Abstract
The regulation of cellular membrane dynamics is crucial for maintaining proper cell growth and division. The Cdc48-Npl4-Ufd1 complex is required for several regulated membrane-associated processes as part of the ubiquitin–proteasome system, including ER-associated degradation and the control of lipid composition in yeast. In this study we report the results of a genetic screen in Saccharomyces cerevisiae for extragenic suppressors of a temperature-sensitive npl4 allele and the subsequent analysis of one suppressor, GET3/ARR4. The GET3 gene encodes an ATPase with homology to the regulatory component of the bacterial arsenic pump. Mutants of GET3 rescue several phenotypes of the npl4 mutant and transcription of GET3 is coregulated with the proteasome, illustrating a functional relationship between GET3 and NPL4 in the ubiquitin–proteasome system. We have further found that Get3 biochemically interacts with the trans-membrane domain proteins Get1/Mdm39 and Get2/Rmd7 and that Δget3 is able to suppress phenotypes of get1 and get2 mutants, including sporulation defects. In combination, our characterization of GET3 genetic and biochemical interactions with NPL4, GET1, and GET2 implicates Get3 in multiple membrane-dependent pathways.
INTRACELLULAR membranes such as the nuclear envelope and the compartments of the secretory pathway are the key feature that distinguishes the eukaryotic cell from bacteria. Along with the evolution of membrane-bound organelles came the need for intricate and highly regulated mechanisms to control the composition and movements of these membranes in response to environmental changes and during complex cellular events such as the cell cycle or meiosis (Howe and McMaster 2001; reviewed in Albertson et al. 2005).
The highly conserved Cdc48-Npl4-Ufd1 complex is an important regulator of several membrane-associated cellular processes in eukaryotic cells. In one critical role, the Cdc48-Npl4-Ufd1 complex participates in the clearing of aberrantly folded proteins from the endoplasmic reticulum (ER) through ER-associated degradation (ERAD) (Bays et al. 2001). During ERAD, misfolded ER proteins are retrotranslocated to the cytosol where they are ubiquitinated and degraded by the proteasome (reviewed in Romisch 2005). The increased level of certain ER proteins that occurs if this process is prevented can cause drastic alterations in the organization of the ER compartment (Wright et al. 2003). The Cdc48-Npl4-Ufd1 complex also functions to regulate intracellular membranes in yeast through a pathway that controls production of unsaturated fatty acids. Specifically, the Cdc48-Npl4-Ufd1 complex directs the ubiquitin–proteasome-dependent cleavage and activation of two ER-membrane anchored transcription factor precursors, Spt23 and Mga2 (Hoppe et al. 2000; Hitchcock et al. 2001). Once released from the membrane, Spt23 and Mga2 activate transcription of the OLE1 gene (Zhang et al. 1999; Chellappa et al. 2001), which encodes a fatty acid desaturase enzyme (Stukey et al. 1990). Thus, not only is the Cdc48-Npl4-Ufd1 complex important for the proteasome-dependent degradation and processing of protein substrates at the ER membrane, but it is also required for the regulation of cellular unsaturated fatty acid (UFA) content, and in turn membrane fluidity, through transcriptional control of OLE1. A further requirement has been shown for Cdc48-Npl4-Ufd1 in postmitotic nuclear membrane assembly in higher eukaryotes (Hetzer et al. 2001).
Forward genetic screens in the yeast Saccharomyces cerevisiae have been critical in elucidating NPL4 function in ERAD (Bays et al. 2001) and in OLE1 regulation (Hoppe et al. 2000; Hitchcock et al. 2001). As such, we sought to learn more about NPL4 function and membrane dynamics by performing a screen for extragenic suppressors of a temperature-sensitive npl4 mutant. In this study, we present the full panel of genes that we identified as extragenic suppressors of an npl4 mutation. In addition, we present extensive characterization of one gene identified as an npl4 suppressor, the GET3/ARR4 gene. GET3 encodes a highly conserved ATPase with homology to ArsA, the regulatory component of the bacterial arsenic export pump (Boskovic et al. 1996), and to a human protein of unknown function, hASNA-I (Kurdi-Haidar et al. 1996). Get3 has been suggested to have roles in cellular resistance to stress (Shen et al. 2003), in metal ion homeostasis (Metz et al. 2006), and, in complex with Get1 and Get2, in protein sorting via the secretory pathway (Schuldiner et al. 2005).
Here we present evidence that GET3 displays functional interactions with NPL4 in the context of the ubiquitin–proteasome pathway. We further illustrate interactions of GET3 with GET1 and GET2 during sporulation, a process in which dynamic new growth of cellular membranes is required for proper spore and spore wall formation. In particular, we have demonstrated that Get3 can play an antagonistic role in both settings, leading to the proposal that Get3 is a functionally conserved regulator of membrane-associated proteins.
MATERIALS AND METHODS
Yeast strains and manipulations:
Standard yeast methods and media were used (Guthrie and Fink 1991). The genotypes of all strains used in this study are provided in Table 1. The npl4-1 mutant strains are FY23-backcrossed strains derived from PSY825 and PSY826, which were previously described (DeHoratius and Silver 1996). Null alleles and C-terminal tags were integrated using PCR-based techniques (Baudin et al. 1993; Knop et al. 1999). CPY* was integrated into wild-type (WT), npl4-1, and Δget3 npl4-1 strains using two-step gene replacement (Adams et al. 1997) of plasmid pRS306-prc1-1 as described (Knop et al. 1996).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| PSY3322 | MATα ura3-52 leu2Δ1 trp1Δ63 | This study |
| PSY3323 | MATα ura3-52 leu2Δ1 trp1Δ63 npl4-1 | This study |
| PSY3383 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 cue1:Tn [LEU2] | This study |
| PSY3384 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 vps27:Tn [LEU2] | This study |
| PSY3385 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 srn2:Tn [LEU2] | This study |
| PSY3386 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 get3:Tn [LEU2] | This study |
| PSY3091 | MATaura3-52 leu2Δ1 trp1Δ63 npl4-1 spt23:Tn [LEU2] | Hitchcock et al. (2001) |
| PSY3387 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 ifh1:Tn [LEU2] | This study |
| PSY3388 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 prp6:Tn [LEU2] | This study |
| PSY3389 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 cbp80:Tn [LEU2] | This study |
| PSY3390 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 Δget3∷HIS3 | This study |
| PSY3391 | MATα ura3-52 leu2Δ1 trp1Δ63 Get3tn-3HA [KanMX] | This study |
| PSY3392 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4-1 Δget3∷HIS3 | This study |
| PSY3393 | MATα ura3-52 leu2Δ1 trp1Δ63 Δget1∷KanMX | This study |
| PSY3394 | MATα ura3-52 leu2Δ1 trp1Δ63 Δget2∷KanMX | This study |
| PSY3395 | MATα ura3-52 leu2Δ1 trp1Δ63 Δget1∷KanMX npl4-1 | This study |
| PSY3396 | MATα ura3-52 leu2Δ1 his3Δ200 npl4Δ∷NPL4-sGFP [URA3] Get3-tevProA [KanMX] | This study |
| PSY3066 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 npl4Δ∷NPL4-sGFP∷URA3 | This study |
| PSY3397 | MATα ura3-52 leu2Δ1 trp1Δ63 Get3-EGFP [KanMX] | This study |
| PSY3398 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 Get3-EGFP [KanMX] Δget1∷LEU2 | This study |
| PSY3399 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 Get3-EGFP [KanMX] Δget2∷HIS3 | This study |
| PSY3400 | MATα ura3-52 leu2Δ1 his3Δ200 Get3-EGFP [KanMX] Δget1∷LEU2 Δget2∷HIS3 | This study |
| PSY3164 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δget3∷KanMX | This study |
| PSY1930 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Winzeler et al. (1999) |
| PSY3402 | MATα ura3-52 leu2Δ1 trp1Δ63 CPY* | This study |
| PSY3403 | MATα ura3-52 leu2Δ1 trp1Δ63 npl4-1 CPY* | This study |
| PSY3404 | MATaura3-52 leu2Δ1 his3Δ200 trp1Δ63 Δget3∷HIS3 CPY* | This study |
| PSY3405 | MATα ura3Δ leu2Δ Δget3∷LEU2 Δget2∷KanMX | This study |
| PSY3406 | MATα ura3-52 leu2Δ1 his3Δ200 Δget3∷HIS3 Δget1∷KanMX | This study |
| PSY3407 | MATa/MATα ho∷hisG/ho∷hisG lys2/lys2 ura3/ura3 leu2/leu2 his3/his3 trp1ΔFA/trp1ΔFA | Benjamin et al. (2003) |
| PSY3408 | MATa/MATα ho∷hisG/ho∷hisG lys2/lys2 ura3/ura3 leu2/leu2 his3/his3 trp1ΔFA/trp1ΔFA Δget3∷KanMX/Δget3∷KanMX | This study |
| PSY3409 | MATa/MATα ho∷hisG/ho∷hisG lys2/lys2 ura3/ura3 leu2/leu2 his3/his3 trp1ΔFA/trp1ΔFA Δget2∷TRP1/Δget2∷TRP1 | This study |
| PSY3410 | MATa/MATα ho∷hisG/ho∷hisG lys2/lys2 ura3/ura3 leu2/leu2 his3/his3 trp1ΔFA/trp1ΔFA Δget3∷KanMX/Δget3∷KanMX Δget2∷TRP1/Δget2∷TRP1 | This study |
Screens for extragenic suppressors of npl4-1:
npl4-1 cells were mutagenized with either ethyl methanesulfonate (EMS) or an mTn-lacZ/LEU2 transposon library (Burns et al. 1994). Methods for the EMS screen and cloning of UBC7 and CUE1 have been described (Hitchcock et al. 2003). Suppressing mutations in DOA4 and UBP3 were identified by transformation with a CEN-based URA3-marked plasmid library containing yeast genomic DNA fragments (Rose et al. 1987) and subsequent analysis of the rescuing plasmids pPS2915, pPS2918, and pPS2931. Transposon mutagenesis was performed essentially as described (Seifert et al. 1986). Two micrograms of NotI-digested DNA were transformed into npl4-1 cultures, and colonies able to grow at the nonpremissive temperature of 30° were observed at the rate of 5 × 10−5. The location of insertion was identified for nine of these colonies by vectorette PCR, as described (Kumar and Snyder 2000).
ERAD assay:
Cells were grown to OD600 = 0.5–1.0, pelleted, and resuspended in 3 ml YPD containing 100 ug/ml cyclohexamide. Samples were removed at each time point and prepared as described (Johnson et al. 1995) for immunoblotting with anti-CPY (Molecular Probes, Eugene, OR). Equal protein loading was verified by subsequent staining with amido black.
Northern blot analysis:
Cultures were grown in YPD at 25° to log phase and then split such that half continued growth at 25° while half were shifted to 37° for 2 hr. All samples were frozen and total RNA was prepared in parallel. Northern blotting and analysis were performed as described (Hitchcock et al. 2001).
Cell fractionation:
Get3-EGFP cells were grown at 30° to logarithmic phase in YPD. Subcellular fractionation and solubilization of the P13 fraction were performed essentially as described (Munoz-Centeno et al. 1999). To determine the fractionation profile of Get3, equal cell volumes were analyzed by Western blot with antibodies to GFP (Seedorf et al. 1999) and Sec62.
Affinity purification and mass spectrometry:
Get3-TEV-protein A and control cells expressing protein A from plasmid pPS1973 were grown in media lacking leucine to logarithmic phase. Microsomes were prepared essentially as described (Baker et al. 1990). After homogenization of spheroplasts, the membrane fraction was isolated by centrifugation at 13,000 rpm for 15 min. The pellet was resuspended and washed in B88 buffer (20 mm HEPES, pH 6.8, 150 mm KOAc, 5 mm Mg(OAc)2, 250 mm sorbitol). Equal protein amounts were resuspended in IPPT-150 buffer (10 mm Tris pH 8.0, 150 mm NaCl, 0.5% Triton X-100) supplemented with 0.5 mm DTT, 0.5 mm PMSF, and protease inhibitors, and proteins were extracted by rocking at 4° for 1–5 hr.
Affinity purification was performed overnight at 4° using 30 μl of IgG Sepharose bead slurry (Pharmacia/Amersham, Piscataway, NJ), 50 μl of 80% glycerol, and 500 μl of extracted proteins. After washing with IPPT-150 buffer (as above except 0.1% Triton), cleavage with TEV protease (GIBCO, Grand Island, NY) was performed in IPPT-150 supplemented with 0.5 mm EDTA and 1 mm DTT at 14°. After SDS–PAGE, silver-stained bands were excised from the gel and analyzed by MALDI-TOF mass spectrometry at the Southern Alberta Mass Spectrometry Centre (Calgary, AB, Canada).
Transcriptional profiling and genomic analysis:
Expression profiling was carried out in triplicate, with swapping of fluor orientation, from Δget3 and WT cells grown at 30° in YPD to early logarithmic phase. Total RNA preparation, cDNA preparation, and hybridization were performed as described (Casolari et al. 2004). Up- and downregulated genes were defined as genes with fold change ≥1.5 and P-value ≤0.05 as determined by Rosetta Resolver. Analysis for enrichment of functional classes was performed using FuncAssociate (Berriz et al. 2003). Data mining for GET3 transcription was performed using Cluster and TreeView software from Eisen et al. (1998) and tools available on the Saccharomyces Genome Database (Dolinski et al. 2006).
Sporulation experiments:
Synchronous sporulation of homozygous SK1 diploid cells was performed using previously described methods (Padmore et al. 1991; Huang et al. 2005). Samples were monitored throughout sporulation to ensure good synchrony and efficiency. Each sample was fixed and stained with DAPI before observation by light and fluorescence microscopy (Grether and Herskowitz 1999). For quantification, images were recorded of 36-hr samples using a Nikon microscope equipped with a DAPI filter (Chroma Technology, Brattleboro, VT) and a 100× DIC (Nomarski) objective. At least 200 cells were scored for each strain. Spore viability was determined by tetrad dissection after sporulation for 2–4 days on 1% potassium-acetate plates.
RESULTS
Identification of npl4-1 extragenic suppressors:
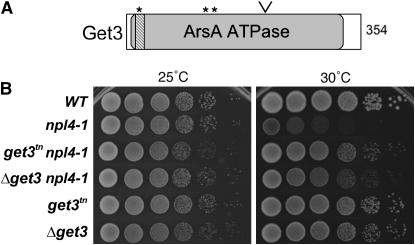
Several phenotypes affecting cellular membranes have been observed in the npl4-1 mutant allele, including defects in ERAD, and in nuclear envelope structure and nucleocytoplasmic transport likely due to the misregulation of OLE1 (DeHoratius and Silver 1996; Bays et al. 2001). To learn more about these effects, we conducted a genetic screen for extragenic suppressors of the temperature-sensitive lethality of this npl4 mutant at 30°. npl4-1 cells were mutagenized with either EMS or transposon-mutagenized yeast genomic DNA, and mutants capable of growth at 30° were isolated, characterized, and cloned. This analysis resulted in the identification of 11 genes whose mutation was capable of rescuing growth at the nonpermissive temperature (Table 2). Suppressing mutations isolated by EMS mutagenesis fell into five distinct complementation groups, four of which were identified by various techniques (materials and methods). An additional 7 genes with suppressing mutations were isolated as transposon insertions and were cloned by vectorette PCR. One gene, CUE1, was isolated by both techniques. None of the isolated suppressors restored wild-type growth to the npl4-1 mutant or were able to rescue at higher temperatures (Figure 1B and data not shown), suggesting that in each case the deficits of this mutation were only partially corrected.
TABLE 2.
Suppressors of npl4-1
| Gene | Cellular activity | Allele(s) | |
|---|---|---|---|
| ERAD | |||
| YMR264W | CUE1a,c | Docking of Ubc7 in ERAD | Transposon after AA72, EMS (5) |
| YMR022W | QRI8/UBC7a,c | Ubiquitin-conjugation to ERAD substrates | EMS (16) |
| Secretion | |||
| YNR006W | VPS27 | Sorting of ubiquitinated proteins in MVB pathway | Transposon after AA260 |
| YLR119W | SRN2/VPS37 | Sorting of ubiquitinated proteins in MVB pathway | Transposon after AA44 |
| YDR069C | DOA4c | Deubiquitination in endosome to vacuole transport | EMS (8) |
| YER151C | UBP3c | Deubiquitination in vesicle transport | EMS (2) |
| Transcription/RNA modulation | |||
| YKL020C | SPT23b | Activation of OLE1 transcription | Transposon after AA710 |
| YLR223C | IFH1 | Chromatin assembly and silencing | Transposon after AA155 |
| YBR055C | PRP6 | Pre-mRNA splicing (U4/U6-U5 snRNP) | Transposon after AA793 |
| YMR125W | STO1/CBP80 | Nuclear mRNA cap-binding protein | Transposons (2) in noncoding regions |
| YDL100C | GET3c | Homolog of bacterial arsenite transporter | Transposon after AA215 |
Suppressors generated by EMS or transposon mutagenesis are listed by functional class. Several were isolated multiple times, as indicated. For mutants isolated from the transposon screen, the integration site is listed.
Previously published in Hitchcock et al. (2003).
Previously published in Hitchcock et al. (2001).
Null allele was tested and also found to rescue growth of npl4-1.
Figure 1.—
get3 mutants suppress the temperature-sensitive lethality of npl4-1. (A) Diagram of the Get3 protein. The site of truncation in the get3tn allele is shown (∨) along with the p-loop ATP-binding site (striped box). Predicted myristoylation sites are marked with asterisks. (B) get3 mutants rescue npl4-1 growth at 30°. Wild-type (WT), npl4-1, get3, and double-mutant strains were grown to log phase and then serially diluted and plated to rich media at 25° and 30° for 2 days.
Mutations in genes affecting several pathways, including ERAD, secretion, and gene expression, were able to ameliorate the temperature-sensitive lethality of npl4-1 (Table 2). Two of the suppressors are active in ERAD (CUE1 and UBC7) (Biederer et al. 1997) and have already been reported by our lab as suppressors of npl4-1 (Hitchcock et al. 2003). All four suppressors in the secretory pathway class have also been implicated in ubiquitin-dependent processes: DOA4 and UBP3 have direct roles in deubiquitination at secretory vesicles (Amerik et al. 2000; Cohen et al. 2003a), whereas VPS27 and SRN2/VPS37 are involved in the sorting and degradation of ubiquitinated proteins via the multivesicular body (MVB) pathway (Katzmann et al. 2001; Bilodeau et al. 2002). Genes in the third class of npl4-1 extragenic suppressors affect transcription (SPT23, IFH1) (Zhang et al. 1999; Dula and Holmes 2000; Schawalder et al. 2004; Wade et al. 2004) or have roles in RNA processing (PRP6, STO1) (Abovich et al. 1990; Das et al. 2000). The rescue of OLE1 transcription in npl4-1 cells by truncations of SPT23 and MGA2 has been reported previously (Hitchcock et al. 2001); however, the links between the remaining members of class 3 and Npl4 and/or ubiquitin–proteasome function are unclear.
In addition to these genes with known functions, we identified a suppressing mutation in the YDL100C open reading frame (ORF), which had no reported functions at the time of the screen. YDL100C has since been named ARR4 for its homology to the bacterial ArsA protein (Shen et al. 2003) and, more recently, GET3 (Schuldiner et al. 2005). We chose to pursue the characterization of this gene as a means to gain insight into both Get3 and Npl4 function.
get3 mutants suppress npl4-1-associated phenotypes:
The get3 mutant isolated as a suppressor of npl4-1 is a truncation produced by transposon insertion and is designated get3tn. The transposon insertion adds two amino acids and a premature stop codon after K215 of Get3, generating a truncated protein product consisting of the first 60% of Get3 (Figure 1A). This region contains the P-loop ATP-binding site and several predicted myristoylation sites. We observed that both get3tn and the null allele Δget3 were able to suppress npl4-1 temperature-sensitive lethality at 30° (Figure 1B). However, phenotypic differences between the two alleles suggested that the isolated transposon insertion does not generate a null allele; specifically, Δget3 is a weaker suppressor of npl4-1 and has a more marked growth defect in cells that are wild type for NPL4 than does the get3tn allele (Figure 1B).
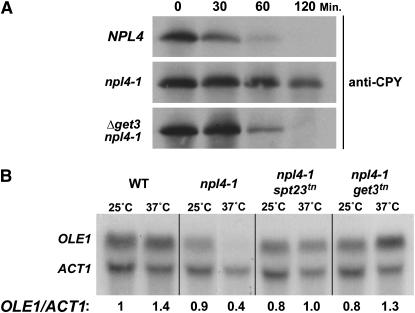
We next tested whether mutants of get3 rescued npl4-1 phenotypes in addition to temperature-sensitive lethality, including a diminished capacity for ERAD and misregulation of OLE1 transcription (Figure 2). To test for function of the ERAD pathway in get3 npl4-1 cells, a mutant allele of carboxypeptidase Y (CPY*) that is subject to rapid degradation via the ERAD system was integrated into the npl4-1 and get3 mutant strains. Cells expressing CPY* were treated with cyclohexamide to halt protein synthesis, and levels of this protein remaining at various time points after treatment were determined by Western blot analysis with anti-CPY antibodies. As expected, CPY* was degraded rapidly in wild-type cells with a half-life of ∼30 min (Figure 2A). In contrast, the half-life of CPY* was significantly extended in npl4-1 cells, as has been previously reported (Bays et al. 2001). Interestingly, the get3 null and transposon alleles both displayed partial rescue of the npl4-1 ERAD defect (Figure 2A and data not shown), decreasing the half-life of the CPY* protein in npl4-1 cells to <1 hr. We found that the Δget3 allele does not have a strong defect or enhancement of ERAD in cells that are wild type for NPL4 (supplemental material at http://www.genetics.org/supplemental/).
Figure 2.—
get3 mutants suppress npl4-1 phenotypes. (A) Δget3 rescues ERAD-mediated CPY* degradation in npl4-1 cells. Wild-type (NPL4), npl4-1, and npl4-1 Δget3 mutant cells were grown to log phase and protein synthesis was halted by treatment with cyclohexamide. Samples were collected 30, 60, and 120 min after cyclohexamide treatment, separated by SDS–PAGE, and subjected to Western blot analysis with anti-CPY antibodies. (B) get3tn restores OLE1 transcription in npl4-1 cells. Northern analysis was performed on total RNA isolated from wild type (WT), npl4-1, or npl4-1 with the suppressing mutations spt23tn or get3tn, using OLE1- and ACT1-specific DNA probes. Cells were either continuously grown at 25° or shifted to 37° for 2 hr (labeled as 25° and 37°, respectively) prior to RNA purification. The ratio of OLE1/ACT1 signal for each sample is given (at the bottom) relative to that for the WT 25° sample.
To determine whether get3 mutation was able to rescue OLE1 transcription in npl4-1 cells, we performed Northern blot analysis using probes for OLE1, and for ACT1 (actin) as a loading control. Total RNA was analyzed from npl4-1 and get3 single- and double-mutant cells, shifted to the nonpermissive temperature of 37° for 2 hr. After shift to 37°, OLE1 transcript levels were observed to increase in wild-type cells by ∼40% and decrease in the npl4-1 mutant by ∼50% compared to the unshifted samples (Figure 2B; Hitchcock et al. 2001). We found that the transposon allele of get3 was able to rescue OLE1 transcription to wild-type levels (Figure 2B). It appears that this effect is specific to the get3tn allele, since Δget3 does not restore OLE1 transcription in npl4-1 cells; however, Δget3 does block the elevation of OLE1 transcript levels seen at 37° in NPL4 cells (supplemental material at http://www.genetics.org/supplemental/). We then compared the ability of get3tn to rescue OLE1 transcription with that of a dominant activating truncation of Spt23 (spt23tn), which encodes a transcription factor that activates OLE1 gene expression (Zhang et al. 1999). Surprisingly, we found that the rescue of OLE1 transcript level in npl4-1 is more robust in the get3 mutant than in the spt23 mutant.
GET3 is coregulated with the proteasome and the Cdc48-Npl4-Ufd1 complex:
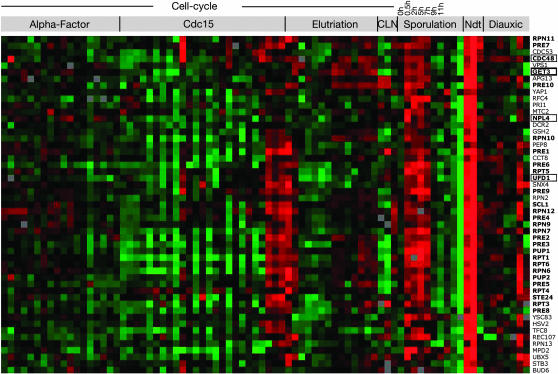
Given that genes with similar function often exhibit transcriptional coregulation, we used genomic analyses of available transcriptional profiling data to examine GET3 expression and coregulation with other yeast genes under various growth conditions. First, the transcriptional profiles of all yeast genes under conditions including specific cell cycle stage, diauxic shift, and sporulation were clustered as described (materials and methods; Eisen et al. 1998). The genes showing the most similar expression with GET3 under these conditions, shown in Figure 3, include a significant number of genes encoding proteasomal components (P < 0.001 by FuncAssociate) and those encoding the Cdc48-Npl4-Ufd1 complex. Supporting this result, we found that GET3 also clustered with proteasome-encoding genes using a different computational method (Jelinsky et al. 2000). These genomic data fit well with the genetic interaction we found between GET3 and NPL4 and provide evidence indicating functional connections between GET3 and the ubiquitin–proteasome pathway.
Figure 3.—
GET3 is coregulated with genes encoding components of the proteasome and the Cdc48-Npl4-Ufd1 complex. GET3 (thick box) is similarly expressed with many protein-degradation genes (boldface type) and genes encoding the Cdc48/Npl4/Ufd1 complex (thin boxes). The data set of expression profiling under various conditions from Eisen et al. (1998) was analyzed by hierarchical clustering using Average Link correlation (uncentered), and the GET3-containing cluster from analysis with TreeView software is shown. Treatments analyzed include cell cycle time courses, the diauxic shift, a time course during sporulation, and the altered expression of NDT80.
The strongest coregulation observed between GET3, NPL4, and the proteasome components occurs under conditions relating to sporulation (Figure 3). Expression of nearly all the genes in this cluster is elevated early in sporulation and under altered expression of NDT80 (Figure 3), which encodes a master regulator for transcriptional activation of middle sporulation-specific genes (Xu et al. 1995; Chu and Herskowitz 1998; Hepworth et al. 1998). Looking more specifically at the regulation of GET3 during sporulation in existing data sets for genomewide expression during sporulation in SK1 cells (Chu et al. 1998; Primig et al. 2000), we found that GET3 mRNA expression is slightly elevated shortly after induction of sporulation. The set of 20 genes with the most similar expression pattern to GET3 during sporulation, according to the Saccharomyces Genome Database (Dolinski et al. 2006), includes genes encoding several proteasome subunits (data not shown) and is significantly enriched for genes involved in ubiquitin-dependent protein catabolism (P < 0.001 by FuncAssociate).
Get3 membrane localization depends on Get1 and Get2:
We next examined the subcellular localization and biochemical characteristics of the Get3 protein. For these analyses, DNA sequence encoding an EGFP tag was integrated in frame at the 3′ end (translated C terminus) of the genomic locus of GET3; the resulting GET3-EGFP strain exhibits no growth defect (data not shown), indicating that the EGFP tag does not disrupt Get3 function. Upon examination by live-cell fluorescence microscopy, Get3-EGFP was observed at the ER/nuclear membrane and in the cytoplasm (Figure 4A). This finding was consistent with published reports for Get3 localization in rich media (Huh et al. 2003; Schuldiner et al. 2005).
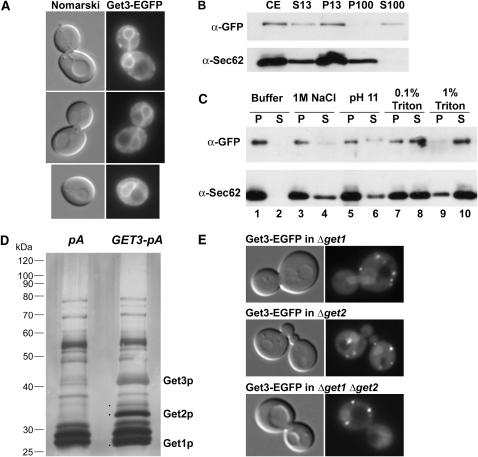
Figure 4.—
ER-membrane localization of Get3 requires Get2 and Get1. (A) Get3-EGFP localizes to the nuclear/ER membrane in rich media, by live cell fluorescence microscopy. Corresponding Nomarski image of cells is shown to the left. (B) Get3-EGFP cofractionates with both soluble and membrane-bound fractions. Cellular extract (CE) was separated into pellet and supernatant fractions following centrifugation at 13,000 rpm (P13 and S13, respectively). The S13 fraction was then subjected to ultracentrifugation at 100,000 rpm and separated into pellet and supernatant fractions (P100 and S100). Corresponding volumes from each isolated fraction were separated by SDS–PAGE and Western blotted with anti-GFP or anti-Sec62 antibodies as indicated. (C) The P13-associated fraction of Get3-EGFP is tightly membrane associated. The P13 fraction as in B was washed with either buffer alone (lanes 1 and 2) or buffer with 1 m NaCl (lanes 3 and 4), 0.2 m Na2CO3 pH 11 (lanes 5 and 6), 0.1% Triton X-100 (lanes 7 and 8), or 1% Triton X-100 (lanes 9 and 10). The samples were then recentrifuged and separated into pellet (P, odd lanes) or supernatant fractions (S, even lanes), which were analyzed by Western blotting as above. (D) Get3 biochemically copurifies with Get2 and Get1. Solubilized membranes from cells expressing Get3-TEV-proteinA (GET3-pA) or protein A alone (pA) were incubated with IgG Sepharose beads. After extensive washing, bound proteins were treated with TEV protease. Proteins released by this treatment were separated by SDS–PAGE and visualized by silver staining. Specific bands marked with a dot were excised for analysis by MALDI-TOF mass spectrometry and two of these bands were identified as Get2 and Get1 as labeled. The protein band corresponding to Get3 is also indicated. (E) Get3-EGFP mislocalizes in the absence of GET2 and/or GET1. Get3-EGFP was visualized by live-cell fluorescence microscopy in Δget1, Δget2, or Δget1 Δget2 cells grown in rich media to logarithmic phase. Images are representative of analysis of more than one clone for each genotype.
To biochemically confirm that a population of Get3 associates with cellular membranes, cells expressing Get3-EGFP were lysed under gentle, nondetergent conditions and fractionated by two successive rounds of centrifugation (materials and methods). Proteins copurifying with the isolated membrane and soluble fractions were separated by SDS–PAGE and probed with anti-GFP antibodies to visualize Get3-EGFP and with antibodies to Sec62, an integral ER-membrane protein. This analysis revealed that the majority of Get3 protein sedimented with the nuclei and ER membranes in the P13 membrane fraction (Figure 4B), consistent with our observation of a membrane-associated population of Get3-EGFP by fluorescence microscopy. We also observed a population of Get3 that remained soluble, even upon ultracentrifugation (S100 fraction), confirming our visualization of a cytoplasmic pool of Get3 by microscopy (Figure 4B).
On the basis of sequence predictions and its homology to ArsA (Boskovic et al. 1996), the Get3 protein is not expected to contain a trans-membrane domain, raising the question of how Get3 associates with cellular membranes. To assess the nature and extent of Get3 membrane association, cellular membranes purified from Get3-EGFP-expressing cells (P13 fraction) were resuspended in various buffers, incubated for 10 min on ice, and repelleted. Proteins released into the soluble phase (S) as well as those remaining in the membrane pellet (P) were analyzed by Western blot as above. We observed that Get3-EGFP remained tightly membrane associated in the presence of high salt and high pH; only treatment with a detergent was able to solubilize Get3 to the supernatant (Figure 4C).
On the basis of our microscopic and biochemical analysis of Get3 subcellular localization, we hypothesized that Get3 was likely to interact with one or more proteins embedded in the ER/nuclear membrane. To identify these potential proteins, we affinity purified protein A-tagged Get3 (Get3-ProA) from solubilized cell membranes by binding to IgG Sepharose. Proteins copurifying with Get3-ProA were eluted by TEV protease cleavage, separated by SDS–PAGE, and detected by silver staining. Two protein bands that were specifically observed in the Get3-ProA purification, and not in a negative control purification from cells expressing protein A alone, were present in similar quantities as Get3-ProA itself (Figure 4D). These were identified by MALDI-TOF mass spectrometry to be Get1 and Get2. A fainter band running at 35 kDa was also observed, but unfortunately this interactor was of insufficient quantity to identify. Further increasing our confidence that Get1 and Get2 represent true Get3-binding partners, these two proteins were also found among the proteins purified with Get3 in a large-scale study (Ho et al. 2002). Despite their colocalization at the ER/nuclear membrane (Hitchcock et al. 2001; Huh et al. 2003), no physical interactions were observed between Get3 and members of the Npl4 complex by Western blot (data not shown).
To test the possibility that Get3 membrane localization is mediated by interaction with the trans-membrane domain-containing proteins Get1 and Get2, we monitored Get3-EGFP localization in Δget1 and Δget2 single- and double-mutant cells. Strikingly, Get3-EGFP was completely absent from the ER membrane in deletions of either GET1 or GET2 (Figure 4E). Instead it was found in punctate sites in the cytoplasm, which are likely Golgi compartments as suggested by Schuldiner et al. (2005). Conversely, we found that the localization of Get1-EGFP and Get2-EGFP was unaffected by the absence of Get3 (supplemental material at http://www.genetics.org/supplemental/). Thus, under these conditions the ER-membrane localization of Get3 is dependent upon the presence of both Get2 and Get1; however, even in the absence of Get1 and Get2, Get3 appears to retain some capacity to interact with other cellular membranes.
Sporulation genes are misregulated in a get3 mutant:
As a complementary approach to identify pathways affected by GET3, transcriptional profiling of Δget3 cells was performed. Total RNA was purified from WT and Δget3 cells, differentially labeled, and competitively hybridized to microarrays spotted with cDNAs representing ∼6200 predicted yeast ORFs. Statistical analysis of WT vs. Δget3 signal for each ORF led to the identification of 265 genes whose transcription is upregulated and 345 genes whose transcription is downregulated in Δget3 relative to WT cells, as defined in materials and methods (complete data set available in supplemental material at http://www.genetics.org/supplemental/). The most striking finding from this analysis was that the set of genes upregulated in Δget3 is significantly enriched for genes whose expression increases during sporulation (P = 3.4 × 10−12, by Fisher's exact test). Table 3 shows the breakdown of these sporulation- and Δget3-induced genes into stages of transcriptional activation during sporulation, on the basis of the classifications made by Chu et al. (1998). Although a large fraction of genes involved in all stages of sporulation was induced in Δget3 cells, enrichment of genes involved in the middle and early I stages was the most significant (P = 9.83 × 10−8 and P = 9.87 × 10−4, respectively; Table 3).
TABLE 3.
Misregulation of sporulation genes in Δget3
| Class | No. genes up in Δget3 | No. genes in class | P-value |
|---|---|---|---|
| Metabolic | 2 | 51 | 0.39 |
| Early I | 7 | 61 | 9.87E-04 |
| Early II | 5 | 56 | 0.013 |
| Early–mid | 4 | 86 | 0.19 |
| Middle | 18 | 156 | 9.83E-08 |
| Mid–late | 3 | 58 | 0.19 |
| Late | 2 | 9 | 0.022 |
| Sporulation | 41 | 477 | 3.40E-12 |
The numbers of genes upregulated during each stage of sporulation (as defined by Chu et al. 1998) and in Δget3 are listed. P-values for significance of overlap were determined using Fisher's exact test.
Given the genetic interactions between GET3 and NPL4, we sought to compare the set of genes with altered transcription in Δget3 mutant cells with those whose transcription is altered in npl4-1 mutant cells (Auld et al. 2006). Surprisingly, the genes with altered transcription in Δget3 and npl4-1 strains showed no significant similarity, except in the activation of genes encoding heat-shock and other stress response proteins (data not shown). Interestingly, however, the transcription of GET3 was significantly induced in npl4-1 (P = 4.33 × 10−3). In all, this transcriptional profiling analysis supports the existence of a functional connection between GET3 and NPL4, but also suggests that GET3 may function independently of NPL4 in affecting the expression of sporulation-specific genes.
Sporulation phenotypes of GET complex and npl4-1 mutants:
We found the misregulation of sporulation genes in Δget3 to be of particular interest given GET3's strong transcriptional coregulation with NPL4 and other ubiquitin–proteasome system genes during this process (see above and Figure 3). Furthermore, both the Get3 interactors Get1 and Get2 (Figure 4 and Schuldiner et al. 2005) have been implicated in sporulation by a large-scale study (Enyenihi and Saunders 2003). Taken together, these data suggested that the GET complex may have a role, perhaps in conjunction with Npl4 and the ubiquitin–proteasome system, in sporulation. To investigate this model, diploid yeast cells homozygous for npl4-1, Δget3, Δget2, or Δget1 (in the synchronously sporulating SK1 strain background) were induced to sporulate, and meiotic divisions were assayed by microscopic analysis of DAPI-stained samples throughout the time course. The terminal phenotype was then recorded by fluorescence and Nomarski microscopy 36 hr after induction. We found that the timing and occurrence of meiotic divisions in the GET complex deletion strains were not significantly altered compared to those in a wild-type strain (supplemental material at http://www.genetics.org/supplemental/). In fact, no strong sporulation defect was apparent in the Δget3 and npl4-1 SK1 strains, even after characterization of the terminal sporulation phenotype (Figure 5B and data not shown).
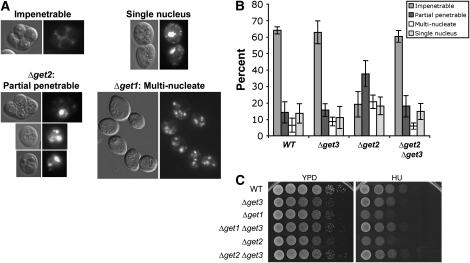
Figure 5.—
Δget3 rescues phenotypes of Δget2 and Δget1 cells. (A) Representative sporulated cells displaying wild-type or defective terminal sporulation phenotypes. Synchronously sporulated SK1 cells were fixed 36 hr after shift to sporulation medium (SPM) and nuclei were stained with DAPI. Nomarski (left) and DAPI (right) images representative of the indicated terminal (36 hr) sporulation phenotypes are shown. (B) Δget3 rescues terminal sporulation defects displayed by Δget2 cells. Synchronously sporulated wild-type (WT), Δget3, Δget2, and Δget3 Δget2 homozygous diploid yeast (SK1 background) were stained with DAPI, and cells falling into each of the four categories shown in A were counted. The average percentage of each phenotype is graphed with error bars depicting the standard deviation over three separate experiments. At least 200 cells were counted for each strain per experiment. (C) Δget3 suppresses the HU sensitivity of Δget2 and Δget1 cells. Wild-type (WT), Δget3, Δget1, Δget2, and double-mutant strains were grown to log phase and then serially diluted and plated to YPD or media containing 150 mm hydroxyurea at 25° for 2 days.
In contrast to Δget3 and npl4-1 cells, we observed a striking defect in the terminal sporulation phenotype of Δget2 and Δget1 mutant cells (Figure 5, A and B). Whereas the majority of wild-type cells generated tetrads with four spores “impenetrable” to DAPI when stained using the conditions described (materials and methods), only 20% of Δget2 cells exhibited this phenotype. Instead, the majority of Δget2 diploid cells produced “partial penetrable” tetrads, which contain one or more spores that are refractile, but immature looking and penetrable to DAPI (Figure 5, A and B). The Δget2 strain also exhibited a mild increase in “multinucleate” cells, those with multiple or fragmented nuclei but with no refractile spores. A similar, though more severe, phenotype was observed in the Δget1 strain (Figure 5A): only ∼10% of Δget1 cells produced complete tetrads after 36 hr in sporulation medium, whereas ∼40% displayed the multinucleate phenotype. A significant number (∼30%) of Δget1 diploids also formed abnormal refractile spores similar to those seen in Δget2, suggesting that these genes function within the same process. All strains tested had a similar percentage of “single-nucleus” cells that have not undergone sporulation, indicating that the fraction of cells initiating sporulation was not significantly different in the various strains (Figure 5B and data not shown). Tetrad dissection was performed to analyze the viability of spores produced in Δget1 and Δget2 strains. Overall, 79.7% of Δget2 spores (287/360 spores) and 86.3% of Δget1 spores (335/388) from tetrads with normal appearance were viable, compared with 99.5% viability of WT spores (199/200).
These observed phenotypes of Δget1 and Δget2 spores (Figure 5A) are similar to those previously described for spo11-1 mutant diploids, which undergo the meiotic divisions but fail to package all of their spores correctly (Klapholz et al. 1985). Since Spo11 is important in catalyzing double-strand breaks necessary for recombination during meiosis (Keeney et al. 1997), this similarity suggested that meiotic recombination may be affected in GET complex mutants. To test this hypothesis, the frequency of heteroallele recombination in Δget1 and Δget2 was analyzed by a return-to-growth time-course experiment during the first 8 hr after induction of sporulation. Interestingly, we observed no significant defect in either mutant (data not shown). An alternative explanation for these phenotypes, in keeping with previous proposals of spore wall defects in Δget1 (Enyenihi and Saunders 2003), is that the GET complex is important for spore packaging. For further characterization of the mutant phenotypes, we also examined the localization of Don1-GFP, a marker for the leading edge of the prospore membrane (Knop and Strasser 2000), in our mutant strains. Prospore membrane growth appeared normal as well, suggesting that GET1 and GET2 function at a later step in spore development.
Δget3 suppresses phenotypes of Δget2 and Δget1:
Given that get3 mutants are capable of suppressing phenotypes of npl4-1 cells, we asked whether Δget3 might also be able to suppress the sporulation defect displayed by Δget2 cells. To this end, we generated a homozygous diploid Δget2 Δget3 double mutant in the SK1 strain background and analyzed its terminal sporulation phenotype as described above. Strikingly, we found that the deletion of get3 completely rescued the significant sporulation defect of Δget2 (Figure 5B). The sporulation defect of Δget1 was also rescued by Δget3 (data not shown).
To investigate the extent of these epistatic interactions, we sought to determine whether Δget3 could rescue other defects displayed by Δget2 and/or Δget1. Previous studies have demonstrated that Δget2 is sensitive to the DNA-damaging agents hydroxyurea (HU) and methyl methanesulfonate (MMS) (Zewail et al. 2003). To test whether Δget3 is able to rescue the HU sensitivity of Δget2 cells, we compared the growth of single- and double-deletion strains of the GET complex on rich media plates containing HU. As expected, Δget2 cells were sensitive to HU, showing a growth defect of at least two orders of magnitude as compared to wild-type cells (Figure 5C). We found that Δget1 cells were similarly sensitive to HU, whereas Δget3 cells were only slightly more sensitive than wild-type cells (less than one order of magnitude, Figure 5C). Interestingly, the HU sensitivity of both Δget2 and Δget1 cells was rescued by Δget3, such that the double mutants displayed the Δget3 phenotype (mild HU sensitivity, Figure 5C). In all, this genetic analysis has revealed that Δget3 suppresses both the terminal sporulation phenotype of Δget2 cells and the HU sensitivity of Δget2 and Δget1 cells.
DISCUSSION
In this study we have characterized the yeast GET3/ARR4, a highly conserved gene encoding an ATPase whose bacterial homolog regulates arsenic transport (Rosen et al. 1995). We isolated a truncation of get3 in a genetic screen for suppressors of an npl4 mutant, along with mutations in several other genes involved in ubiquitin-dependent events in protein trafficking through the secretory system. We have demonstrated that mutants of get3 rescue multiple defects in this npl4 mutant and that GET3 exhibits transcriptional coregulation with genes encoding proteasome components. These results combine to suggest that GET3 can antagonize the pathways of Cdc48-Npl4-Ufd1 complex activity in the ubiquitin–proteasome system. Further characterization of GET3, independent of its interaction with NPL4, included biochemical and localization studies demonstrating that the ER-membrane localization of Get3 depends on the presence of both Get1 and Get2. Transcriptional profiling and phenotypic analyses then illustrated a role for the GET complex in sporulation. Intriguingly, Get3 appears to have a negative role in this process, as evidenced by our discovery that Δget3 can reverse the sporulation phenotypes of get1 and get2 mutants. In combination, we have demonstrated that GET3 can modulate pathways requiring either the Npl4 complex or the GET complex.
Several lines of evidence from this and other studies suggest that Get3 may act in a regulatory capacity in these pathways. First, the Get3 protein itself is not required for the processes of sporulation or ERAD (Figure 5B and supplemental Figure S2 at http://www.genetics.org/supplemental/), yet it modulates these pathways in mutants of the GET complex genes and npl4, respectively (Figures 5B and 2A). This type of interaction implies a regulatory relationship that could be mediated by the ATPase domain of Get3 (Schuldiner et al. 2005; Shen et al. 2003). The physical interaction between Get3, Get2, and Get1 (Figure 4D) supports the possibility of a direct action of Get3 on these proteins. Finally, the homology of Get3 to a bacterial ATPase that regulates the channel for transport of arsenic across the plasma membrane (Boskovic et al. 1996) supports the idea that the yeast GET3 may have a conserved function with its bacterial homolog in the regulation of membrane-associated proteins.
A screen for suppressors of npl4:
The set of genes isolated as suppressors of npl4-1 reveals several insights into Npl4 function and membrane dynamics (Table 2). First, CUE1 and UBC7 are important for the ubiquitination step of ERAD (Biederer et al. 1997) and deletions in these genes reverse the accumulation of ubiquitinated proteins at the ER membrane in npl4-1 cells (Hitchcock 2003); thus our isolation of these particular ERAD components suggests this accumulation as one reason for temperature-sensitive lethality in npl4-1 cells. Additional suppression likely occurs in these mutants through increased half-life of the Ole1 protein (Hitchcock 2003), which is a documented ERAD substrate (Braun et al. 2002). Second, the isolation of a class of secretory pathway genes suggests intimate connections between the phenotypes of the npl4 mutant and secretory function. The mechanism of rescue by these suppressors is not clear but could be caused by a reduction in ER protein load or by the diversion of more proteins to alternative degradation systems such as the vacuole (Spear and Ng 2003). Finally, the involvement of many of these secretory pathway suppressors in ubiquitin-dependent events illustrates the importance of degradation pathways in maintaining proper function of the membranes of the secretory system.
It is more difficult to interpret potential mechanisms of npl4 phenotype suppression by the RNA-processing genes PRP6 and CBP80. The npl4 mutants were initially isolated on the basis of their ability to block nuclear transport (DeHoratius and Silver 1996). Thus these RNA-processing mutants may alleviate an RNA transport defect, now presumed to be caused by the membrane defects in npl4-1 cells; alternatively, they may increase the half-life of OLE1 or npl4-1 transcripts.
GET3 and the ubiquitin–proteasome system:
Several plausible explanations can be envisioned for get3-mediated suppression of npl4-1 phenotypes. One possibility is that this suppression is due to activity of the GET complex in retrograde transport from the Golgi to the ER (Schuldiner et al. 2005) as part of the secretory pathway. The identification in our screen of other genes affecting secretion, including one (UBP3) that also participates in this type of retrograde transport (Cohen et al. 2003b), supports this model. However, our data suggest that Get3 has a more specific function relating to the ubiquitin–proteasome system and that it is this function that explains the ability of get3 mutants to partially rescue npl4-1 cells. First, given our understanding of the transcriptional regulation of OLE1, it is difficult to explain how an ER–Golgi transport defect would rescue the OLE1 transcription defect of npl4-1 cells more potently than a dominant activating truncation of the OLE1 transcription factor Spt23 (Figure 2B). Second, the coregulation of GET3 with genes encoding the Npl4 complex and components of the proteasome (Figure 3) suggests a functional interaction between GET3 and this pathway. Finally—and most convincingly—GET1 and GET2, which are required along with GET3 for retrograde transport, are not coregulated with the proteasome (Figure 3 and data not shown) and are unable, when deleted, to suppress npl4-1 temperature sensitivity (data not shown). If the ability of Δget3 to suppress npl4-1 were due to its activity in the secretory pathway, it would be expected that GET1 and GET2 would display identical genetic interactions with NPL4. Thus, our finding that GET3 has a unique ability to antagonize NPL4 activities indicates a functional connection between Get3 and the ubiquitin–proteasome system, independent of its interactions and functions with the GET complex.
The GET complex in sporulation:
In our studies of Get3 as part of a complex with Get1 and Get2 we have focused on its role in sporulation, a line of study suggested by our transcriptional profiling of Δget3 and by documented sporulation defects of Δget1/mdm39 and Δget2/rmd7 (Enyenihi and Saunders 2003). This function of Get3 is likely distinct from its interactions with the ubiquitin–proteasome system, as discussed above.
Several hypotheses can be proposed for the mechanism of GET complex function in sporulation. The first possibility is that the sporulation defects we observed in Δget1 and Δget2 mutants are a result of defects in retrograde protein transport in the secretory pathway. Given the requirement of the GET complex for proper localization of proteins within the ER and Golgi (Schuldiner et al. 2005), and the importance of these compartments to prospore membrane and spore wall formation (Neiman 2005), these structures could be affected in the get1 and get2 mutant strains. The increased HU sensitivity and DAPI penetrability of Δget2 spores may reflect membrane or spore wall defects, supporting this model.
The sporulation phenotypes we observed are somewhat different from the previously described defects in spore wall formation (Δget1) and meiotic division (Δget2) (Enyenihi and Saunders 2003). While preliminary data suggest that there may be spore wall defects, we observed no defect in meiotic divisions (supplemental material at http://www.genetics.org/supplemental/). It is possible that this discrepancy is due to strain differences, particularly in the timing and efficiency of sporulation. Our detailed analysis of Δget1 and Δget2 mutants in synchronously sporulated SK1 cells, especially the observed increases in partial penetrable tetrads, fragmented nuclei, and nonviable spores, suggests that the GET complex may act at a postmeiosis step to allow proper spore packaging (Figure 5 and results).
Our genetic analysis of the sporulation phenotypes of GET complex mutants provides evidence that the sporulation defects of Δget2 may not be due to the secretory activity of the complex. In the secretory pathway, the presence of GET3 is required for the retrograde transport of proteins meant to reside in the ER (Schuldiner et al. 2005). In contrast, GET3 itself is not required for sporulation, but this process cannot occur correctly when the complex is disrupted by the absence of GET2 (Figure 5B). The rescue of sporulation and HU-sensitive phenotypes of Δget1 and Δget2 in Δget3 illustrates that in addition to cooperating with Get2 and Get1 in the secretory pathway, in other cases Get3 is capable of antagonizing the function of Get1 and Get2.
The evolution of GET3 function:
The bacterial homolog of GET3, ArsA, is the cytoplasmic regulatory subunit of the pump required to export arsenic through the plasma membrane (Rosen et al. 1995). One potential hypothesis suggested by our work is that this activity as a regulatory ATPase for membrane-associated proteins is conserved in the yeast protein Get3. Through our analysis of the GET3 gene as a suppressor of npl4-1 we have shown that it can antagonize the pathways of Npl4 complex activity, while experiments independent of this interaction have shown a similar capacity for Get3 with Get1 and Get2. Thus, in combination, our studies of GET3 have illustrated its role in two different cellular pathways: proteasome-dependent events at the ER membrane through Cdc48-Npl4-Ufd1 and sporulation through Get1 and Get2. Interestingly, a recent study has shown a similar interaction between Get3 and the intracellular CLC chloride-transport protein, Gef1 (Metz et al. 2006), suggesting metal ion homeostasis as yet a third pathway subject to potential regulation by Get3.
In summary, Get3 has been implicated, through this and other studies, in such seemingly disparate cellular pathways as the ubiquitin–proteasome system, secretion, and sporulation. We propose the regulation of intracellular membrane composition and organization to be the fundamental connection among these activities, and future studies of Get3 will undoubtedly further our understanding of its role in the membrane dynamics of the eukaryotic cell.
Acknowledgments
The authors gratefully acknowledge Y. Ye and T. Rapaport for plasmids and antibodies and M. Snyder for the transposon library. We thank D. Drubin, S. Komili, and O. Johnstone for critical reading of the manuscript and the members of the Silver laboratory for stimulating discussion. This work was supported by a training grant from the National Cancer Institute to K.L.A. and A.L.H., a grant from the National Institutes of Health (NIH) to L.S.H., and grants from the NIH to P.A.S.
References
- Abovich, N., P. Legrain and M. Rosbash, 1990. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol. Cell. Biol. 10: 6417–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Albertson, R., B. Riggs and W. Sullivan, 2005. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 15: 92–101. [DOI] [PubMed] [Google Scholar]
- Amerik, A. Y., J. Nowak, S. Swaminathan and M. Hochstrasser, 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11: 3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld, K. L., C. R. Brown, J. M. Casolari, S. Komili and P. A. Silver, 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell 21: 861–871. [DOI] [PubMed] [Google Scholar]
- Baker, D., L. Wuestehube, R. Schekman, D. Botstein and N. Segev, 1990. GTP-binding Ypt1 protein and Ca2+ function independently in a cell-free protein transport reaction. Proc. Natl. Acad. Sci. USA 87: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute and C. Cullin, 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays, N. W., S. K. Wilhovsky, A. Goradia, K. Hodgkiss-Harlow and R. Y. Hampton, 2001. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12: 4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, K. R., C. Zhang, K. M. Shokat and I. Herskowitz, 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz, G. F., O. D. King, B. Bryant, C. Sander and F. P. Roth, 2003. Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502–2504. [DOI] [PubMed] [Google Scholar]
- Biederer, T., C. Volkwein and T. Sommer, 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278: 1806–1809. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P. S., J. L. Urbanowski, S. C. Winistorfer and R. C. Piper, 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4: 534–539. [DOI] [PubMed] [Google Scholar]
- Boskovic, J., A. Soler-Mira, J. M. Garcia-Cantalejo, J. P. Ballesta, A. Jimenez et al., 1996. The sequence of a 16,691 bp segment of Saccharomyces cerevisiae chromosome IV identifies the DUN1, PMT1, PMT5, SRP14 and DPR1 genes, and five new open reading frames. Yeast 12: 1377–1384. [DOI] [PubMed] [Google Scholar]
- Braun, S., K. Matuschewski, M. Rape, S. Thoms and S. Jentsch, 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus et al., 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Chellappa, R., P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula et al., 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276: 43548–43556. [DOI] [PubMed] [Google Scholar]
- Chu, S., and I. Herskowitz, 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Cohen, M., F. Stutz, N. Belgareh, R. Haguenauer-Tsapis and C. Dargemont, 2003. a Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 5: 661–667. [DOI] [PubMed] [Google Scholar]
- Cohen, M., F. Stutz and C. Dargemont, 2003. b Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J. Biol. Chem. 278: 51989–51992. [DOI] [PubMed] [Google Scholar]
- Das, B., Z. Guo, P. Russo, P. Chartrand and F. Sherman, 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20: 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoratius, C., and P. A. Silver, 1996. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell 7: 1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski, K., R. Balakrishnan, K. R. Christie, M. C. Costanzo, S. S. Dwight et al., 2006. Saccharomyces Genome Database (http://www.yeastgenome.org/).
- Dula, M. L., and S. G. Holmes, 2000. MGA2 and SPT23 are modifiers of transcriptional silencing in yeast. Genetics 156: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi, A. H., and W. S. Saunders, 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether, M. E., and I. Herskowitz, 1999. Genetic and biochemical characterization of the yeast spo12 protein. Mol. Biol. Cell 10: 3689–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Hepworth, S. R., H. Friesen and J. Segall, 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M., H. H. Meyer, T. C. Walther, D. Bilbao-Cortes, G. Warren et al., 2001. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3: 1086–1091. [DOI] [PubMed] [Google Scholar]
- Hitchcock, A. L., 2003. Regulation of ubiquitinated membrane proteins. Ph.D. Thesis, Harvard University, Cambridge, MA.
- Hitchcock, A. L., H. Krebber, S. Frietze, A. Lin, M. Latterich et al., 2001. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12: 3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock, A. L., K. Auld, S. P. Gygi and P. A. Silver, 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100: 12735–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., K. Matuschewski, M. Rape, S. Schlenker, H. D. Ulrich et al., 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586. [DOI] [PubMed] [Google Scholar]
- Howe, A. G., and C. R. McMaster, 2001. Regulation of vesicle trafficking, transcription, and meiosis: lessons learned from yeast regarding the disparate biologies of phosphatidylcholine. Biochim. Biophys. Acta 1534: 65–77. [DOI] [PubMed] [Google Scholar]
- Huang, L. S., H. K. Doherty and I. Herskowitz, 2005. The Smk1p MAP kinase negatively regulates Gsc2p, a 1,3-beta-glucan synthase, during spore wall morphogenesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jelinsky, S. A., P. Estep, G. M. Church and L. D. Samson, 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., P. C. Ma, I. M. Ota and A. Varshavsky, 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270: 17442–17456. [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., M. Babst and S. D. Emr, 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88(3): 375–384. [DOI] [PubMed] [Google Scholar]
- Klapholz, S., C. S. Waddell and R. E. Esposito, 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and K. Strasser, 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19: 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., A. Finger, T. Braun, K. Hellmuth and D. H. Wolf, 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15: 753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and M. Snyder, 2000. Genome-wide transposon mutagenesis in yeast, pp. 13.3.1–13.3.15 in Current Protocols in Molecular Biology, edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. John Wiley & Sons, New York. [DOI] [PubMed]
- Kurdi-Haidar, B., S. Aebi, D. Heath, R. E. Enns, P. Naredi et al., 1996. Isolation of the ATP-binding human homolog of the arsA component of the bacterial arsenite transporter. Genomics 36: 486–491. [DOI] [PubMed] [Google Scholar]
- Metz, J., A. Wachter, B. Schmidt, J. M. Bujnicki and B. Schwappach, 2006. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J. Biol. Chem. 281: 410–417. [DOI] [PubMed] [Google Scholar]
- Munoz-Centeno, M. C., S. McBratney, A. Monterrosa, B. Byers, C. Mann et al., 1999. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell 10: 2393–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore, R., L. Cao and N. Kleckner, 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway et al., 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Romisch, K., 2005. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 21: 435–456. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., P. Novick, J. H. Thomas, D. Botstein and G. R. Fink, 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237–243. [DOI] [PubMed] [Google Scholar]
- Rosen, B. P., H. Bhattacharjee and W. Shi, 1995. Mechanisms of metalloregulation of an anion-translocating ATPase. J. Bioenerg. Biomembr. 27: 85–91. [DOI] [PubMed] [Google Scholar]
- Schawalder, S. B., M. Kabani, I. Howald, U. Choudhury, M. Werner et al., 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061. [DOI] [PubMed] [Google Scholar]
- Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati et al., 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., M. Damelin, J. Kahana, T. Taura and P. A. Silver, 1999. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19: 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, H. S., E. Y. Chen, M. So and F. Heffron, 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83: 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., C. M. Hsu, B. K. Kang, B. P. Rosen and H. Bhattacharjee, 2003. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals 16: 369–378. [DOI] [PubMed] [Google Scholar]
- Spear, E. D., and D. T. Ng, 2003. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell 14: 2756–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey, J. E., V. M. McDonough and C. E. Martin, 1990. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 265: 20144–20149. [PubMed] [Google Scholar]
- Wade, J. T., D. B. Hall and K. Struhl, 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wright, R., M. L. Parrish, E. Cadera, L. Larson, C. K. Matson et al., 2003. Parallel analysis of tagged deletion mutants efficiently identifies genes involved in endoplasmic reticulum biogenesis. Yeast 20: 881–892. [DOI] [PubMed] [Google Scholar]
- Xu, L., M. Ajimura, R. Padmore, C. Klein and N. Kleckner, 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewail, A., M. W. Xie, Y. Xing, L. Lin, P. F. Zhang et al., 2003. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. USA 100: 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Y. Skalsky and D. J. Garfinkel, 1999. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]