Abstract
It is unknown whether homologous loci underlie the independent and parallel wing pattern radiations of Heliconius butterflies. By comparing the locations of color patterning genes on linkage maps we show that three loci that act similarly in the two radiations are in similar positions on homologous chromosomes.
WHEN different taxa independently evolve the same trait, do they use the same genes to do it? Available data suggest that the answer is “sometimes.” Multiple studies have found that the same gene plays a role in the evolution of convergent morphological characters. The independent evolution of albinism in multiple populations of the Mexican tetra (Astyanax mexicanus) has resulted from mutations in the gene Oca2 (Protas et al. 2006); repeated loss of the pelvic skeleton and reductions in armor plates in threespine sticklebacks (Gasterosteus aculeatus) have been linked to Pitx1 (Shapiro et al. 2004) and Eda (Colosimo et al. 2005), respectively; loss of larval trichomes and convergent evolution of abdominal pigmentation in different species of Drosophila result from regulatory changes of the genes svb/ovo (Sucena et al. 2003) and bab2 (Gompel and Carroll 2003), respectively; and melanism in a wide variety of animals is associated with the gene MC1R (Mundy 2005). However, convergent evolution does not necessarily require the same genetic mechanisms. For instance, different genes are responsible for the evolution of melanism in different populations of rock pocket mice (Hoekstra and Nachman 2003) and while expression of the gene yellow is sometimes correlated with Drosophila pigmentation (Wittkopp et al. 2002a,b), other times it is not (Wittkopp et al. 2003). In theory, an ideal system to study the genetic basis of convergent evolution would contain multiple independent instances of convergence across multiple distinct traits. The Neotropical butterfly genus Heliconius is such a system.
Heliconius butterflies are distasteful, warningly colored, and mimetic. As a classic example of Müllerian mimicry, different Heliconius species, while all distasteful, gain protection by resembling one another and thereby distributing the cost of educating naïve predators. In Heliconius, intrageneric wing pattern mimicry has resulted from two parallel radiations: the genus consists of two major clades, and each mimetic wing pattern is shared by at least one species from each of these two lineages (Figure 1). Average pairwise mtDNA divergence between species of the two Heliconius clades is 9.148% (SD=0.521%), which suggests that the two lineages separated ∼4 million years ago, assuming an evolutionary rate of 1.1–1.2% per lineage per million years (Brower 1994). Hence, since sharing a common ancestor as recently as 4 million years ago, the two Heliconius groups have radiated onto the same myriad of wing pattern phenotypes at both the racial and the species levels. Decades of crossing experiments have identified the phenotypic effects of discrete color patterning loci and have verified the homology of many of these loci among species and races of the same clade (Turner and Crane 1962; Turner 1971; Sheppard et al. 1985; Nijhout and Wray 1988; Mallet 1989; Nijhout et al. 1990; Linares 1996; Jiggins and McMillan 1997; Gilbert 2003; Naisbit et al. 2003). However, virtually nothing is known about color pattern homology between the clades because they cannot be interbred. Comparative genetic mapping provides our first insight into the potential homology of wing patterning loci in the two lineages.
Figure 1.—
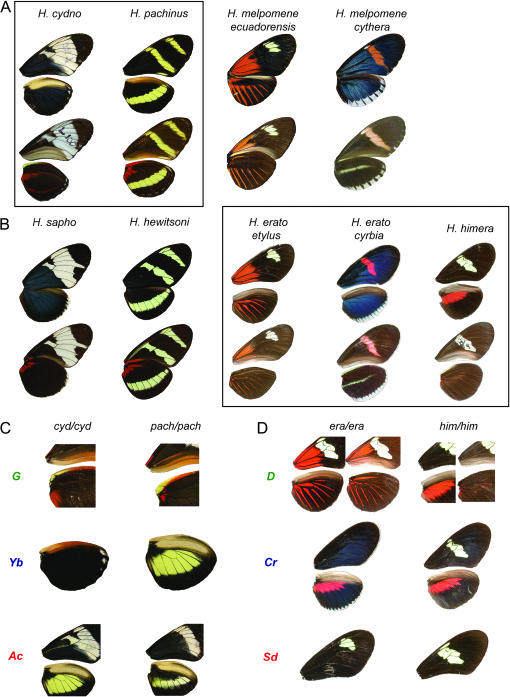
Mimicry and mimicry genes in Heliconius butterflies. (A) Four phenotypes (dorsal, top; ventral, bottom) from three species belonging to one major Heliconius clade. H. cydno and H. pachinus (shown in a box) were crossed to study color pattern genetics from this clade. (B) Five phenotypes from four species belonging to the second Heliconius clade are shown below their respective comimics; H. himera does not have a corresponding comimic. Crosses between H. himera and each of the two H. erato races (shown in a box) were used to study color pattern genetics in this clade (Tobler et al. 2005; Kapan et al. 2006). (C) Effects of alternative alleles at three color-patterning loci that distinguish H. cydno and H. pachinus: G controls whether the ventral base of the fore and hind wings is red or brown, Yb controls the presence or absence of melanic scales on the hind wing, and Ac controls the presence or absence of melanic scales on the proximal portions of both the fore and the hind wings. (D) Effects of alternative alleles at three color-patterning loci that distinguish H. erato and H. himera: D controls the distribution of red/orange scales on the wings (DetDet vs. DhiDhi shown), Cr controls the placement of melanic scales on the fore and hind wings (CrcyCrcy vs. CrhiCrhi shown), and Sd controls the portion of the forewing that is covered in melanic scales (SdetSdet vs. SdhiSdhi shown).
To address this issue, we compared the genomic positions of three color-patterning loci that act similarly in the two radiations. Using two hybrid crosses between different Heliconius erato races and the closely related species H. himera, Tobler et al. (2005) and Kapan et al. (2006) localized the positions of three large-effect color-patterning loci. The D locus controls the distribution of red/orange scales on the wings, the Cr locus controls the placement of melanic scales on the fore and hind wings that “shutter” or limit the distribution of yellow and white scales on the wings, and the Sd locus controls the portion of the forewing that is covered in melanic scales (Figure 1; Sheppard et al. 1985; Mallet 1989; Jiggins and McMillan 1997; Kapan et al. 2006). To evaluate the potential for homology between the two radiations, we identified the positions of three color-patterning loci that act similarly to those mapped in H. erato, on a map derived from a hybrid cross between two species from the other Heliconius radiation, H. cydno and H. pachinus. The G locus controls the distribution of red scales on the wings, which in H. pachinus is limited to small basal spots on the ventral side (Figure 1). In other crosses this locus has been shown to segregate with the presence vs. the absence of much larger regions of red, such as the red forewing band of H. melpomene rosina (Naisbit et al. 2003). The Yb locus [also know as Cs (Nijhout et al. 1990)] controls the presence or absence of melanic scales on the hind wing, which prevent or allow the expression of underlying yellow or white scales (Figure 1; Gilbert 2003; Jiggins et al. 2005). Finally, the Ac locus [also known as Ps (Nijhout et al. 1990)] controls the presence or absence of melanic scales on the proximal portions of both the fore and the hind wings (Figure 1). The hind wing action of Ac is unique to H. pachinus (Nijhout et al. 1990; Gilbert 2003).
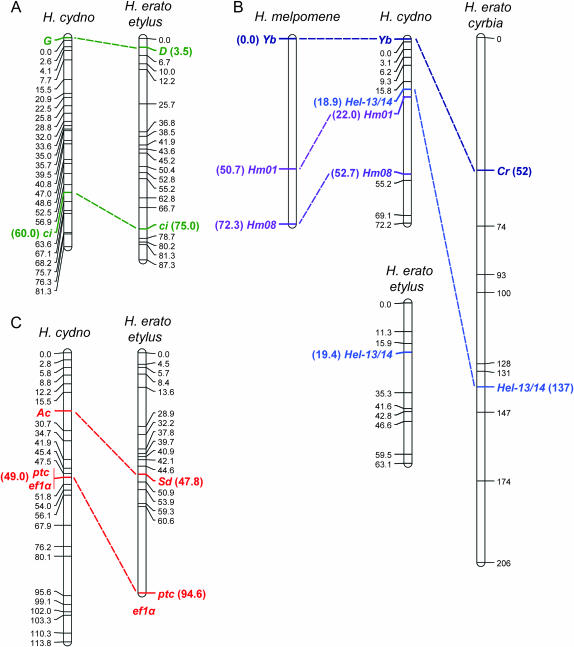
Comparing the actions of the loci between the two groups reveals that in a broad sense they act similarly. Both D and G influence the distribution of red, both Cr and Yb control the placement of melanic scales that influence the distribution of yellow and white, and both Sd and Ac influence the size of the nonmelanic region revealed on the forewing (although the H. pachinus Ac allele has a similar effect on the hind wing). Our comparison of their genomic locations revealed that all three loci map to homologous chromosomes in the two lineages. Both D and G are syntenic with the locus cubitus interruptous (ci), both Cr and Yb are syntenic with the microsatellite locus Hel-13/14, and both Sd and Ac are syntenic with the loci patched (ptc) and elongation factor 1 α (ef1α) (Figure 2). Furthermore, Yb in H. cydno is syntenic with microsatellite loci Hm01 and Hm08, which are linked to the Yb locus in closely related H. melpomene (Jiggins et al. 2005; Joron et al. 2006). Assuming that the polarity of the aligned chromosomes is correct (Figure 2), the ∼95% confidence intervals for the placement of potentially homologous loci, relative to the anchor loci, overlap for all comparisons.
Figure 2.—
Comparative genetic mapping of mimicry genes in Heliconius. (A) H. cydno color-patterning gene G and H. erato D are both syntenic with anchor locus ci. (B) H. cydno color-patterning gene Yb and H. erato Cr are both syntenic with anchor locus Hel-13/14. The H. erato cyrbia linkage group containing Hel-13/14 (Tobler et al. 2005) is substantially longer than that of H. erato etylus (Kapan et al. 2006), suggesting that distances may be artificially inflated in the former. H. cydno Yb is also syntenic with microsatellite loci Hm01 and Hm08, which are linked to Yb in crosses between races of sister species H. melpomene (Jiggins et al. 2005; Joron et al. 2006). (C) H. cydno color-patterning gene Ac and H. erato Sd are both syntenic with anchor loci ptc and ef1α. Although ef1α is syntenic with ptc and Sd in H. erato etylus, its exact position is unknown (Kapan et al. 2006). H. erato maps were estimated using backcross or F2 broods derived from crosses between H. himera and either H. erato cyrbia (for the linkage group with Cr) as described in Tobler et al. (2005) or H. erato etylus (for linkage groups with D and Sd) as described in Kapan et al. (2006). The H. melpomene Yb linkage group was mapped using F2 broods derived from crosses between H. m. cythera and H. m. melpomene as described in Jiggins et al. (2005) and Joron et al. (2006). For H. cydno, we used segregation data from 33 H. pachinus (female) × H. cydno (male) F1 hybrids (pseudotestcross design) to map amplified fragment length polymorphism (AFLP) loci, microsatellite loci, and single-copy nuclear loci that were heterozygous in the H. cydno male parent (Kronforst et al. 2006). We typed ci, Hel-13/14, Hm01, and Hm08 using PCR product length variation and ptc and eflα using single-nucleotide polymorphisms. We then scored 65 F2 offspring for allelic variation at color-patterning loci and a subset of the mapped markers and used these data to locate the positions of color-patterning loci G, Yb, and Ac. Completely linked AFLP loci in opposite phases were used to score each F2 individual as marker present (homozygous or heterozygous for the H. cydno allele) or marker absent (homozygous for the H. pachinus allele) while AFLP loci without a linked marker in the opposite phase were scored as marker present or missing data. Using likelihood we assigned each H. cydno color-patterning locus to the interval with the highest probability, but we did not estimate approximate positions within intervals. We also used likelihood to estimate placement support limits (∼95% confidence intervals) for each of the loci in both studies (Kapan et al. 2006). Mapping was performed with Joinmap 3.0 (Van Ooijen and Voorrips 2001) and Mapmaker/Exp 3.0 (Lincoln et al. 1993) using the Haldane mapping function and maps were drawn with MapChart 2.1 (Voorrips 2002). Markers are labeled with their position, in centimorgans, and homologous loci are highlighted.
These results provide tentative support for the positional homology of wing patterning loci in the two Heliconius radiations. All species studied here have n = 21 chromosomes (Brown et al. 1992), so the probability of randomly assigning all three H. cydno loci to homologous chromosomes identified in H. erato is very small (1/213 = 0.0001). However, aside from the relationship between G and D the positional correspondence between potentially homologous loci does not appear to be strong. There are several possible explanations for this. First, given that we have identified homologous chromosomes on the basis of one or two shared markers in each case, the possibility remains that the chromosomes, and the color-patterning loci, are not homologous at all. Second, the chromosomes may be homologous but the color-patterning loci may not be. Third, the overall amount of recombination differs between species in the two lineages: H. erato's total map length is 1430 cM (Kapan et al. 2006) while that of H. cydno's sister species, H. melpomene, is 1616 cM (Jiggins et al. 2005). Thus, for any given comparison we expect different amounts of recombination and different distances, between linked, homologous loci in the two radiations. Finally, there is undoubtedly error associated with estimating recombination frequencies from the relatively small data sets that have been analyzed so far, which may contribute to the lack of strong positional correspondence between potentially homologous loci. Current and future research that incorporates larger sample sizes and more homologous anchor loci will better refine the positions of major color-patterning genes in the two radiations and will help identify the extent to which synteny has been conserved. Indeed, Joron et al. (2006) recently utilized this approach to show that the Yb locus of H. melpomene, the Cr locus of H. erato, and the P locus of H. numata all lie within ∼1 cM of one another.
Studies of wing pattern development in other butterflies suggest that future, fine-scale association studies, physical mapping, and comparative analyses of gene expression will likely verify the homology of color-patterning loci between the two Heliconius clades. Brunetti et al. (2001) compared the expression patterns of three transcription factors (Engrailed/Invected, Distal-less, and Spalt) during development of eyespot wing patterns across four butterfly species representing two different families and three genera within the family Nymphalidae. They found that the expression patterns differed among species but often correlated with adult wing pattern. Similarly, Reed and Serfas (2004) surveyed Notch and Distal-less expression among eight moth and butterfly species and found that both genes were generally associated with intervein midline/eyespot development in butterflies. While there is no evidence that Heliconius wing patterning is homologous to these eyespot determination systems (Reed and Gilbert 2004), these studies do suggest that wing-patterning genes are likely to be conserved over evolutionary time.
Acknowledgments
We thank Joan Strassmann and Dave Queller for use of lab facilities and reviewers for comments on the manuscript. This work was supported by National Science Foundation grant DEB 0415718.
References
- Brower, A. V. Z., 1994. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 91: 6491–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. S., T. C. Emmel, P. J. Eliazar and E. Suomalainen, 1992. Evolutionary patterns in chromosome numbers in neotropical Lepidoptera I: chromosomes of the Heliconiini (family Nymphalidae: subfamily Nymphalinae). Hereditas 117: 109–125. [DOI] [PubMed] [Google Scholar]
- Brunetti, C. R., J. E. Selegue, A. Monteiro, V. French, P. M. Brakefield et al., 2001. The generation and diversification of butterfly eyespot color patterns. Curr. Biol. 11: 1578–1585. [DOI] [PubMed] [Google Scholar]
- Colosimo, P. F., K. E. Hosemann, S. Balabhadra, G. Villarreal, M. Dickson et al., 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Gilbert, L. E., 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic “tool box” from synthetic hybrid zones and a theory of diversification, pp. 281–318 in Ecology and Evolution Taking Flight: Butterflies as Model Systems, edited by C. L. Boggs, W. B. Watt and P. R. Ehrlich. University of Chicago Press, Chicago.
- Gompel, N., and S. B. Carroll, 2003. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424: 931–935. [DOI] [PubMed] [Google Scholar]
- Hoekstra, H. E., and M. W. Nachman, 2003. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 12: 1185–1194. [DOI] [PubMed] [Google Scholar]
- Jiggins, C. D., and W. O. McMillan, 1997. The genetic basis of an adaptive radiation: warning colour in two Heliconius species. Proc. R. Soc. Lond. Ser. B 264: 1167–1175. [Google Scholar]
- Jiggins, C. D., J. Mavarez, M. Beltrán, W. O. McMillan, J. S. Johnston et al., 2005. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron, M., R. Papa, M. Beltrán, N. Chamberlain, J. Mavárez et al., 2006. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. (in press). [DOI] [PMC free article] [PubMed]
- Kapan, D. D., N. S. Flanagan, A. Tobler, R. D. Reed, J. A. Gonzalez et al., 2006. Localization of Müllerian mimicry genes on a dense linkage map of Heliconius erato. Genetics 173: 735–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst, M. R., L. G. Young, D. D. Kapan, C. McNeely, R. J. O'Neill et al., 2006. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl. Acad. Sci. USA 103: 6575–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares, M., 1996. The genetics of the mimetic coloration in the butterfly Heliconius cydno weymeri. J. Hered. 87: 142–149. [Google Scholar]
- Lincoln, S. E., M. J. Daly and E. S. Lander, 1993. MAPMAKER/EXP. Whitehead Institute for Biomedical Research, Boston.
- Mallet, J., 1989. The genetics of warning colour in Peruvian hybrid zones of Heliconius erato and H. melpomene. Proc. R. Soc. Lond. Ser. B 236: 163–185. [Google Scholar]
- Mundy, N. I., 2005. A window on the genetics of evolution: MCIR and plumage colouration in birds. Proc. R. Soc. Lond. Ser. B 272: 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit, R. E, C. D. Jiggins and J. Mallet, 2003. Mimicry: developmental genes that contribute to speciation. Evol. Dev. 5: 269–280. [DOI] [PubMed] [Google Scholar]
- Nijhout, H. F., and G. A. Wray, 1988. Homologies in the colour patterns of the genus Heliconius (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. 33: 345–365. [Google Scholar]
- Nijhout, H. F., G. Wray and L. E. Gilbert, 1990. An analysis of the phenotypic effects of certain color pattern genes in Heliconius (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. 40: 357–372. [Google Scholar]
- Protas, M. E., C. Hersey, D. Kocchanek, Y. Zhou, H. Wilkens et al., 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38: 107–111. [DOI] [PubMed] [Google Scholar]
- Reed, R. D., and L. E. Gilbert, 2004. Wing venation and Distal-less expression in Heliconius butterfly wing pattern development. Dev. Genes Evol. 214: 628–634. [DOI] [PubMed] [Google Scholar]
- Reed, R. D., and M. S. Serfas, 2004. Butterfly wing pattern evolution is associated with changes in a Notch/Distal-less temporal pattern formation process. Curr. Biol. 14: 1159–1166. [DOI] [PubMed] [Google Scholar]
- Shapiro, M. D., M. E. Marks, C. L. Peichel, B. K. Blackman, K. S. Nereng et al., 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428: 717–723. [DOI] [PubMed] [Google Scholar]
- Sheppard, P. M., J. R. G. Turner, K. S. Brown, W. W. Benson and M. C. Singer, 1985. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos. Trans. R. Soc. Lond. B 308: 433–610. [Google Scholar]
- Sucena, E., I. Delon, I. Jones, F. Payre and D. L. Stern, 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424: 935–938. [DOI] [PubMed] [Google Scholar]
- Tobler, A., D. Kapan, N. S. Flanagan, C. Gonzalez, E. Peterson et al., 2005. First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity 94: 408–417. [DOI] [PubMed] [Google Scholar]
- Turner, J. R. G., 1971. The genetics of some polymorphic forms of the butterflies Heliconius melpomene (Linnaeus) and H. erato (Linneaus), 2: the hybridization of subspecies from Surinam and Trinidad. Zoologica 56: 125–157. [Google Scholar]
- Turner, J. R. G., and J. Crane, 1962. The genetics of some polymorphic forms of the butterflies Heliconius melpomene Linnaeus and H. erato Linneaus, I: major genes. Zoologica 47: 141–152. [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Voorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., J. R. True and S. B. Carroll, 2002. a Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development 129: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., K. Vaccaro and S. B. Carroll, 2002. b Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. L. Williams, J. E. Selegue and S. B. Carroll, 2003. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. USA 100: 1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]