Abstract
We examined mismatch repair (MMR)-defective diploid strains of budding yeast grown for ∼160 generations to determine whether decreases in spore viability due to the uncovering of recessive lethal mutations correlated with an increase in gross chromosomal rearrangements (GCRs). No GCRs were detected despite dramatic decreases in spore viability, suggesting that frameshift and/or other unrepaired DNA replication lesions play a greater role than chromosomal instability in decreasing viability in MMR-defective strains.
THE mismatch repair (MMR) system contributes to genome stability by repairing DNA replication errors and suppressing recombination between divergent sequences. Long-term-growth experiments in several organisms have indicated that the absence of MMR leads to decreased fitness due to the accumulation of mutations (Funchain et al. 2000; Wloch et al. 2001; Zeyl and DeVisser 2001; Degtyareva et al. 2002; Hoffman et al. 2004). In Saccharomyces cerevisiae, the effects of MMR deficiency include an increase in the accumulation of recessive lethal mutations, as revealed by the progressive decline in the viability of meiotic progeny of repeatedly subcultured diploid strains (Williamson et al. 1985; Reenan and Kolodner 1992; Prolla et al. 1994; Wloch et al. 2001; Heck et al. 2006). Analyses of single loci in Escherichia coli, yeast, and human cells have shown that MMR-defective mutants display increased rates of gross chromosomal rearrangements (GCRs), including duplications, deletions, and translocations (Petit et al. 1991; Chen et al. 2001; Myung et al. 2001). In contrast, cytogenetic and microarray-based comparative genomic hybridization (CGH) analyses have suggested that human tumor cells with MMR defects contain fewer GCRs than most other solid tumors (e.g., Snijders et al. 2003).
These studies encouraged us to utilize a controlled system to examine whether chromosomal rearrangements accompany the progressive decrease in spore viability observed in MMR-defective diploid yeast strains. We used a microarray-based CGH analysis and pulsed-field gel electrophoresis (PFGE) (Dunham et al. 2002) to examine the effects of extended growth of MMR-defective diploid yeast strains on genome stability. Specifically, a temperature-sensitive mlh1 allele, mlh1-7 (K67A, D69A; Argueso et al. 2003), was used to examine mutation accumulation in replicate diploid lines over 160 generations. The temperature-sensitive MMR phenotype of mlh1-7 was confirmed using a lys2-A14 reversion assay in haploid strains (Tran et al. 1997). At 26°, the lys2-A14 reversion rates in mlh1-7 and mlh1Δ strains were 32- and 2550-fold higher, respectively, than those in wild type (7.0 × 10−7); at 35°, they were 955- and 3650-fold higher, respectively. Use of a conditional allele allowed us to bypass the meiotic defect of an mlh1 mutation by inducing meiosis and sporulation at the permissive temperature (Hunter and Borts 1997). It also allowed us to minimize further mutation accumulation through growth at the permissive temperature in cultures grown for the purpose of preparing chromosomal DNA for GCR analysis.
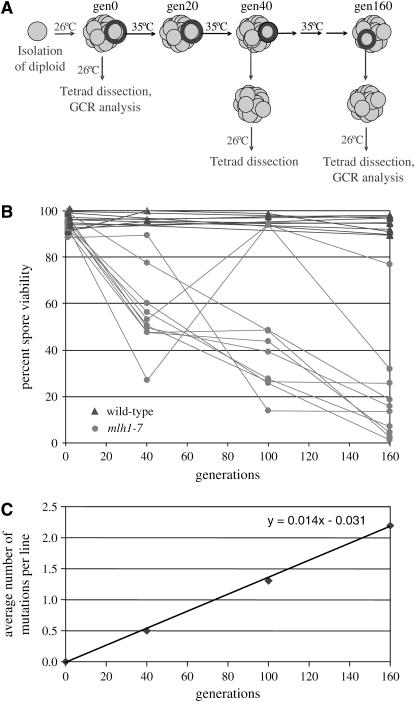
Ten wild-type and 10 mlh1-7 diploid strains were isolated at the permissive temperature, 26°, and then subcultured repeatedly on rich media at the nonpermissive temperature, 35° (Figure 1A). Spore viability was assessed after 0, 40, 100, and 160 generations of growth at 35° by sporulating diploids at the permissive temperature and dissecting at least 40 complete tetrads. Control experiments performed in parallel at 26° indicated that there was little difference between wild type and mlh1-7 in the accumulation of mutations affecting spore viability. At generation 0, wild-type and mlh1-7 spore viabilities were 95.2 and 94.0%, respectively; after 160 generations at 26°, they were 94.1 and 92.7%, respectively. These small differences in viability were statistically significant at generation 0 (P = 0.043) but not at generation 160 (P = 0.11; for significance of the difference between two independent proportions, see http://faculty.vassar.edu/lowry/VassarStats.html).
Figure 1.—
Mutation accumulation in wild-type and mlh1-7 strains. (A) Schematic of the mutation accumulation procedure. Wild-type (EAY102 MATa his3 cyhs ade2 ura3 trp1/EAY104 MATα leu2 cyhr ura3 trp1) and mlh1-7 (EAY1491 MATa his3 cyhs ade2 ura3 trp1 mlh1-7∷KanMX4/EAY1492 MATα leu2 cyhr ura3 trp1 mlh1-7∷KanMX4) S288c diploid strains were isolated by mating the indicated haploid strains at the permissive temperature (26°) and then by selecting for diploids on complete media lacking leucine and histidine. The resulting single colonies were frozen in glycerol stocks; these isolates were designated as generation 0. Portions of each single colony were also struck out on YPD plates to obtain single colonies at 26° and 35°, and stable diploid lines were propagated at 26° or 35° by serial transfer of randomly chosen colonies on YPD plates. Colonies were frozen every transfer (∼20 generations). To determine spore viability at generation 0, cells were patched from frozen stocks onto YPD plates, grown overnight at 26°, and then plated on sporulation media. To determine spore viability and analyze DNA at later generations, cells from the frozen stocks were first struck to single colonies at the permissive temperature to reduce mutation accumulation and obtain a uniform population of cells. These single colonies were frozen, and samples were taken from frozen stocks to determine spore viability and isolate genomic DNA for pulsed-field electrophoresis and CGH. All tetrads were dissected on minimal complete plates. Colonies visible to the naked eye after 4 days at 26° were scored as viable. (B) Spore viability of 10 mlh1-7 lines (solid circles) was assessed after 0, 40, 100, and 160 generations at 35°. Spore viability of 10 wild-type lines (solid triangles) was assessed after 0 and 160 generations at 35°; that of three of the lines was also assessed after 40 and 100 generations. A total of 40–100 tetrads was dissected for each wild-type and mlh1-7 line at the indicated generation. (C) Relationship between the average number of recessive lethal mutations per mlh1-7 isolate and generation. See text for details.
Wild-type spore viabilities after 40, 100, and 160 generations at 35° remained high, while decreases in spore viability were observed for all 10 mlh1-7 lines (Figure 1B). In two cases, mlh1-7 cells sampled at a later generation had higher spore viabilities than at an earlier generation. This apparent increase is likely a matter of chance; the cells chosen to continue growth of the culture had fewer mutations affecting spore viability than those sampled for measurement of spore viability at a given step. After 160 generations at the restrictive temperature, mlh1-7 spore viabilities ranged from 1.1 to 76.9%, with a median value of 14.5%. The mean viability declined at a rate of 0.0042/generation (Figure 1B; mean spore viability = −0.0042 × generation + 0.85), with the variance increasing from 0 to 100 generations. The variance decreased at generation 160, presumably because of the high percentage of lethals. At 40, 100, and 160 generations, spore viabilities of some mlh1-7 lines grown at 35° clustered near 50 and 25%. This can be explained by a single recessive lethal mutation in the former case and by two unlinked recessive lethal mutations in the latter case.
We used estimates of the number of recessive lethal mutations in each line to calculate the rate of such mutations in mlh1-7 strains grown at 35°. These estimates were made using the following assumptions: spore viability >50% = 0 recessive lethal mutations; 30–50% = 1; 15–30% = 2; <15% = 3. We observed a linear relationship (average number of mutations = 0.014 × generation −0.031) between the average number of mutations per isolate and generation, with the slope of the line, 0.014, corresponding to the mutation rate per generation (Figure 1C).
While analysis of segregation patterns allows us to infer the number and rate of mutations affecting spore viability, it does not reveal the type of mutation. To address this question, we used pulsed-field gel electrophoresis to look for alterations in chromosome size in all 10 of the generation 160 mlh1-7 strains as well as in wild-type and mlh1-7 generation 0 controls. The electrophoretic karyotypes were identical among all strains, indicating an absence of gross deletions, amplifications, or translocations (data not shown). We also examined these samples using array CGH to investigate whether impaired MMR results in alteration of DNA copy number (Dunham et al. 2002). Using CGH and PFGE, Dunham et al. (2002) performed an unbiased genomewide analysis and identified GCRs that were >100 kb. We used Agilent yeast 11k 60-mer oligonucleotide microarrays that contain a single probe for each open reading frame. Although CGH has been used to detect single-gene copy-number change, accurate identification usually requires at least a twofold DNA copy-number change. Thus, in a diploid genome, an increased copy number of a single probe, resulting in a hybridization ratio of 3:2, or a decreased copy number resulting in a ratio of 1:2, cannot be reliably detected. Initial tests of the Agilent arrays using self–self hybridizations confirmed the unreliability of single probe measurements (data not shown). Thus, we considered the running average intensity over nine probes (Dunham et al. 2002), which limits our resolution to GCRs that involve nine or more genes (probes), but reduces noise in the data. While this survey has limitations in terms of resolution, it provides an unbiased genomewide analysis for GCRs that is distinct from studies involving a selection for GCRs (e.g., Myung et al. 2001; Lemoine et al. 2005).
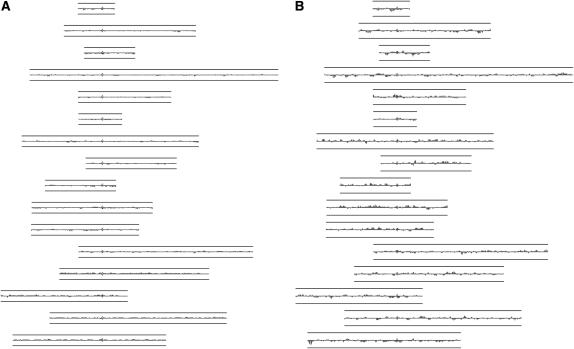
For each gene i, we averaged the log2(Cy5/Cy3) value for (k − 1)/2 genes to the left and right of i. We set k to 9 and imposed a heuristic requirement of five contiguous probes reporting a log2(Cy5/Cy3) value ≥0.58 or ≤−1 as evidence of a GCR. No chromosomal segments fulfilled this requirement, suggesting that no changes in DNA copy number were present in samples from the 10 independently derived generation 160 mlh1-7 lines. Representative results from one experiment in which DNA from generation 0 and generation 160 mlh1-7 strains were cohybridized to an array are shown in Figure 2.
Figure 2.—
Analysis of DNA copy number by array CGH as described by Dunham et al. (2002). The images, generated using JavaTreeView (http://jtreeview.sourceforge.net/), show log2-transformed array data averaged over nine genes for each of the 16 yeast chromosomes, aligned by centromeres (I–XVI in order, from top to bottom). (A) Control: an mlh1-7 generation 0 strain cohybridized with DNA from an isogenic wild-type strain. (B) An mlh1-7 generation 0 strain cohybridized with an mlh1-7 generation 160 strain. Lines above and below each chromosome correspond to a log2 value of ±1 (or a DNA copy-number ratio of 2:1) and the height of each individual feature along the chromosome corresponds to its log2 ratio as measured on the array. In diploids, an extra copy of a locus results in a 3:2 copy-number ratio, or a log2 value of 0.58.
Although the increased rates of GCRs reported previously for MMR mutants are moderate compared to those of mutants in some other DNA repair pathways (Petit et al. 1991; Myung et al. 2001), we reasoned that GCRs could also arise as a secondary effect of the MMR defect through mutations accumulating in other pathways. However, our analyses using CGH and pulsed-field gel electophoresis did not reveal GCRs in any of the strains. It is possible that our sample set was too small to detect an increased rate of GCR formation. Moreover, it is possible that the strains contained chromosomal defects not detectable using Agilent 11k yeast arrays or PFGE, either because they were too small or because they did not change chromosome size or DNA copy number (e.g., an inversion). Alternatively, the postreplicative MMR defect, which has been shown to confer an increase in base substitution, frameshift, and DNA slippage events, fully accounts for the reduced spore viability observed. In the following paragraph, we provide evidence within the context of DNA slippage of how such a postreplicative MMR defect could yield the observed decrease in viability.
Numerous studies have shown that loss of MMR results in a large increase in the rate of frameshift mutations within mono- and dinucleotide repeats, with longer repeats exhibiting greater instability (Tran et al. 1997; Harfe and Jinks-Robertson 2000). In human cancers resulting from MMR deficiency, including hereditary nonpolyposis colorectal cancer and some sporadic cancers, instability of repeat sequences within genes that control cell growth is suspected to contribute to cancer progression (Peltomaki 2001; Chung and Rustgi 2003). To investigate the extent to which repeat instability could be contributing to spore lethality in our system, we searched the yeast genome for mononucleotide repeats in coding DNA and used previously reported mutation rates to estimate probabilities of frameshift mutations (Table 1; Tran et al. 1997; Harfe and Jinks-Robertson 2000). Twenty-two essential genes contain mononucleotide runs of at least 10 nucleotides. We calculated that the probability of at least one of these genes incurring a −1 frameshift within a run is 89% after 160 generations in yeast lacking MMR. In contrast, although there are ∼600 seven-nucleotide homopolymer runs in essential genes, the probability of mutation in one or more of these runs is <30%. Thus, mutations in a small subset of essential genes may be major contributors to the reduced spore viability in our MMR-deficient strains. We did not pursue a similar analysis for the probability of base substitution mutations in essential genes that confer a null phenotype because the rate of such substitutions in MMR-defective mutants appears to be highly dependent on the type and orientation of the mismatch as well as sequence context (Earley and Crouse 1998; Marsischky and Kolodner 1999).
TABLE 1.
Frameshift mutation probability
| Repeats in coding DNAa | Repeats in essential coding DNAa | Mutation rateb
|
Mutation probability, generation 160c
|
|||
|---|---|---|---|---|---|---|
| Repeat | Wild type | MMR− | Wild type | MMR− | ||
| ≥10 A/T | 143 | 22 | 4.7 × 10−8 | 3.1 × 10−4 | 0.00033 | 0.89 |
| ≥10 G/C | 0 | 0 | 7.0 × 10−8 | 7.5 × 10−4 | 0.0 | 0.0 |
| 7 A/T | 2998 | 561d | 3.8 × 10−9 | 1.6 × 10−6 | 0.00068 | 0.25 |
| 7 G/C | 81 | 18 | 1.9 × 10−8 | 7.6 × 10−6 | 0.00011 | 0.043 |
Number of repeats in haploids (http://www.yeastgenome.org).
Rates of frameshifts (mutations per generation) within indicated repeats in wild-type and MMR null strains, taken or estimated from Tran et al. (1997) and Harfe and Jinks-Robertson (2000). Note that rates at specific loci may vary, depending on factors such as chromosomal location and sequence context (Harfe and Jinks-Robertson 2000; Hawk et al. 2005). The probability of incurring mutations in a given set of sequences within a given number of generations was calculated assuming a Poisson distribution, as one minus the probability of incurring no mutations, which was calculated as p(x = 0) = e−(μ)(no. of loci) (no. of generations), where μ is mutations per locus per generation.
Probability of incurring frameshift mutations within essential genes containing the indicated type of repeat.
Calculated on the basis of the percentage of genes in the genome that are essential (18.7%).
In addition to decreased replication fidelity, the chromosomal instability associated with MMR deficiency could also contribute to cancer susceptibility. Recent analysis of MMR-defective human cell lines by array CGH, however, showed that these lines displayed fewer copy-number changes than MMR-proficient lines (Snijders et al. 2003). Consistent with this, our results suggest that base substitution, frameshift, and DNA slippage events associated with defects in MMR are likely to play a greater role than GCRs in disrupting genes with important cellular functions.
Acknowledgments
We thank Charles Aquadro, Stephane Bentolila, Ann Bernard, Andrew Clark, Maitreya Dunham, Chad Myers, and Jen Wanat for technical advice and comments. This work was supported by National Institutes of Health (NIH) grant GM53085 (E.A.), a Center Grant from the National Institute of General Medical Sciences (NIGMS) (P50 GM071508), a research grant from the NIGMS (R01 GM046406, D.B.), and a Genetics and Development NIH training grant (J.A.H.).
References
- Argueso, J. L., A. W. Kijas, S. Sarin, J. A. Heck, M. Waase et al., 2003. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 23: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., S. H. Bigner and P. Modrich, 2001. High rate of CAD gene amplification in human cells deficient in MLH1 or MSH6. Proc. Natl. Acad. Sci. USA 98: 13802–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, D. C., and A. K. Rustgi, 2003. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann. Intern. Med. 138: 560–570. [DOI] [PubMed] [Google Scholar]
- Degtyareva, N. P., P. Greenwell, E. R. Hofmann, M. O. Hengartner, L. Zhang et al., 2002. Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc. Natl. Acad. Sci. USA 99: 2158–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown et al., 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, M. C., and G. F. Crouse, 1998. The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95: 15487–15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funchain, P., A. Yeung, J. L. Stewart, R. Lin, M. M. Slupska et al., 2000. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154: 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics 156: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk, J. D., L. Stefanovic, J. C. Boyer, T. D. Petes and R. A. Farber, 2005. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA 102: 8639–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, J. A., J. L. Argueso, Z. Gemici, R. G. Reeves, A. Bernard et al., 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, P. D., J. M. Leonard, G. E. Lindberg, S. R. Bollman and J. B. Hays, 2004. Rapid accumulation of mutations during seed-to-seed propagation of mismatch repair-defective Arabidopsis. Genes Dev. 18: 2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, K. Lobachev and T. D. Petes, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Marsischky, G. T., and R. D. Kolodner, 1999. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274: 26668–26682. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta, C. Chen and R. D. Kolodner, 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Peltomaki, P., 2001. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 10: 735–740. [DOI] [PubMed] [Google Scholar]
- Petit, M. A., J. Dimpfl, M. Radman and H. Echols, 1991. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics 129: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla, T. A., D. M. Christie and R. M. Liskay, 1994. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 14: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan, R. A., and R. D. Kolodner, 1992. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132: 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders, A. M., J. Fridlyand, D. A. Mans, R. Segraves, A. N. Jain et al., 2003. Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene 22: 4370–4379. [DOI] [PubMed] [Google Scholar]
- Tran, H. T., J. D. Keen, M. Kricker, M. A. Resnick and D. A. Gordenin, 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, M. S., J. C. Game and S. Fogel, 1985. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1–1 and pms1–2. Genetics 110: 609–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloch, D. M., K. Szafraniec, R. H. Borts and R. Korona, 2001. Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae. Genetics 159: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl, C., and J.A. DeVisser, 2001. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics 157: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]