and tables.
RESULTS
Isolation of repair knockouts and mutation accumulation:
The predicted coding sequence R10E4.5 in the C. elegans genome was identified as the ortholog of the human NTH1 gene (encodes a DNA glycosylase involved in BER) through phylogenetic analysis in a previous study (Denver et al. 2003); the K07G5.2 predicted coding sequence was previously characterized (Park et al. 2002) as the C. elegans ortholog of human XPA (involved in NER damage recognition), which we confirmed through conventional BLASTP and TBLASTN searches and phylogenetic analyses (data not shown). After identification, R10E4.5 and K07G5.2 were submitted to the C. elegans Gene Knockout Consortium as targets for generating deletion alleles for this study. Deletion alleles were isolated for each of these genes and in both cases the deletion included significant portions of exon sequence. The deletion for R10E4.5 (named allele ok724 carried by strain RB877) spanned 792 bp and resulted in the complete elimination of exons 2 and 3 in addition to most of exon 4. The K07G5.2 deletion (named allele ok698 carried by strain RB864) was 912 bp and resulted in the complete elimination of exons 3, 4, and 5. R10E4.5 was given the C. elegans gene name nth-1 and K07G5.2 was already named xpa-1 at the time of this study. For both deletion alleles, multiple highly conserved residues known to be necessary for DNA damage binding were eliminated. Thus it is highly likely that each of these deletions resulted in a null allele. After obtaining strains RB877 and RB864, we backcrossed each to the N2 genome six times prior to initiating MA experiments (see materials and methods for details).
Fifty MA lines were initiated for each of the backcrossed nth-1 and xpa-1 C. elegans strains and propagated across an average of 39 (nth-1) and 40 (xpa-1) generations through single-hermaphrodite descent. This treatment ensured that all but the most deleterious mutations accumulated in the repair-deficient MA line genomes across generations in an effectively neutral fashion. At the end of MA procedures, worm populations from each of the nth-1 and xpa-1 MA lines were harvested for DNA extractions and frozen stocks. All 50 nth-1 MA lines survived to the end of the MA procedures whereas one xpa-1 was declared extinct before the initiation of molecular mutation surveys. The nth-1 MA lines were named N-01–N-50, and the xpa-1 MA lines were named A-01–A-50.
Mutations detected in the nth-1 and xpa-1 MA lines:
We sequenced 20,469 bp of nuclear DNA, distributed across 24 PCR product loci (supplemental Table 1 at http://www.genetics.org/supplemental/), from each of 50 nth-1 and 49 xpa-1 C. elegans MA lines to survey for mutations in each repair-deficient background. The randomly selected loci sequenced were the same ones used for a previous study on mutation processes in msh-2 and msh-6 MA lines (Denver et al. 2005). Fifteen total mutations were observed in the nth-1 MA lines and 24 total mutations were observed in the xpa-1 MA lines (Table 1). In the nth-1 MA lines, 13 base substitutions were observed and two insertion/deletions (indels) were observed. In the xpa-1 MA lines, 17 base substitutions and seven indels were detected. Among base substitutions, transition (Ts) mutations slightly outnumbered transversion (Tv) changes in the nth-1 MA lines (Ts:Tv = 8:5) whereas roughly equal numbers of Ts and Tv substitutions were observed in the xpa-1 MA lines (Ts:Tv = 8:9). Among indels, the two detected in the nth-1 MA lines were both insertions; four insertions and three deletions were observed in the xpa-1 MA lines. All indels observed were single-nucleotide changes. For both the nth-1 and xpa-1 MA lines, no mutations were observed at any of the 15 homopolymeric nucleotide runs ≥8 bp in length that were present in the 24 target loci.
TABLE 1.
Mutations observed in the nth-1 and xpa-1 MA lines
| Chr. | Locus | Locus pos. | Mut. | Context | Line(s) | Cod. |
|---|---|---|---|---|---|---|
| nth-1 | ||||||
| I | ZK337 | 25,889 | A → G | TCACG → TCGCG | N-20 | EX (R: H → R) |
| I | ZK337 | 26,580 | T → C | TCTCT → TCCCT | N-24 | EX (R: L → P) |
| I | ZK337 | 25,984 | C → G | AACGT → AAGGT | N-34 | EX (R: R → G) |
| I | ZK337 | 25,648 | T → G | AGTGC → AGGGC | N-35 | EX (S) |
| II | C17G10 | 23,624 | T → C | GGTCT → GGCCT | N-46 | IG |
| II | C17G10 | 23,629 | T → C | TTTCC → TTCCC | N-46 | IG |
| II | M106 | 19,396 | A → G | TAAAG → TAGAG | N-06 | IG |
| II | M106 | 19,453 | A → C | CCAAA → CCCAA | N-08 | IG |
| V | W05B10 | 20,887 | +G | CTCT → CTGCT | N-04 | IG |
| X | F19C6 | 3,776 | T → C | CATTT → CACTT | N-49 | IN |
| X | F19C6 | 3,997 | T → G | TCTAA → TCGAA | N-08 | IN |
| X | C07A4 | 11,775 | G → A | ACGTT → ACATT | N-25 | IN |
| X | C07A4 | 11,638 | A → G | GGAGA → GGGGA | N-35 | EX (S) |
| X | F59F5 | 22,175 | A → C | AAATT → AACTT | N-24 | IN |
| X | F59F5 | 21,782 | +T | TACT → TATCT | N-41 | IN |
| xpa-1 | ||||||
| II | C17G10 | 24,313 | C → T | TTCAA → TTTAA | A-04 | IG |
| II | M106 | 19,445 | C → G | GGCGG → GGGGG | A-48 | IG |
| II | M106 | 19,440 | A → C | GGAGA → GGCGA | A-08 | IG |
| II | M106 | 19,481 | C → G | GCCAA → GCGAA | A-24 | IG |
| II | M106 | 19,554 | T → G | GGTGC → GGGGC | A-37 | IG |
| II | M106 | 19,557 | A → G | GCACA → GCGCA | A-37 | IG |
| II | Y48G9A | 119,086 | C → G | CCCAG → CCGAG | A-41 | IG |
| V | B0240 | 2,189 | (T)6 → (T)7 | TC(T)6CA → TC(T)7CA | A-46 | IN |
| V | B0240 | 2,447 | A → G | GGAGT → GGGGT | A-05 | IN |
| V | W05B10 | 21,606 | T → A | AGTGA → AGAGA | A-17 | IG |
| V | WO5B10 | 21,579 | G → A | ATGAA → ATAAA | A-35, A-46 | IG |
| V | Y113G7A | 4,758 | +C | CAAC → CACAC | A-41 | IG |
| X | R03E9 | 21,601 | −A | ATAGA → ATGA | A-16 | EX (FS) |
| X | R03E9 | 22,136 | (G)4 → (G)5 | GA(G)4CA → GA (G)5CA | A-43 | EX (FS) |
| X | R03E9 | 21,839 | +T | AGAT → AGTAT | A-50 | IN |
| X | F19C6 | 3,530 | T → C | TTTCC → TTCCC | A-32 | IN |
| X | F59F5 | 22,287 | T → A | ATTTG → ATATG | A-03 | EX (R: N → Y) |
| X | F59F5 | 22,421 | T → G | CATCG → CAGCG | A-03 | EX (R: H → P) |
| X | F59F5 | 21,815 | G → A | AAGTT → AAATT | A-28 | IN |
| X | F59F5 | 21,825 | T → G | AATAC → AAGAC | A-28 | IN |
| X | F59F5 | 21,785 | G → A | TTGTT → TTATT | A-31 | IN |
| X | F59F5 | 21,860 | (C)3 → (C)2 | AG(C)3AC → AG(C)2AC | A-13, A-21 | IN |
Chr. indicates the chromosome in which the mutation was found. Locus refers to the sequenced C. elegans cosmid or yeast artificial chromosome in which the mutation was found (PCR loci surveyed for mutation are named according to C. elegans cosmids, fosmids, and yeast artificial chromosomes; see supplemental Table 1 for details). Mut. refers to the observed mutation. Context provides information regarding the bases surrounding the observed mutation with respect to the (+) strand of C. elegans chromosomes. Line indicates the specific MA line in which the mutation was detected. Cod. refers to the coding context of the sequence in which the mutation was found: EX, exon, IG, intergenic, and IN, intron. For exon base substitution mutations, S indicates a silent base substitution and R indicates a replacement base substitution (amino acid change is indicated after R). EX (FS) denotes a mutation predicted to result in a frameshift in an open reading frame.
The mutation data were then used to estimate per generation total mutation rates for each of the nth-1 and xpa-1 MA lines. A higher total mutation rate of 6.0 (±1.2) × 10−7/bp/generation was observed in the xpa-1 MA lines, as compared to the lower total rate, 3.7(±0.9) × 10−7/bp/generation, observed in the nth-1 MA lines (Figure 1A). Although a slightly elevated mutation rate specific to base substitutions was observed in the xpa-1 MA lines as compared to the nth-1 MA lines, the difference between the indel mutation rates between the two sets of MA lines was more drastic (Figure 1B).
Figure 1.—
Mutation rates observed in DNA repair-deficient MA lines. (A) Genotype-specific total mutation rates (mutations per base pair per generation) for repair-deficient MA lines. (B) Mutation rates specific for base substitutions (shaded bars) and indels (open bars). Indels at mononucleotide runs ≥8 bp in length were excluded for all rate calculations. Error bars indicate SEM as described in materials and methods. Rates for msh-2 and msh-6 are from Denver et al. (2005) and rates for N2 (WT) are from Denver et al. (2004).
Mutational distributions across loci and MA lines:
We next considered the distribution patterns of mutations across the 24 nuclear loci and among the 50 (nth-1) or 49 (xpa-1) MA lines surveyed in this study. Observed distribution patterns were compared to Poisson expectations, which were used to predict random distribution patterns. In both the nth-1 and xpa-1 MA lines, the observed distributions of mutations across the 24 surveyed loci deviated drastically from Poisson expectations (χ2 = 21.1, P < 0.001 for nth-1; χ2 = 576.1, P < 0.001 for xpa-1) (see Figure 2). Furthermore, we detected two instances in the xpa-1 MA lines where the same exact nucleotide site was found to have mutated in two distinct MA lines: loci W05B10 (lines A-35 and A-46) and F59F5 (lines A-13 and A-25)—see Table 1. In contrast, the distribution of mutations across nth-1 MA lines did not deviate from Poisson expectations (χ2 = 2.1, P < 0.5) and the distribution across xpa-1 MA lines was even closer to Poisson expectations (χ2 = 0.6, P < 0.5).
Figure 2.—
Distributions of mutations across loci. The observed distributions (solid bars) of mutations across the 24 surveyed loci were compared to expectations based on Poisson distributions (hatched bars). Mutational distributions in the (A) nth-1 MA lines, (B) xpa-1 MA lines, (C) msh-2 MA lines, and (D) msh-6 MA lines are shown (data for the latter two are from Denver et al. 2005). Significant deviations from Poisson expectations were observed for the nth-1, xpa-1, and msh-6 MA lines, but not for the msh-2 MA lines (see text). Arrows indicate loci defined here as mutational hotspots.
DISCUSSION
Mutation rates in DNA repair-deficient MA lines:
The findings reported here on mutation rates and spectra in BER- and NER-deficient MA lines, in conjunction with previous data from MMR-deficient MA lines (Denver et al. 2005), provide critical insights into the relative roles of the three excision DNA repair pathways in maintaining genome stability in the C. elegans germline. Mutational observations from the long-term set of WT C. elegans MA lines (Denver et al. 2004) serve as a unique and essential baseline for gauging the relative roles of the three excision repair pathways (BER, MMR, and NER) in preventing mutation accumulation. Total average mutation rates in each of the four DNA repair-deficient backgrounds (nth-1, xpa-1, msh-2, and msh-6) were significantly greater than the WT C. elegans mutation rate (Figure 1A), as expected. The total rate observed in the nth-1 MA lines was ∼17-fold greater than in WT, whereas the xpa-1 rate was ∼28-fold greater than the WT rate. This contrasts with the ∼48-fold increase over WT in mutation rate observed in the MMR-deficient MA lines. Consistent with these mutational estimates, the majority of MMR-deficient MA lines were unable to be propagated beyond 20 generations of MA, whereas the majority of BER- and NER-deficient MA lines survived to 40 generations. Roughly speaking, the above observations suggest that, on average, MMR plays a greater role than NER, which in turn plays a greater role than BER, in preventing mutation accumulation across generations in C. elegans.
The above generalization, however, assumes that each of the three repair pathways was completely eliminated in each respective knockout surveyed, which is almost certainly not the case. For instance, despite the central role of xpa-1 in NER damage surveillance (King et al. 2001), other protein factors such Rpa-1 and Xpc-1 play key essential roles in NER-mediated damage recognition (Saijo et al. 2004) and it is possible that there is some residual repair ongoing in the xpa-1 MA lines involving rpa-1, xpc-1, and other factors. Likewise, a diverse array of BER DNA glycosylases exist, other than nth-1 orthologs, capable of repairing varying damage types. The C. elegans genome is peculiar among eukaryotes, however, as nth-1 is only one of two DNA glycosylases encoded: oxoguanine DNA glycosylases, formamidopyrimidine glycosylases, and methyladenine glycosylases are not detected in the C. elegans genome using a variety of BLAST searching approaches (Denver et al. 2003; our unpublished data). Nonetheless, the other DNA glycosylase detected in C. elegans (ung-1, a uracil DNA glycosylase) is presumably active in the nth-1 MA lines, rendering our assessment of BER's role (based solely on nth-1) in preventing mutation accumulation an almost certain underestimate. Since the C. elegans genome lacks an ortholog of human MSH3 (Denver et al. 2005), it is highly likely that MMR is completely impaired in both the msh-2 and msh-6 MA lines. A more comprehensive understanding of the relative roles of the three excision repair pathways in maintaining C. elegans genome stability requires further studies involving MA lines derived from additional knockouts such as ung-1 and xpc-1.
The indel mutation rates were much more variable among the four sets of repair-deficient MA lines as compared to the base substitution mutation rates, which were overall much more similar to one another among the repair-deficient sets of MA lines (Figure 1B). The highest indel mutation rates were detected in the MMR-deficient MA lines and the lowest indel rate was detected in the nth-1 MA lines. The high indel rate in the MMR-deficient MA lines, as compared to the other repair-deficient MA lines, is consistent with its well-characterized role in correcting postreplicative DNA loop-outs. Likewise, the low relative indel rate in the nth-1 MA lines is consistent with the widespread perception that the primary role of BER is correcting base damage, thereby primarily preventing base substitution mutations. Fifteen homopolymeric nucleotide runs were found to be invariant in the nth-1 and xpa-1 MA lines whereas they displayed markedly high mutation rates in the MMR-deficient MA lines (Denver et al. 2005), as predicted. In the xpa-1 MA lines, however, four of the seven total indels detected were at shorter mononucleotide runs (3–6 bp), indicating that NER plays an important role in maintaining the stability of short, but not long, homopolymer repeats. In a NER-deficient human cell line system, ∼20% of the indels observed in a reporter gene were at similar short mononucleotide runs (King et al. 2001).
In S. cerevisiae, MMR-deficient strains reproducibly display strong mutator phenotypes in standard single-gene reversion assays (Harfe and Jinks-Robertson 2000). Strains deficient in BER or NER, alone, did not display mutator phenotypes in most previous surveys (Scott et al. 1999; Swanson et al. 1999; Gellon et al. 2001); one study, however, reported that BER-deficient yeast strains do show evidence of mutator phenotypes (Alseth et al. 1999). Our findings that average mutation rates are most greatly elevated in MMR-deficient C. elegans backgrounds (Figure 1) are generally consistent with the above observations and suggest that the relative roles of the three excision repair pathways in preventing mutation accumulation might be highly conserved in eukaryotes. In S. cerevisiae and mammalian cell lines, however, the disparities between MMR-deficient and BER- or NER-deficient mutator phenotypes are generally much greater than that observed here for C. elegans. This observation could reflect fundamental, between-species differences in the relative roles of different excision repair pathways in maintaining genome stability. However, fair comparisons to S. cerevisiae and mammalian cells would require large-scale mutation surveys in these systems, of a scale comparable to the current study, rather than relying on comparisons to the existing data from conventional reporter gene approaches. Furthermore, this observed disparity might be a consequence of our analysis targeting cross-generational, germline mutation processes whereas most yeast and mammalian cell culture surveys consider only mutations observed across mitotic (i.e., somatic) cell divisions.
Distributional heterogeneities of mutations in the genome:
Although the mutation rates discussed above, averaged across all surveyed loci, provide some basic insights into the relative roles of BER, MMR, and NER in preventing C. elegans mutation accumulation, our data also provide an opportunity to consider patterns of among-locus variation in mutation processes and associated mutational hotspots. For both the nth-1 and xpa-1 sets of MA lines, the distribution of mutations across the 24 surveyed nuclear loci deviated significantly from Poisson expectations, suggesting that there is substantial heterogeneity in the abilities of DNA repair pathways to prevent mutation accumulation across different regions of the genome (Figure 2, A and B). This pattern of distributional heterogeneity across loci was also observed in the msh-6, but not the msh-2, MA lines (see Figure 2, C and D). For the each of the three genotypes (nth-1, xpa-1, and msh-6) where significant deviations from Poisson expectations were observed, there were substantially greater numbers of loci containing zero mutations as well as loci containing four or more mutations, which we consider here putative mutational hotspots. Correspondingly, the observed numbers of loci containing a single mutation were substantially lower than Poisson expectations for all three genotypes, as well.
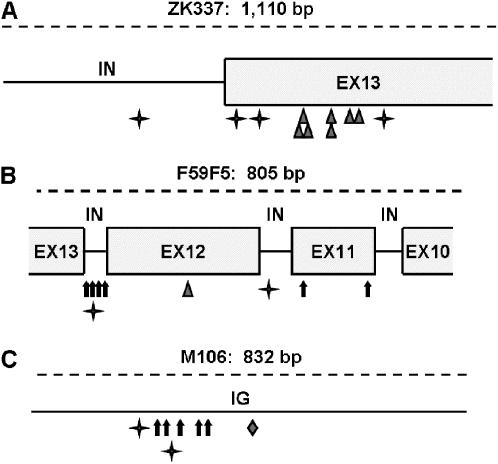
We now point out four genotype/locus combinations (nth-1/ZK337, msh-6/ZK337, xpa-1/F59F5, and xpa-1/M106) that each contained significantly greater numbers of mutations than Poisson distributions predicted at any given locus—we classify these here as genotype-specific mutational hotspots (Figure 3). The mutation rates specific to these four genotype/locus combinations were all approximately two orders of magnitude greater than the respective corresponding average mutation rates observed across all loci (supplemental Table 2 at http://www.genetics.org/supplemental/). The two xpa-1-specific hotspot loci displayed highly similar mutation patterns, dominated by base substitutions, and were also highly similar to the xpa-1 mutation spectra observed across all 24 loci. Although the ZK337 locus was observed to be a mutational hotspot both in nth-1 and in msh-6 backgrounds, the mutation spectra observed in these two sets of lines differed substantially: base substitutions were observed exclusively in the nth-1 MA lines (Table 1) whereas small insertions were observed exclusively at this locus in the msh-6 MA lines (Denver et al. 2005). For the nth-1 MA lines, this pattern was consistent with observations at other loci, but the predominance of insertions at ZK337 in the msh-6 MA lines differed significantly from patterns observed at other loci where base substitutions dominated over indels. The remarkably different mutation patterns observed at ZK337 in nth-1 and msh-6 MA lines might reflect distinctive underlying mutagenic forces (i.e., base damage vs. replication slippage errors) leading to the highly elevated mutation numbers observed in each genetic background. An alternative explanation might be that this locus is prone to a singular type of DNA damage that has distinctive mutagenic consequences due to differential damage processing mechanisms in nth-1 vs. msh-6 backgrounds.
Figure 3.—
Three mutation hotspots. Schematics of the three loci characterized as repair pathway-specific mutational hotspots are shown. Dashed lines represent the PCR product locus surveyed; below dashed lines, boxes indicate exon (EX) sequence and solid lines indicate noncoding sequence—intron (IN) or intergenic (IG). Four-pointed stars indicate nth-1 mutations, triangles indicate msh-6 mutations, arrows indicate xpa-1 mutations, and the diamond indicates an msh-2 mutation. (A) The ZK337 locus—exon and intron sequence is from the ZK337.1 gene that encodes a protein in the α-2-macroglobulin family. (B) The F59F5 locus—exon and intron sequence is from the F59F5.6 gene that encodes a liprin-α protein. (C) The intergenic M106 locus.
The distribution of mutations within the ZK337 locus also differed dramatically between those observed in the msh-6 MA lines (all clustered in a 128-bp subsegment) vs. those in the nth-1 MA lines (more randomly distributed across the entire locus) (see Figure 3A). Intralocus clustering was also observed for the two hotspot loci specific to xpa-1 backgrounds. At the F59F5 locus, four of the six observed mutations occurred in a short 75-bp stretch of intronic sequence; the other two mutations were observed in upstream exon sequence (Figure 3B). It is also worth noting that one of the two mutations observed at this locus in the nth-1 MA lines occurred in this short intronic stretch. Similarly, the five xpa-1 mutations observed at the intergenic M106 locus were observed in a 117-bp stretch of DNA, and the two nth-1 mutations observed at this locus occurred in this same locus subsection (Figure 3C). These observations suggest that such mutational hotspots may mostly be very small, on the order of 50–150 bp; however, the broader overall within-locus mutational distributions of nth-1 mutations at the ZK337 locus and xpa-1 mutations at the F59F5 locus (when all mutations are considered) limit our ability to generalize. Also, the occurrence of a few nth-1 mutations at the xpa-1 hotspots might indicate that these particular regions are mutational “warmspots” in BER-deficient backgrounds. More comprehensive studies targeting larger continuous regions of the genome are required to better understand the overall abundances, ranges, and genomic distributions of these mutational hotspots.
We also detected two nucleotide sites in the xpa-1 MA lines that each experienced two identical mutations in distinct MA lines (Table 1), an observation that deviated significantly from expectations based on a random distribution of mutations across the 20,469 nucleotide sites surveyed in each MA line (χ2 = 129.5, P < 0.001). We thus interpret these two sites as xpa-1-specific, nucleotide site-specific mutational hotspots. Although the entire F59F5 locus was previously described as a mutational hotspot, the F59F5 nucleotide site hotspot still deviated significantly from expectations based on a random distribution of the seven mutations observed in the xpa-1 MA lines across F59F5 nucleotide sites (χ2 = 14.9, P < 0.001). These findings indicate that these two nucleotide site-specific hotspots are particularly prone to mutagenic effects specifically countered by NER.
One potential concern with the approach reported here is that, in a given defined repair-deficient background, a mutation might occur at another locus that affects mutation processes (e.g., another DNA repair gene), which would then alter the rate and pattern of mutation accumulation observed. If such occurrences were to predominate in our system, we would expect them to be reflected by the presence of individual MA lines that suffered a significantly greater number of mutations than the among-line average. The Poisson analyses of mutational distributions across MA lines revealed that none of the four sets of repair-deficient MA lines displayed among-line distribution patterns that deviated from random expectations. Thus, it is highly unlikely that the mutation spectra reported here were significantly affected by such “second-site” mutations. It is also possible, however, that a second-site mutation in another DNA repair gene might have occurred during backcrossing that might complicate interpretation of our results. Future studies targeting additional excision repair genes and the initiation of repair-deficient genotype-specific MA lines from multiple backcrossed lines will ameliorate this issue.
Conclusion:
The findings reported here offer unique insights into the relative roles of the three excision DNA repair pathways in preventing mutation accumulation across different regions of the C. elegans genome. Our approach taking advantage of MA line methodologies provided an opportunity to directly calculate germline mutation rates, rather than mutation frequencies. Mutation rates were found to vary significantly among repair-deficient different genotypes (Figure 1), suggesting a hierarchy in the relative importance of the three excision repair pathways in maintaining C. elegans genome stability: MMR over NER over BER (with the important caveat that there are likely varying levels of residual repair in some repair-deficient backgrounds). By surveying large numbers of loci endemic to the C. elegans genome rather than one or a few reporter genes, we were also able to investigate and detect significant heterogeneity in mutation rates and spectra across different broad genomic regions in addition to different specific nucleotide sites. The observed among-locus heterogeneity in repair-deficient genotype-specific mutation rates suggests the possible presence of pathway-specific DNA repair territories for each of the three excision DNA repair pathways. Similarly, a recent survey employing reporter loci introduced in different regions of the S. cerevisiae genome suggests substantial intragenomic variation in yeast MMR efficiency (Hawk et al. 2005). Key questions remain as to the abundance, range, and distributions of repair pathway-specific mutational hotspots throughout the genome and the extent to which this phenomenon is observed in different species.
Acknowledgments
We thank the C. elegans Gene Knockout Consortium for generating and providing the deletion mutants used in this study. Our gratitude is also extended to Lawrence Washington at the Indiana Molecular Biology Institute for DNA sequencing. D.R.D. was supported by the National Institutes of Health (NIH) (F32 GM66652) and funding for this work was provided by the NIH (R01 GM36827) to M.L.
References
- Al Tassan, N., N. H. Chmiel, J. Maynard, N. Fleming, A. L. Livingston et al., 2002. Inherited variants of MYH associated with somatic G:C→A:T mutations in colorectal tumors. Nat. Genet. 30: 227–232. [DOI] [PubMed] [Google Scholar]
- Alani, E., 1996. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 17: 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth, I., L. Eide, M. Pirovano, T. Rognes, E. Seeberg et al., 1999. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 19: 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and E. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Baer, C. F., F. Shaw, C. Steding, M. Baumgartner, A. Hawkins et al., 2005. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc. Natl. Acad. Sci. USA 102: 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, H., S. Jones, X. Guan, T. M. Wilson, J. R. Sampson et al., 2005. Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Res. 33: 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson et al., 2002. Combined functional genomics map of the C. elegans DNA damage response. Science 295: 127–131. [DOI] [PubMed] [Google Scholar]
- Buermeyer, A., S. M. Deschenes, S. K. Baker and R. M. Liskay, 1999. Mammalian DNA mismatch repair. Annu. Rev. Genet. 33: 533–564. [DOI] [PubMed] [Google Scholar]
- Bruner, S. D., H. M. Nash, W. S. Lane and G. L. Verdine, 1998. Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol. 8: 393–403. [DOI] [PubMed] [Google Scholar]
- Cadet, J., A. G. Bourdat, C. D'Ham, V. Duarte, D. Gasparutto et al., 2000. Oxidative base damage to DNA: specificity of base excision repair enzymes. Mutat. Res. 462: 121–128. [DOI] [PubMed] [Google Scholar]
- Cleaver, J. E., 2005. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5: 564–573. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., S. L. Swenson and M. Lynch, 2003. An evolutionary analysis of the helix- hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 20: 1603–1611. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., S. Feinberg, S. Estes, W. K. Thomas and M. Lynch, 2005. Mutation rates, spectra, and hotspots in mismatch repair-deficient Caenorhabditis elegans. Genetics 170: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, M. C., and G. F. Crouse, 1998. The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95: 15487–15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M., A. D'Souza, G. Moulder, S. McKay, B. Shen et al., 2002. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 30: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J. A., and P. C. Hanawalt, 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435: 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin, C. J., P. M. Richardson, H. M. Fourcade, N. M. Hammon, M. J. Pollard et al., 2001. High-throughput plasmid purification for capillary sequencing. Genome Res. 11: 1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon, L., R. Barbey, P. Auffret van der Kemp, D. Thomas and S. Boiteux, 2001. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol. Genet. Genomics 265: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Gragg, H., B. D. Harfe and S. Jinks-Robertson, 2002. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 8756–8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken, Y., P. Sung, L. Prakash and S. Prakash, 1996. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 6: 1185–1187. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Hawk, J. D., L. Stefanovic, J. C. Boyer, T. D. Petes and R. A. Farber, 2005. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA 102: 8639–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D. G., J. D. Thompson and T. J. Gibson, 1994. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266: 383–402. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and B. Charlesworth, 2005. Genetic instability of C. elegans comes naturally. Trends Genet. 21: 67–70. [DOI] [PubMed] [Google Scholar]
- King, N. M., G. G. Oakley, M. Medvedovik and K. Dixon, 2001. XPA protein alters the specificity of ultraviolet light-induced mutagenesis in vitro. Environ. Mol. Mutagen. 37: 329–339. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kunz, B. A., L. Kohalmi, X. Kang and K. A. Magnusson, 1990. Specificity of the mutator effect caused by disruption of the RAD1 excision repair gene of Saccharomyces cerevisiae. J. Bacteriol. 172: 3009–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark, S. C., L. E. Sandercock, H. A. Luchman, A. Baross, W. Edelmann et al., 2002. Elevated mutation frequencies and predominance of G:C to A:T transition mutations in Msh6−/− small intenstinal epithelium. Oncogene 21: 7126–7130. [DOI] [PubMed] [Google Scholar]
- Park, H. K., J. S. Yook, H. S. Koo, I. S. Choi and B. Ahn, 2002. The Caenorhabditis elegans XPA homolog of human XPA. Mol. Cells 14: 50–55. [PubMed] [Google Scholar]
- Pothof, J., G. van Haaften, K. Thijssen, R. S. Kamath, A. G. Fraser et al., 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 17: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, A. M., D. L. Cheo, L. B. Meira, M. S. Greenblatt, J. P. Bond et al., 2000. Genotype-specific Trp53 mutational analysis in ultraviolet B radiation-induced skin cancers in Xpc and Xpc Trp53 mutant mice. Cancer Res. 60: 1571–1579. [PubMed] [Google Scholar]
- Saijo, M., T. Matsuda, I. Kuraoka and K. Tanaka, 2004. Inhibition of nucleotide excision repair by anti-XPA monoclonal antibodies which interfere with binding to RPA, ERCC1, and TFIIH. Biochem. Biophys. Res. Commun. 321: 815–822. [DOI] [PubMed] [Google Scholar]
- Schwartz, E. M., I. Antoshechkin, C. Bastiani, T. Bieri, D. Blasiar et al., 2004. Wormbase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32: D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A. D., M. Neishabury, D. H. Jones, S. H. Reed, S. Boiteux et al., 1999. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast 15: 205–218. [DOI] [PubMed] [Google Scholar]
- Swanson, R. L., N. J. Morey, P. W. Doetsch and S. Jinks-Robertson, 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, B. S., and K. M. Vasquez, 2003. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 38: 1–13. [DOI] [PubMed] [Google Scholar]
- Vassilieva, L. L., A. M. Hook and M. Lynch, 2000. The fitness effects of spontaneous mutation in Caenorhabditis elegans. Evolution 54: 1234–1246. [DOI] [PubMed] [Google Scholar]