Abstract
Drosophila melanogaster males lack recombination and have evolved a mechanism of meiotic chromosome segregation that is independent of both the chiasmatic and achiasmatic segregation systems of females. The teflon (tef) gene is specifically required in males for proper segregation of autosomes and provides a genetic tool for understanding recombination-independent mechanisms of pairing and segregation as well as differences in sex chromosome vs. autosome segregation. Here we report on the cloning of the tef gene and the molecular characterization of tef mutations. Rescue experiments using a GAL4-driven pUAS transgene demonstrate that tef corresponds to predicted Berkeley Drosophila Genome Project (BDGP) gene CG8961 and that tef expression is required in the male germ line prior to spermatocyte stage S4. Consistent with this early prophase requirement, expression of tef was found to be independent of regulators of meiotic M phase initiation or progression. The predicted Tef protein contains three C2H2 zinc-finger motifs, one at the amino terminus and two in tandem at the carboxyl terminus. In addition to the zinc-finger motifs, a 44- to 45-bp repeat is conserved in three related Drosophila species. On the basis of these findings, we propose a role for Tef as a bridging molecule that holds autosome bivalents together via heterochromatic connections.
THE pairing and subsequent segregation of homologous chromosomes to opposite poles at meiosis I are key events required to ensure the equal distribution of chromosomes to gametes. In most organisms, these events are intimately associated with meiotic recombination. During prophase, a recombination-associated proteinaceous structure called the synaptonemal complex (SC) assembles along the chromosome arms of paired chromosomes, establishing a physical connection between homologs. This structure disassembles later in prophase, after which homolog connections are relegated to sites of reciprocal exchange, or chiasmata, that serve to maintain homolog adhesion until their resolution at anaphase I. Chiasmata are stabilized by sister chromatid cohesion proteins (cohesins), and connections made by cohesins between sisters distal to crossover events act to prevent resolution of chiasmata. The regulated destruction of cohesins along chromosome arms initiates anaphase I by allowing resolution of chiasmata and the resulting coordinated separation of homologs (Buonomo et al. 2000). In addition to physically uniting paired homologs, chiasmata also balance opposing poleward forces across the bivalent, and the resulting tension is an important component of a cell cycle checkpoint that monitors proper homolog orientation and alignment prior to anaphase I (Nicklas 1997).
Not all organisms, however, depend on recombination for meiotic chromosome segregation. In the female fruit fly Drosophila melanogaster, alternative genetic pathways exist to segregate chromosomes that have not undergone reciprocal exchange. A distributive, or achiasmate, pathway depends on heterochromatic homologies to segregate nonexchange homologs, while a second achiasmate pathway exists for segregating nonhomologous chromosomes (Hawley et al. 1992). Male Drosophila completely lack meiotic recombination, yet do not appear to use the same achiasmate systems as do females, as mutations in components of the female systems do not affect chromosome segregation in the male. Thus, males appear to have evolved a separate system for pairing and/or homolog adhesion.
A comparison between the female achiasmate and male segregational systems reveals both similarities and differences. Heterochromatic homologies play an important role in both sexes. In females, genetic studies of chromosome rearrangements (Karpen et al. 1996) and cytological studies involving in situ hybridization to heterochromatin sequences (Dernburg et al. 1996) indicate a role of heterochromatin in partner choice and adhesion. Male sex chromosome pairing also relies on heterochromatic homology; the XY pairing sites lie within the rDNA repeats (McKee et al. 1992), which are located in the heterochromatin of both the X and Y chromosomes. Observations of meiosis in males using the LacO/LacI-GFP system to visualize specific loci suggest that heterochromatic pairing may also be critical for maintaining autosomal adhesion. In G2, paired homologs move into discrete nuclear compartments and euchromatic associations between both homologs and sister chromatids are dissolved or relaxed. Associations between homologous centromeres are also lost in G2, and as chromosomes condense and move toward the metaphase plate during prometaphase, neither homologous centromeres nor euchromatic loci appear tightly associated. It is unclear how the bivalents are held together at this point, but it has been suggested that adhesion is maintained either through heterochromatic associations or via topoisomerase-resolvable entanglements (Vazquez et al. 2002). Whatever these associations are, they must be sufficient to balance poleward forces in lieu of chiasmata. In the absence of chiasmata, the role of cohesins in regulating homolog adhesion/separation in males is also unclear, but if they are involved in regulating anaphase onset it seems that their activity may be limited to heterochromatic regions.
Recently, a number of male-specific meiotic mutants have been described that identify genes involved in regulating homolog pairing and adhesion (Hirai et al. 2004; Wakimoto et al. 2004). Two of these, mod(mdg4)in meiosis (MNM) and stromalin in meiosis (SNM), have been characterized at the molecular and cytological level (Thomas et al. 2005). These genes are required for maintaining adhesion between paired homologs at late prophase. MNM and SNM proteins have been colocalized to the sex chromosome pairing sites at this stage, suggesting that they act in an adhesion complex to maintain associations between paired homologs. MNM has also been localized to a small number of foci on each autosomal bivalent, and this latter localization is dependent on an additional gene, teflon (tef) (Thomas et al. 2005). Mutations in tef result in precocious separation of autosomal bivalents during late prophase and early prometaphase of meiosis I, whereas the sex bivalent is unaffected by tef (Tomkiel et al. 2001). Together, these genetic and cytological observations suggest that there are separate protein complexes that mediate homolog adhesion in males: an autosomal complex that is tef-dependent, and a sex chromosome-specific complex that is not. Toward a further understanding of the nature of the differences between autosome and sex chromosome pairing and adhesion, we have cloned and characterized the tef gene. On the basis of our results, we suggest a direct physical role of the Tef protein in maintaining adhesion between paired autosomes.
MATERIALS AND METHODS
Stocks:
Drosophila stocks were grown at 25° on standard media consisting of cornmeal, molasses yeast, and agar. All D. melanogaster mutations and chromosomes are described in Lindsley and Zimm (1992) or in FlyBase (http://flybase.bio.indiana.edu). The wild-type D. pseudoobscura strain MV2-25 was obtained from the Tucson stock center.
Assay for nondisjunction:
Tests for sex and fourth chromosome nondisjunction were performed as previously described (Tomkiel et al. 2001). Males homozygous for the recessive fourth chromosome mutation sparkling-poliert (spa pol) and bearing a yellow (y) X chromosome and In(1)YsX.YL, y+ were mated to tester y w sn3; C(4) ci ey females. Errors in reductional segregation of sex chromosomes were detected as y w sn sons (resulting from nullo-XY sperm) or y+ daughters (resulting from diplo-XY sperm). Reductional or equational fourth chromosome nondisjunction was detected as spa progeny (from diplo-4 sperm) or ci ey progeny (from nullo-4 sperm).
Genomic PCR:
Genomic DNA was isolated from 40 adult flies for each genotype, as described (http://www.fruitfly.org/about/methods/index.html). PCR amplification was performed using the following primer pairs corresponding to D. melanogaster predicted gene CG8961: 5′-CCTCCGCAGTAATGGTAA-3′ and 5′-CTTCTGAGCCTTCGATGG-3′, 5′-GGTGACTATAGTTCACTG-3′ and 5′-GGTACACAGTTCAGTACG-3′, 5′-CATACCGAGCTCATCTC-3′ and 5′-CAAGTGCTTACTCCAACCGC-3′, 5′-ACAGCACAAAGATCCGCA-3′ and 5′-GTAAGCAAAACTAACAGG-3′, and 5′-CTGGCGAACAGCGATATA-3′ and 5′-ATCCAGAGCAGATGGTGA-3′.
Amplification of genomic DNA corresponding to the homologous region from D. pseudoobscura strain MV2-25 was performed using the following primer pairs: 5′-CACATATGTCCTCGTTTCTTGAT-3′ and 5′-TTTCAAGCGATCGCATGATACGCT-3′, 5′-AGCGTATCATGCGATCGCTTGAAA-3′ and 5′-GATTGTATGGTACGTATG-3′, and 5′-CATACGTACCATACAATC-3′ and 5′-TAGCGGCCGCTTGGCACGTTATGGCTTA-3′.
Inverse PCR to analyze transgene insertions was performed as described (http://www.fruitfly.org/about/methods/index.html).
RT–PCR:
Total RNA was isolated from tissues or whole organisms using the RNeasy mini kit (QIAGEN, Palo Alto, CA). Reverse transcription (RT) was performed using Superscript II (Invitrogen, Carlsbad, CA) using 1 μg total RNA and 10 pmol of a gene-specific primer at 42° as per manufacturer's instructions. Full-length teflon cDNA was recovered by RT of total RNA isolated from 1- to 3-day-old adult male testis. For D. melanogaster tef, RT was performed using 5′-TTGCGGCCGCAGAATGAGCGTTTCGCAA-3′. The cDNA was amplified by PCR using the same reverse primer in combination with the 5′-TAGGATCCATATGTCTAAGTTTCTGGA-3′ forward primer. The resulting product was subcloned into pT7-7 (Tabor and Richardson 1985) at the NdeI and NotI sites and verified by DNA sequencing.
For D. pseudoobscura tef cDNA, RT was performed as above using a mixture of the three reverse primers above, and the resulting cDNA was amplified by PCR using the following primer pairs: 5′-ATGTCCTCGTTTCTTGATATTC-3′ and 5′-GAGATGACCGTTAGCGAAGA-3′, 5′-GCCACCCATCAATATAAAAC-3′ and 5′-CCTGTTTACCTCCTCGGTGA-3′, and 5′-GCGGCCAAGTTTAGCAAAAT-3′ and 5′-TGGCACGTTATGGCTTAAGA-3′. PCR cycling conditions were 40 cycles of 30 sec at 94°, 1 min at 51°, and 1 min at 72°. The resulting three overlapping cDNA fragments, each spanning at least one intron, were assembled into a contig.
For tef expression analysis, RT–PCR was performed using primers 5′-GCCCAATGAAACCTGAAC-3′ and 5′-GGTACACAGTTCAGTAGC-3′. These primers flanked an intron in the tef gene, allowing differentiation between PCR products resulting from amplification of cDNA vs. possible contaminant genomic DNA. For fat-spondin expression analysis, 5′-CGGCAAGACGTATAATTTACT-3′ and 5′-CAGGCGCAGCATTCCTT-3′ primers were used.
Creation of transgenic flies:
Full-length teflon cDNA was PCR amplified using 5′-TAGGATCCATATGTCTAAGTTTCTGGA-3′ forward and 5′-TTGCGGCCGCAGAATGAGCGTTTCGCAA-3′ reverse primers. A BamHI–NotI fragment of the resulting PCR product was cloned into the BglII- and NotI-digested pUAST vector (van Roessel and Brand 2000). The gene encoding enhanced green fluorescent protein (EGFP, Clontech, Palo Alto, CA) was amplified using 5′-TTGCGGCCGCTCTGGTGAGCAAGGGCGA-3′ and 5′-CCTCTAGATTACTTGTACAGCTCG-3′ forward and reverse primers. PCR cycling conditions for both reactions were as follows: 30 cycles of 94° for 30 sec, 46° for 30 sec, 72° for 30 sec. The amplified EGFP gene was cloned into BglII- and NotI-digested pUAST-tef vector. The resulting pUAST-tef∷GFP construct produces a fusion protein containing full-length Teflon protein with EGFP fused to its carboxyl terminus. The start methionine codon is absent from the GFP gene, ensuring GFP production only as part of a Tef fusion protein. The plasmid was purified using a DNA purification kit (QIAGEN) and verified by DNA sequencing. All sequencing was performed by MWG (High Point, NC).
Pooled 0- to 1-hr embryos were dechorionated in 50% bleach, rinsed in water, then aligned on double-stick tape. Embryos were dehydrated in chambers containing dri-rite for 5–10 min, then covered with halocarbon oil prior to injection. pUAST-tef∷GFP along with the transposase source P{SB2.1} (DeCicco and Spradling 1984) were injected through Femtotip II glass needles (Eppendorf, Hamburg, Germany) at a concentration of 500 μg/μl each into 1956 embryos. From 183 surviving larvae, 20 adults (11 males and 9 females) were obtained. From these, 16 were fertile (9 males and 7 females). Twenty-four transgenic individuals were obtained from a single G0 adult.
Genomic DNA was extracted from adult flies using the Quick Fly Genomic DNA Prep (http://www.fruitfly.org/about/methods/inverse.pcr.html). The tef∷GFP transgene was amplified by PCR as above using the same EGFP primers as above and the following teflon primer pairs: 5′-TAGGATCCATATGTCTAAGTTTCTGGAC-3′ and 5′-CGGCAATCCAACCGCTT-3′, and 5′-GCCCAATGAAACCTGAAC-3′ and 5′-CACTGAAACCGAACTTGG-3′. These tef-specific primers were designed to bridge the two introns in the teflon gene such that only the transgene cDNA was amplified. PCR fragments were gel purified using the QIAquick Spin columns (QIAGEN) and were sequenced to verify the integrity of the transgene.
Confocal microscopy:
Testes from 0- to 3-day-old adults were dissected in Schneider's Drosophila tissue culture media (Sigma, St. Louis), transferred to 95% ethanol for 1 min, then stained in 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml for 1 min. Tissues were either splayed open or mounted whole in 50% glycerol and examined at ×40–60 using an Olympus Fluoview FV500 confocal laser scanning microscope.
RESULTS
Molecular mapping of tef:
We determined the location of the tef gene using a combination of deletion mapping, complementation testing, and DNA sequencing. The tef gene had previously been mapped to salivary chromosome bands 53F–54A (Tomkiel et al. 2001). We used P element-mediated male recombination (Preston et al. 1996) to generate deletions in this vicinity, starting with a P element inserted at 53E, P{w[+mC]=lacW}·GstS1[k08805]. From 42 male recombination events, a single deletion, P803Δ15, was recovered that extended proximally from the P-insertion site. The presence of the w[+mC] marker gene indicated that the deletion chromosome retained the original P insertion. Flanking sequences were recovered by inverse PCR and DNA sequencing revealed that 64,420 base pairs had been deleted from the proximal side of the P element. This deletion was used in a complementation test with five EMS-induced tef alleles that had been isolated from the Zuker collection (Koundakjian et al. 2004) by screening for mutations that elevated the frequency of fourth chromosome loss (Wakimoto et al. 2004). Four of these were previously described (Tomkiel et al. 2001). The fifth allele, tefZ3455, is described here. Assays of meiotic nondisjunction in males heterozygous for P803Δ15 and each of these tef alleles revealed that this deletion fails to complement the tef male meiotic defect. The frequency of nondisjunction in tefZ5684/P803Δ15 males does not significantly differ from that previously reported for tefZ5684 homozygotes (Table 1) (Tomkiel et al. 2001), indicating that tefZ5684 is a null allele. We previously found that all heteroallelic combinations of the EMS tef alleles behave similarly (Tomkiel et al. 2001). Together, these observations suggest that all five EMS-induced tef alleles behave as null alleles with respect to meiotic chromosome transmission.
TABLE 1.
Sex and fourth chromosome disjunctional data from crosses of y/y+Y; tef;spapol males to y w sn; C(4)EN ci ey/0 females
| Paternal genotype
|
|||
|---|---|---|---|
| Recovered male gametes | tefz5864/ Df803Δ15 | tefl(2)k15914/ tefl(2)k15914 | tefl(2)k15914/ tefz5864 |
| Y;4 | 160 | 688 | 684 |
| X;4 | 204 | 860 | 484 |
| 0;4 | 0 | 0 | 0 |
| X/Y;4 | 0 | 0 | 0 |
| Y;0 | 57 | 0 | 9 |
| X;0 | 65 | 0 | 12 |
| Y;4/4 | 48 | 1 | 16 |
| X;4/4 | 70 | 0 | 16 |
| 0;0 | 0 | 0 | 0 |
| 0;4/4 | 0 | 0 | 0 |
| X/Y;0 | 0 | 0 | 0 |
| X/Y;4/4 | 0 | 0 | 0 |
| Fourth nondisjunction (%) | 39.7 | 0.1 | 4.3 |
Complementation tests were performed using the tefZ5864 allele and a P element, P{w[+mC]=lacW}l(2)·k15914[k15914], that maps within the sequences deleted by P803Δ15. This insertion failed to complement tef, but trans-heterozygous males exhibited only a low frequency of fourth chromosome nondisjunction (∼4%, Table 1), rather than the near-random segregation observed in males homozygous for tefZ5864. This suggests that this P insertion results in a hypomorphic tef allele, and will henceforth be referred to as tefl(2)k15914. Although this element was originally characterized as a lethal insertion, we were able to establish a viable line of flies homozygous for this insertion. Males homozygous for tefl(2)k15914 are wild type with respect to meiotic chromosome transmission (Table 1), consistent with the interpretation that the insertion results in a nearly wild-type, hypomorphic tef mutation.
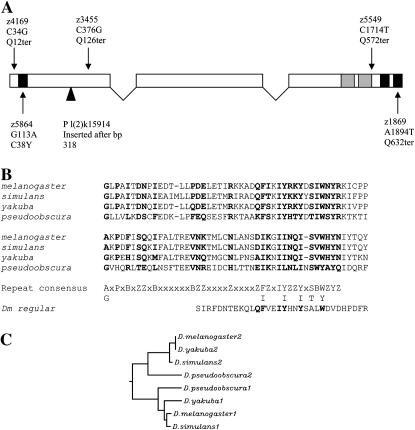
The P insertion of tefl(2)k15914 is located in the first exon of predicted gene CG8961, which is entirely contained with the first intron of a second predicted gene, CG6953, a Drosophila fat-spondin homolog (http://flybase.bio.indiana.edu/). To determine which of these putative genes might be affected in tef mutants, we sequenced PCR-amplified genomic DNA from flies homozygous for each of the five EMS-induced tef alleles. Genomic sequences were compared to that of the original isogenic chromosome on which these mutations were induced. Each mutant allele was found to contain a single base pair substitution in the predicted open reading frame of CG8961. Four of these are nonsense mutations, predicted to cause truncations in the resulting protein, and the fifth was a missense mutation (Figure 1). CG8961 is predicted to encode a 649-amino acid protein that contains three predicted classical C2H2 zinc fingers—one at the amino terminus and two in tandem at the carboxyl terminus. C2H2 zinc fingers are frequently involved in DNA binding (Wolfe et al. 1999), although they may also act as RNA- or protein-binding domains (Shastry 1992; Mackay and Crossley 1998). The tefz5864 allele is a missense mutation predicted to disrupt the amino-terminal zinc finger, changing the first conserved cysteine residue to a tyrosine [C(38)Y] (Figure 1). Sequencing of RT–PCR-recovered CG8961 cDNA from homozygous tefl(2)k15914 males indicated that, despite P insertion within the coding region of the gene, the processed message is wild type. This suggests that the hypomorphic nature of P-insertion alleles may reflect reduced transcription or inefficient splicing to remove P sequences from the CG8961 message.
Figure 1.—
(A) Diagram of D. melanogaster tef transcript and mutations, with solid boxes indicating the positions of consensus sequences for C2H2 zinc-finger motifs and shaded boxes indicating a conserved repeat motif. For each mutation, the allele designation, alteration in DNA sequence, and predicted alteration in amino acid sequence are indicated. The insertion site of the P allele is indicated by a solid triangle. (B) Sequence alignment of a conserved tandem repeat, which resides in amino acid residues 478–573 in D. melanogaster (Z, hydrophilic; B, hydrophobic). The homologous repeat from the D. melanogaster regular gene is indicated below the consensus sequence. (C) ClustalW-generated dendrogram showing the degree of similarity between the two repeat domains identified in the carboxyl half of the predicted Tef protein. Branch lengths are proportional to degree of differences.
Transgene rescue of tef:
We used RT–PCR to isolate wild-type CG8961 cDNA from Oregon-R wild-type flies and sequenced it to confirm the predicted exon/intron structure (Figure 1). To establish that CG8961 indeed corresponded to tef, we created transgenic flies that expressed this cDNA to test for rescue of the tef meiotic defect. We used the GAL4-UAS conditional expression system (Brand and Perrimon 1993) to express full-length CG8961 cDNA as a fusion protein with a carboxy-terminal EGFP. In addition to testing for rescue, this system allowed us to query temporal and spatial requirements for tef expression by utilizing a collection of driver lines that express GAL4 in various male reproductive tissues and at various times with respect to spermatogenesis (Hrdlicka et al. 2002). The GFP fusion also potentially allows visualization of the expressed protein, although efforts to do so have been unsuccessful.
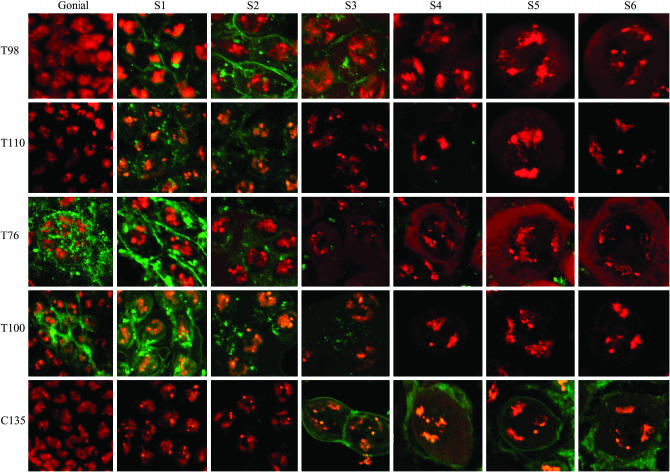
Detailed expression patterns for these GAL4 drivers with respect to spermatocyte development and meiotic progression were not previously reported. We were unable to detect the Tef∷GFP fusion protein and therefore could not directly define the spatial and temporal expression of Tef from these drivers. As an alternative, we characterized the ability of each of the driver lines to express an easily detected pUAS CD8∷GFP reporter gene. The CD8 protein is localized to the cell membrane. This same reporter system was used originally to define the gross expression of these driver lines in living testis (Hrdlicka et al. 2002). To more precisely define expression patterns here, we examined fixed testis that had been stained with the DNA-specific dye DAPI. This allowed use of nuclear morphology to accurately stage expressing cells. Two Gal4 drivers, T155 and C729, caused expression of the GFP reporter exclusively in somatic cells. Among the Gal4 drivers that caused germ-line reporter gene expression, T76, T98, T100, and T110 all drove expression at or prior to the earliest stages of primary spermatocytes (S1–S3) (Cenci et al. 1994). In contrast, C135 showed detectable expression only during later (S4–6) stages of primary spermatocyte development (Figure 2 and Table 2).
Figure 2.—
Testis expression patterns of GFP from a pUAST CD8∷GFP reporter transgene under the control of the indicated GAL4 drivers. DAPI-stained DNA (red) and GFP images (green) of testis from males bearing the indicated GAL4 driver in combination with pUAST CD8∷GFP.
TABLE 2.
Summary of expression patterns
| Cell type assayed for GFP expression
|
Rescue of tef-induced nondisjunction | |||
|---|---|---|---|---|
| GAL4 driver | Cyst cells | Spermatogonia | Spermatocytes | |
| T155 | + | − | − | − |
| C729 | + | − | − | − |
| C135 | + | − | + (S4–6) | − |
| T76 | +/− | + | + (S1–2) | + |
| T100 | − | − | + (S1–3) | + |
| T98 | − | − | + (S1–3) | + |
| T110 | − | + | + (S1–2) | + |
Expression patterns include cyst cell expression patterns reported by Hrdlicka et al. (2002), which were confirmed here (data not shown).
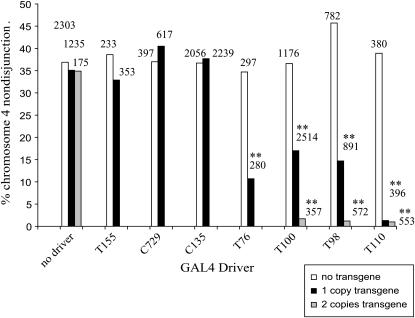
Male flies homozygous for tefz5864, heterozygous or homozygous for a chromosome 2 insertion of the pUAST CG8961∷GFP transgene, and heterozygous for one of the GAL4 driver lines were assayed for fourth chromosome nondisjunction. We monitored segregation of the fourth chromosome because it appears to be more sensitive than the major autosomes to changes in tef gene product, as males bearing the hypomorphic P allele show low frequencies of fourth chromosome nondisjunction, whereas the major autosomes segregate normally (data not shown). These crosses also allowed us to directly calculate nondisjunction frequencies, as viable progeny that are euploid or aneuploid for the fourth chromosome are both recovered from these crosses. Results are shown in Figure 3. The CG8961 transgene alone (in absence of a GAL4 driver) and each GAL4 driver alone (in the absence of the CG8961 transgene) had no effect on fourth chromosome meiotic segregation. Similarly, no rescue was observed in flies with both the CG8961 transgene and either GAL4 driver line T155 or C729. Neither of these lines expresses GAL4 in germ-line cells. In contrast, significant decreases in tef-induced nondisjunction were observed in flies bearing the CG8961 transgene and one of the following GAL4 drivers: T76, T98, T100, or T110. Each of these lines is reported to drive expression in spermatocytes. Nearly complete rescue was seen in flies bearing two copies of the CG8961 transgene in combination with T98, T100, and T110.
Figure 3.—
Complementation of the tef meiotic defect by a pUAS CG8961∷GFP transgene under the transcriptional regulation of various GAL4 drivers. Fourth chromosome nondisjunction frequencies among progeny of tef males bearing the indicated GAL4 driver and transgene combinations are shown. Numbers above bars indicate progeny scored. **Significantly different, P < 0.01.
We also verified that the transgene reduced nondisjunction of the major autosomes both genetically and cytologically. Males homozygous for the tefz5684 and the tef transgene and heterozygous for the T98, T100, or T110 GAL4 driver were mated to C(2)EN females and the number of progeny produced were counted. The only progeny that survive from such a mating are those that receive either two or zero second chromosomes from their father, and therefore progeny numbers are a direct reflection of the frequencies of paternal nondisjunction. From matings of 50 homozygous tef z5684 males lacking the transgene, 145 viable F1 were produced. In contrast, in each case <5 viable progeny were produced from equal numbers of matings of GAL4/+ ; pUAST CG8961∷GFP males bearing the T98, T100, or T110 drivers.
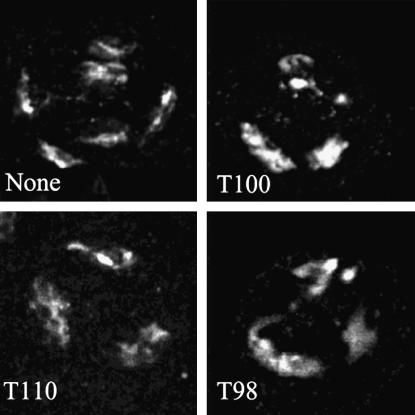
The tef cytological defect observed in late spermatocytes (S6), a separation of autosomal homologs, was also ameliorated by transgene expression in early stage spermatocytes. Autosomal pairing appeared normal in T98, T100, and T110 males bearing two copies of the transgene (Figure 4).
Figure 4.—
Cytological evidence of rescue of the autosomal pairing defect by expression of the UAS tef∷GFP transgene. A representative late prophase (S6) spermatocyte from a male bearing two copies of the UAS tef∷GFP transgene and the indicated GAL4 driver are shown. Each cell is oriented with the sex bivalent uppermost. Note the unpaired major autosomes in the cell lacking a Gal4 driver.
We conclude that predicted gene CG8961 corresponds to tef and that tef expression is required in the germ line to effect proper meiotic chromosome transmission. These observations further suggest that expression of tef is required prior to the S4 stage of primary spermatocyte development to influence chromosome behavior.
Identification of tef homologs in related species:
BLAST homology searches (http://www.ncbi.nlm.nih.gov/) using the full-length Tef primary amino acid sequence failed to identify obvious homologs in non-drosophilids. Significant regions of homology to proteins in other species were limited to the zinc-finger domains. Searches performed using only the zinc-finger domains revealed that the best non-Drosophila matches were to two putative genes from Anopheles gambiae (XM319648 and XM317117), a tumor suppressor gene in Homo sapiens (AF294278), and the transcriptional repressor deltaEF1 in Gallus gallus (D76434). This suggests that Tef, as proposed for MNM and SNM (Thomas et al. 2005), may have evolved to perform a function unique to Drosophila.
Homologs of tef in other Drosophila were identified through BLAST searches of the D. pseudoobscura (http://www.hgsc.bcm.tmc.edu/projects/drosophila/), D. simulans, and D. yakuba databases (http://www.genome.wustl.edu/projects/simulans, ftp://genome.wustl.edu/pub/seqmgr/yakuba). Searches of simulans and yakuba databases prior to their publication were kindly performed by J. C. Yasuhara and B. T. Wakimoto. In all species, the nested arrangement of tef within fat-spondin was found to be conserved. The tef intron/exon structures and predicted Tef proteins in D. simulans and D. yakuba are extremely conserved and are nearly identical to that of D. melanogaster, whereas the D. pseudoobscura gene contains additional introns (see accession AY840221). A ClustalW (http://clustalw.genome.jp/) alignment of all four Tef proteins revealed that they share a similar organization. All contain a single amino terminal zinc-finger motif. D. melanogaster, D. simulans, and D. pseudoobscura also each have two zinc-finger motifs in tandem at the carboxyl end, while D. yakuba lacks the last conserved histidine residue in the second carboxy-terminal zinc-finger motif. In addition to the zinc-finger motifs, this alignment also revealed a conserved 44- to 45-bp motif repeated in tandem adjacent to the carboxy-terminal zinc fingers, ([FI]-X-X-I-[IY]-X(0,1)-[ST]-X-[WY]-X-Y, Figure 1B). Greater similarity was found between the first repeat from all four species than between the first and second repeat within any of the species (Figure 1C). This suggests that the duplication of this motif in Tef occurred in an ancestral species. The conservation between the tandem copies also suggests that this motif may be functionally significant.
Within D. melanogaster, the protein most similar to Tef was found to be Regular (Rgr), a putative zinc-finger-containing transcription factor. The rgr message shows oscillation in abundance in concert with circadian rhythms (Claridge-Chang et al. 2001). Rgr and Tef have a similar amino and carboxyl location of C2H2 zinc fingers. Each protein has a single zinc-finger motif at the amino terminus, but Rgr has seven rather than two at the carboxyl terminus. In addition, Rgr contains homology to a single copy of the Tef tandem repeat, and this homology resides in a similar relative position adjacent to the carboxy-terminal zinc fingers. The overall identity of Rgr to Tef is 22.4% and the similarity is 54.3%. These sequence and structural similarities, considered together with the location of tef within an intron of another gene, suggest that tef may have evolved from an ancient duplication of rgr. Consistent with a more ancient function for Rgr, the Rgr homolog is more highly conserved between D. melanogaster and D. pseudoobscura (72% identical and 77% similarity) than is Tef (35% identical and 43% similarity).
Pattern of tef expression:
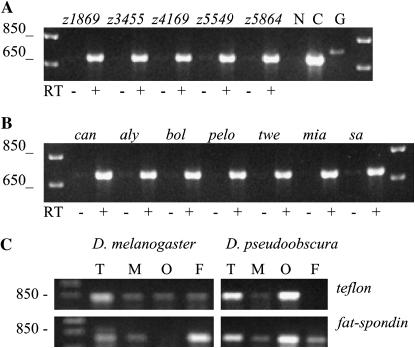
We examined the expression pattern of the tef gene in D. melanogaster by in situ hybridization and RT–PCR. We could not detect tef expression in adult testis by in situ hybridization but could detect its presence by RT–PCR. Using this latter method, we assayed for expression in testis of wild-type and tef males (Figure 5A). We also examined expression in testis of males homozygous for mutations that affect entry into meiotic M phase, including the boule (Eberhart et al. 1996), always early, the transcriptional activating factors cannonball (can), spermatocyte arrest (sa), and meiosis I arrest (mia) (Hiller et al. 2004), and the cell cycle regulators twine (twe) (Alphey et al. 1992) and pelota (pelo) (Eberhart and Wasserman 1995). We detected tef message in males of each genotype, indicating that each of the tef EMS alleles produces transcript and tef expression is not dependent upon initiation of meiotic M (Figure 5B).
Figure 5.—
RT–PCR of tef mRNA agarose gel electrophoresis analysis of RT–PCR products. (A) RT of testis RNA isolated from males homozygous for the indicated tef alleles, amplified with tef-specific primers that flank the first intron. N, no template; C, cDNA template; G, genomic DNA template. (B) RT of testis RNA isolated from males homozygous for mutations that affect entry into or progression through meiosis, amplified using the same primers as in A. (C) RT of total RNA isolated from D. melanogaster or D. pseudoobscura testis (T), male soma minus testis (M), ovaries (O), and female soma minus ovaries (F). Amplifications were done on the same reverse-transcribed RNA samples using primers specific either for tef (top) or for the Drosophila fat-spondin homolog (bottom).
By in situ hybridization in embryos, we observed a uniform distribution of tef message prior to cycle 13, but were unable to detect a signal after cellularization (data not shown). This confirms the embryonic expression pattern for CG8961 previously reported (http://www.fruitfly.org/cgi-bin/ex/insitu.pl). Expression of tef in embryos prior to activation of zygotic transcription suggests a possible role for tef in early development; however, there is no detectable developmental or chromosomal phenotype in embryos from tef null mothers. Furthermore, the severity of the meiotic defect in tef hypomorphic males is not influenced by maternal tef levels (data not shown).
The tef message could also be detected by RT–PCR in 3- to 24-hr embryos, larvae, pupae, and both adult males and females, confirming prior microarray expression analysis (http://genome.med.yale.edu/Lifecycle/).
RT–PCR of the same D. melanogaster RNA samples using primers specific to fat-spondin revealed that its expression was neither reciprocal nor identical to tef. A similar result was obtained in D. pseudoobscura (Figure 5C). This suggests that tef and fat-spondin expression are not coregulated as a consequence of their nested arrangement, although it must be cautioned that this analysis represents an average expression at the tissue level and may not reveal coregulation at the cellular level.
DISCUSSION
Our molecular characterization of tef mutations in combination with transgene rescue experiments reveals that tef corresponds to BDGP predicted gene CG8961. The tef gene, as well as its location within the first intron of a fat-spondin homolog, is conserved in at least three additional Drosophila species. This suggests that tef may have arisen from an ancient duplication and transposition event. The similarity of tef to the rgr gene in D. melanogaster suggests that rgr may be the original ancestral gene. It is interesting to note that rgr was identified as a transcript that is expressed in correlation with circadian rhythms. The Caenorhabditis elegans TIM-1 gene, a paralog of the Drosophila clock gene timeless, provides precedence for a link between circadian rhythm and meiotic chromosome segregation. TIM-1 associates with the cohesin complex and is required for the assembly of cohesin subunits onto meiotic chromosomes (Chan et al. 2003). The homology between tef and rgr may be an indication of an additional relationship between genes involved in circadian rhythm and chromosome segregation.
No obvious tef homologs could be identified by amino acid sequence homology in organisms other than Drosophila. This may reflect that tef evolved specifically in response to a need for meiotic chromosome segregation in the absence of recombination. Conservation of tef in related Drosophila species may reflect a conserved role in segregating noncrossover chromosomes in male meiosis. Like D. melanogaster, D. simulans has been reported to have extremely low rates of male meiotic recombination (Woodruff and Bortolozzi 1976). Male meiotic recombination frequencies have not been reported for D. yakuba or D. pseudoobscura.
Alternatively, proteins analogous to tef may exist but simply not share sufficient primary sequence homology to permit detection. A conserved, repeated motif in Tef is found in a variety of proteins from other species (data not shown), but there is no apparent functional similarity shared by these proteins, and thus the role of this conserved region is enigmatic.
RT–PCR analysis indicated that tef is expressed in a variety of tissues in addition to the male germ line and at various stages of development. The functional significance of this expression is unclear, as there are no detectable somatic phenotypes at any stages of development in tef mutants and no effect on chromosome segregation in females (Tomkiel et al. 2001). The Tef protein may have an additional, redundant function outside of male meiosis, or this expression pattern may be an indirect consequence of fat-spondin regulation, in which the tef gene is nested. That is, establishment of an open chromatin conformation to facilitate fat-spondin expression may also result in tef expression. Such coregulation of nested genes has been observed in mice for the transcription factor Rbpsuhl and the extracellular matrix protein Matn4. Similar to the arrangement of tef within fat-spondin, Rbpsuhl resides within an intron of Matn4 and is coded on the antiparallel strand (Wagener et al. 2001).
The expression of tef in the male germ line is independent of transcriptional regulators of meiosis and is also independent of genes that control meiotic cell cycle progression. This suggests that tef may be expressed early in meiosis. Results of transgene rescue experiments are consistent with this interpretation, as amelioration of the tef segregational defect required transgene expression prior to the S4 stage of spermatocyte development. This requirement suggests Tef is present in wild-type males during the early spermatocyte stages when homologs are paired at both euchromatic and centromeric regions. This is prior to the phenocritical period observed in tef mutants, which occurs at stages S5–6. In wild-type S5–6 spermatocytes, euchromatic and close centromeric associations between homologs are no longer observed, and each bivalent has been sequestered into a unique nuclear domain. It is during these later stages that it has been proposed that the homologs remain associated through heterochromatic connections (Vazquez et al. 2002). In tef mutants, homolog associations are disrupted at S5 and S6 and homologs prematurely separate. Our observations support a model in which Tef functions during pairing stages to ensure bivalent adhesion after euchromatic associations are disrupted. Tef might be loaded onto the bivalent, perhaps at heterochromatin, prior to the dissolution of euchromatic pairing.
This model is well accommodated by the domain organization of the predicted Tef protein. We suggest that the antithetically located zinc fingers in the Tef protein may act as part of a bridging structure, analogous to a synaptonemal complex that physically connects homologs. Tef's terminal zinc fingers may recognize specific DNA sequences or chromatin-associated proteins at one end and mediate self-interaction at the other end, either directly or through an adhesion complex. In this manner, Tef's function might be analogous to the yeast SC protein Zip1 (Sym et al. 1993; Sym and Roeder 1995; Dong and Roeder 2000). This model is supported by our molecular analysis of tef mutants, which revealed that the zinc-finger domains at either end of the protein are required for its function. Either truncation of the carboxy-terminal zinc finger or alteration of a conserved cysteine residue in the amino-terminal finger results in a null mutation. A variation of this model is that, rather than acting as the bridge itself, chromatin-associated Tef protein may play an essential role in tethering an MNM-containing adhesion complex, as suggested by Thomas et al. (2005). This might be considered analogous to the role proposed for the zinc-finger-containing yeast lateral element SC component Hop1 in tethering other SC components (Hollingsworth and Ponte 1997; de los Santos and Hollingsworth 1999).
This model makes the testable prediction that Tef will reside between paired homologous autosomes at late prophase metaphase of meiosis I. It further suggests that Tef may be a useful tool for identifying the cis-acting sequences that mediate autosomal pairing and/or adhesion.
Acknowledgments
We thank Margaret Fuller and Steve Wasserman for providing fly stocks, Dennis Lajeunesse for use of injection facilities, and Jiro Yasuhara and Barbara Wakimoto for assistance with BLAST searches of D. yakuba and D. simulans libraries. This work was funded by National Science Foundation grant MCB-0350707. Confocal microscopy was performed on instrumentation funded by National Science Foundation grant DBI-0319021, the North Carolina Biotechnology grant 2003-IDG-1011, and the University of North Carolina at Greensboro Department of Biology.
References
- Alphey, L., J. Jimenez, H. White-Cooper, I. Dawson, P. Nurse et al., 1992. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69: 977–988. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann et al., 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387–398. [DOI] [PubMed] [Google Scholar]
- De Cicco, D. V., and A. C. Spradling 1984. Localization of a cis-acting element responsible for the developmentally regulated amplification of Drosophila chorion genes. Cell 38: 45–54. [DOI] [PubMed] [Google Scholar]
- Cenci, G., S. Bonaccorsi, C. Pisano, F. Verni and M. Gatti, 1994. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107: 3521–3534. [DOI] [PubMed] [Google Scholar]
- Chan, R. C., A. Chan, M. Jeon, T. F. Wu, D. Pasqualone et al., 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423: 1002–1009. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang, A., H. Wijnen, F. Naef, C. Boothroyd, N. Rajewsky et al., 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., and N. M. Hollingsworth, 1999. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J. Biol. Chem. 274: 1783–1790. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., J. W. Sedat and R. S. Hawley, 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Dong, H., and G. S. Roeder, 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart, C. G., and S. A. Wasserman, 1995. The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development 121: 3477–3486. [DOI] [PubMed] [Google Scholar]
- Eberhart, C. G., J. Z. Maines and S. A. Wasserman, 1996. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 381: 783–785. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., H. Irick, A. E. Zitron, D. A. Haddox, A. Lohe et al., 1992. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 13: 440–467. [DOI] [PubMed] [Google Scholar]
- Hiller, M., X. Chen, M. J. Pringle, M. Suchorolski, Y. Sancak et al., 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131: 5297–5308. [DOI] [PubMed] [Google Scholar]
- Hirai, K., S. Toyohira, T. Ohsako and M. T. Yamamoto, 2004. Isolation and cytogenetic characterization of male meiotic mutants of Drosophila melanogaster. Genetics 166: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., and L. Ponte, 1997. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlicka, L., M. Gibson, A. Kiger, C. Micchelli, M. Schober et al., 2002. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis 34: 51–57. [DOI] [PubMed] [Google Scholar]
- Karpen, G. H., M. H. Le and H. Le, 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. [DOI] [PubMed] [Google Scholar]
- Koundakjian, E. J., D. M. Cowan, R. W. Hardy and A. H. Becker, 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167: 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Mackay, J. P., and M. Crossley, 1998. Zinc fingers are sticking together. Trends Biochem. Sci. 23: 1–4. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., L. Habera and J. A. Vrana, 1992. Evidence that intergenic spacer repeats of Drosophila melanogaster rRNA genes function as X-Y pairing sites in male meiosis, and a general model for achiasmatic pairing. Genetics 132: 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R. B., 1997. How cells get the right chromosomes. Science 275: 632–637. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry, B. S., 1992. Transcription factor IIIA(TFIIIA) in the second decade. J. Cell. Sci. 109: 535–539. [DOI] [PubMed] [Google Scholar]
- Sym, M., and G. S. Roeder, 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym, M., J. A. Engebrecht and G. S. Roeder, 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Thomas, S. E., M. Soltani-Bejnood, P. Roth, R. Dorn, J. M. Logsdon, Jr. et al., 2005. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 123: 555–568. [DOI] [PubMed] [Google Scholar]
- Tomkiel, J. E., B. T. Wakimoto and A. Briscoe, Jr., 2001. The teflon gene is required for maintenance of autosomal homolog pairing at meiosis I in male Drosophila melanogaster. Genetics 157: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel, P., and A. H. Brand, 2000. GAl4-mediated ectopic gene expression in Drosophila, pp. 458–461 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Vazquez, J., A. S. Belmont and J. W. Sedat, 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Wagener, R., B. Kobbe, A. Aszodi, D. Aeschlimann and M. Paulsson, 2001. Characterization of the mouse matrilin-4 gene: a 5′ antiparallel overlap with the gene encoding the transcription factor RBP-l. Genomics 76: 89–98. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B. T., D. L. Lindsley and C. Herrera, 2004. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, S. A., L. Nekludova and C. O. Pabo, 1999. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biomol. Struct. 3: 183–212. [DOI] [PubMed] [Google Scholar]
- Woodruff, R. C., and J. Bortolozzi, 1976. Spontaneous recombination in males of Drosophila simulans. Heredity 37: 295–298. [DOI] [PubMed] [Google Scholar]