Abstract
The PAR proteins play an essential role in establishing and maintaining cell polarity. While their function is conserved across species, little is known about their regulators and effectors. Here we report the identification of 13 potential components of the C. elegans PAR polarity pathway, identified in an RNAi-based, systematic screen to find suppressors of par-2(it5ts) lethality. Most of these genes are conserved in other species. Phenotypic analysis of double-mutant animals revealed that some of the suppressors can suppress lethality associated with the strong loss-of-function allele par-2(lw32), indicating that they might impinge on the PAR pathway independently of the PAR-2 protein. One of these is the gene nos-3, which encodes a homolog of Drosophila Nanos. We find that nos-3 suppresses most of the phenotypes associated with loss of par-2 function, including early cell division defects and maternal-effect sterility. Strikingly, while PAR-1 activity was essential in nos-3; par-2 double mutants, its asymmetric localization at the posterior cortex was not restored, suggesting that the function of PAR-1 is independent of its cortical localization. Taken together, our results identify conserved components that regulate PAR protein function and also suggest a role for NOS-3 in PAR protein-dependent cell polarity.
ASYMMETRIC cell division is a key process for the generation of cell diversity during the development of metazoans (reviewed in Betschinger and Knoblich 2004; Roegiers and Jan 2004). Stem cells, for instance, divide asymmetrically to generate two daughter cells with distinct fates: one daughter committed to differentiation and another that retains the totipotent characteristics of the mother. Asymmetric cell division relies on the partitioning of cell fate determinants and orientation of the mitotic spindle prior to cytokinesis to generate cells of different fates. Accordingly, polarity cues in these cells must be interpreted accurately to ensure proper partitioning of these determinants at cytokinesis. Although establishment and maintenance of cell polarity is crucial for asymmetric cell division in all organisms, the nature of the molecular interactions leading to these processes remains elusive.
In recent years, work in several systems demonstrated that the conserved partitioning-defective (PAR) proteins play a fundamental role in establishing and maintaining polarity in many asymmetrically dividing cell types (reviewed in Etienne-Manneville and Hall 2003; Nance 2005). In Drosophila, PAR proteins are essential for the asymmetric division of neuroblasts and sensory organ precursor cells (Betschinger and Knoblich 2004). Mutations in any gene encoding a PAR protein result in loss of cell polarity and a failure to divide asymmetrically. PAR proteins are also required to establish and maintain polarity in the developing oocyte (Cox et al. 2001; Huynh et al. 2001). In mammalian cells, PAR proteins were shown to be localized at tight junctions and to participate in epithelial cell polarity (Lin et al. 2000; Qiu et al. 2000; Hurd et al. 2003). They are also required for the correct migration of astrocytes after wound healing and for specifying axonal fate in rat hippocampal neurons (Etienne-Manneville and Hall 2001; Shi et al. 2003).
The Caenorhabditis elegans embryo is an excellent model system to study polarity and spindle positioning (Kemphues and Strome 1997). The first division of the embryo is asymmetric, resulting in two cells that are different in size and fate. The seven PAR proteins (PAR-1 to PAR-6 and PKC-3) are found at the cortex of the zygote and are responsible for specifying the antero-posterior axis of polarity (Kemphues and Strome 1997; Tabuse et al. 1998; Cuenca et al. 2003). Embryos mutant for any par gene do not properly establish polarity, undergo symmetric cell divisions, and fail to hatch. The PAR-3, PAR-6, and PKC-3 (hereafter referred to as anterior PARs) proteins are localized at the anterior cortex of the embryo (Etemad-Moghadam et al. 1995; Tabuse et al. 1998; Hung and Kemphues 1999), while PAR-1 and PAR-2 are restricted to the posterior cortex (Guo and Kemphues 1995; Boyd et al. 1996). This localization along the antero-posterior axis is mutually exclusive, as members of the anterior and posterior groups show little overlap in their respective localization and the PAR-2 and PAR-3 proteins exclude each other from their respective cortices (Etemad-Moghadam et al. 1995; Boyd et al. 1996; Cuenca et al. 2003). Therefore, PAR proteins define anterior and posterior cortical domains in the zygote and specify cell polarity and asymmetric cell divisions in the early C. elegans embryo.
While PAR proteins are conserved across species, little is known about the precise molecular mechanisms by which they contribute to the establishment and maintenance of cell polarity. This lack of understanding is partly due to the fact that only a limited number of PAR protein activators and effectors that function during these processes have been identified thus far (Macara 2004). Interestingly, previous work demonstrated that while removing both copies of par-2 leads to the mislocalization of the anterior PARs to the posterior of the embryo, and thus to defects in polarity and embryonic lethality, reducing by half the amounts of PAR-6 in a par-2 mutant background is sufficient to restore viability (Watts et al. 1996) (supplemental Figure 1 at http://www.genetics.org/supplemental/). Similarly, disruption by RNA interference (RNAi) (Fire et al. 1998) of cdc-42, a regulator of the anterior PAR activity, in a par-2 mutant also restores viability (Gotta et al. 2001). These results indicate that genes that are required to regulate the levels, the localization, and/or the activity of anterior PAR components can be identified on the basis of their ability to restore viability in a par-2 mutant background. We therefore sought to find additional genes that are required for polarity by screening for suppressors of the lethality caused by disruption of the polarity gene par-2. Using a systematic, genomewide, RNAi-based screening approach, we identified several potential components of the PAR polarity pathway in C. elegans. Many of these components are present in other species, suggesting that their role in cell polarity might also be conserved. To gain further insights into possible suppression mechanism(s), we studied in more detail the phenotype of embryos mutant for the suppressor gene nos-3, which encodes one of the three C. elegans homologs of the protein Nanos (Kraemer et al. 1999; Subramaniam and Seydoux 1999). In Drosophila, Nanos has been shown to control embryonic polarity by repressing translation of the hunchback mRNA in the posterior of the embryo (Hulskamp et al. 1989; Irish et al. 1989). We find that nos-3 can suppress a strong loss-of-function allele of par-2, indicating that it could function independently of the PAR-2 protein. Surprisingly, PAR-1 was localized in the cytoplasm of these embryos, suggesting that the essential function of PAR-1 is independent of its asymmetric cortical localization. Taken together, these results indicate that the par-2 suppressor screen revealed genes involved in regulating PAR protein function and further demonstrate that nos-3 participates in PAR protein-dependent cell polarity in C. elegans embryos.
MATERIALS AND METHODS
Strains:
All strains were maintained as described by Brenner (1974) and were grown at 15° unless otherwise stated. The alleles used in this study were the following: LGI, rpn-10(tm1180), rpn-10(tm1349); LGII, nos-3(q650), fbf-1(ok91); LGIII, par-2(lw32), par-2(it5ts); and LGX, spat-3(gk22). The wild-type strain was the N2 (Bristol) strain.
The generation of nos-3(q650); par-2(it5ts) double mutants was done as follows. Males mutant for nos-3(q650) were mated with par-2(it5ts) mutant hermaphrodites at permissive temperature and five animals of the resulting F1 cross progeny were singled. These F1 animals were allowed to lay eggs for 24 hr at permissive temperature before being lysed and the presence of the nos-3(q650) allele was confirmed by PCR analysis, using pairs of primers specific for the nos-3 locus. Twenty F2 animals were then singled and allowed to lay eggs for 24 hr at permissive temperature before being transferred to new plates and shifted to restrictive temperature. Animals producing dead embryos at restrictive temperature were considered homozygous for par-2(it5ts). These animals were lysed and genotyped by PCR analysis to identify those bearing the nos-3(q650) allele. F3 animals that had hatched at permissive temperature were then singled, allowed to lay eggs, and genotyped to find those homozygous for nos-3(q650). The presence of par-2(it5ts) was further confirmed by noncomplementation analysis by mating par-2(it5ts) males with nos-3(q650); par-2(it5ts) hermaphrodites at permissive temperature, shifting the F1 cross progeny at restrictive temperature, and scoring for absence of hatched larvae. Double-mutant strains with other par-2 suppressors were done by the same approach and genotyping was done by PCR using pairs of primers specific for each locus.
The generation of nos-3(q650); par-2(lw32) double mutants was done as follows. Males mutant for nos-3(q650) were mated with unc-45(e286) par-2(lw32)/qC1[dpy-19(e1259) glp-1(q339)] mutant hermaphrodites. Males resulting from the F1 cross progeny were then mated with their F1 sister hermaphrodites and 30 F1 wild-type animals resulting from this second cross were singled to individual plates. The animals giving rise to wild-type, Unc, and Dpy animals were lysed and genotyped by PCR analysis to identify those homozygous for the nos-3(q650) allele. The presence of the par-2(lw32) was confirmed by sequence analysis of the par-2 locus. Double-mutant strains with other par-2 suppressors were produced by the same approach and genotyping was done by PCR using pairs of primers specific for each locus.
Genomewide RNAi screen:
The screen for par-2 suppressors was performed using the following procedure. par-2(it5ts) animals were grown in large quantities on solid media at 15° and bleached to collect embryos. These embryos were incubated at 15° with rocking in M9 buffer to allow hatching of L1 larvae. Assays were done in plates containing 96 wells. RNAi clones from the available collection (Kamath et al. 2003) were seeded in individual wells and grown overnight at 37° in LB medium containing 100 μg/ml of carbenicillin. Each well in the 96-well plates was then filled with 75 μl of 3× NGM (containing triple amount of peptone) and 2 μl of overnight bacterial culture was added. The plates were incubated at 37° for 2.5 hr without shaking. Twenty-five microliters of 3× NGM containing 24 mm IPTG was then added to each well (6 mm IPTG final) and the plates were incubated at 37° for 5 hr without shaking. Five to 10 L1 worms were then added to each well (15 μl from a suspension containing approximately eight worms/15 μl of M9 buffer). The plates were incubated for 2.5 days at the permissive temperature of 15° without agitation and were then shifted to the semirestrictive temperature of 22° for 3–4 days until food was depleted and F1 progeny had hatched. The suppression level was estimated by visual inspection under a dissecting scope, and the presence of more swimming L1 larvae compared to the control was scored as positive. Liquid handling was performed using a Beckman Biomek FX robotic system equipped with a 96-channel pipetting head.
Each RNAi clone was scored in duplicate in two independent screens. The first screen identified 1847 RNAi clones showing suppression while the second screen identified 952 RNAi clones. Of these RNAi clones, 136 were identified in both screens and were kept for further analysis. The high number of false positives in these screens might reflect the fact that these were carried out at a semipermissive temperature for embryonic lethality, where minute fluctuations of temperature might have important consequences on the physiology of the animals. RNAi to these 136 clones was then performed on par-2(it5ts) animals in duplicate on solid assays as previously described (Kamath et al. 2003), and 18 clones were found to have a higher proportion of hatching progeny compared with the control at 20° (supplemental Figure 1 and supplemental Table 1 at http://www.genetics.org/supplemental/). RNAi to these 18 clones was then repeated eight additional times at 20° and the 8 clones with a percentage of hatching progeny significantly higher than the controls after counting were considered as suppressors of par-2(it5ts) (supplemental Table 2 at http://www.genetics.org/supplemental/). The molecular identity of these clones was confirmed by DNA sequencing.
Suppression assays:
For mutant animals, L4 animals from each strain were shifted to semirestrictive (20°) or restrictive (25°) temperature for 24 hr. Nine worms were transferred to three plates (three worms per plate) and allowed to lay eggs at the same temperature for 16 hr before being removed. The viability of the progeny was determined after incubating another 24 hr at the same temperature, dividing the number of unhatched embryos by the total number of progeny.
To assess the level of par-2(RNAi) suppression, mutant animals were injected with double-stranded RNA (dsRNA) from par-2, or wild-type animals were co-injected with dsRNA from both par-2 and a given suppressor as described previously, and worms were subsequently incubated for 16–24 hr at 20°. Twelve injected animals were then transferred to three plates (four worms per plate) and allowed to lay eggs for 10 hr. The viability of the progeny was assessed 24 hr after removing the worms. The following alleles were tested and did not suppress par-2(RNAi): LGI, cey-2(ok902), rpn-10(tm1180), rpn-10(tm1349); LGII, lat-2(tm463), lat-2(ok301), nos-1(ok250), fbf-1(ok91); LGIII, dgk-3(gk110); LGIV, fat-2(ok873); LGV, nhr-84(ok792), egl-3(ok979); LGX, ceh-18(mg57), sad-1(ky289), hen-1(tm501), zig-3(tm924).
To test for the involvement of fbf-1 and fbf-2 in par-2 suppression, par-2(it5ts) mutant or fbf-1(ok91); par-2(it5ts) double-mutant L4 animals were placed on plates containing bacteria expressing dsRNAs for either fbf-1 or fbf-2 and incubated at 20° for 48 hr. Nine worms were transferred to three plates (three worms per plate) containing the appropriate dsRNA-expressing bacteria and allowed to lay eggs at the same temperature for 12 hr before being removed. The viability of the progeny was determined as described above.
To assay for sterility, L4 animals from each strain were shifted to permissive (15°) or restrictive (25°) temperature and allowed to develop until some eggs had been laid. These eggs were allowed to hatch and develop at their respective temperature until animals reached adulthood. Sterility was then determined by the presence or absence of embryos in the uterus of these animals, dividing the number of animals without progeny by the total number of animals.
Microscopy:
For the visualization of early embryonic development in live specimens, embryos were obtained by cutting open gravid hermaphrodites using two 25-gauge needles. Embryos were handled individually and mounted on a coverslip coated with 1% poly-l-lysine in 20 μl of egg buffer (Edgar 1995). The coverslip was placed on a 3% agarose pad and the edge was sealed with petroleum jelly. Time-lapse images were acquired by an Orca ER Hamamatsu 16-bit cooled CCD camera (Hamamatsu Photonics, Bridgewater, NJ) mounted on a Zeiss Axiovert 200 microscope (Carl Zeiss AG, Jena, Germany), and the acquisition system was controlled by Openlab software (Improvision, Coventry, UK). Images were acquired at 10-sec intervals using a Plan Apochromat 63×/1.4 NA objective.
For immunofluorescence analysis, embryos were fixed in methanol and stained according to standard procedures. The following primary antibodies were used: rabbit anti-PAR-1 (Gonczy et al. 2001) (1/1000), rabbit anti-PAR-2 (N-terminal antibody generated as described by Boyd et al. 1996) (1/20), rabbit anti-PAR-3 (generated as described in Etemad-Moghadam et al. 1995) (1/100), rabbit anti-PAR-6 (generated as described in Hung and Kemphues 1999) (1/50), mouse OIC1D4 (Developmental Studies Hybridoma Bank, University of Iowa) (1/1) for P-granule staining. Secondary antibodies were Cy3-coupled goat-anti-rabbit (Jackson Immunoresearch) (1/100) and Alexa488-coupled goat-anti-mouse (Molecular Probes, Eugene, OR) (1/500). Images were acquired using a Leica SP2 confocal microscope.
RESULTS
A screen for suppressors of par-2(it5ts) uncovers 11 loci:
Previous reports have demonstrated that the lethal phenotype of par-2 can be suppressed by reducing the levels of either par-6 or cdc-42 (Watts et al. 1996; Gotta et al. 2001) (supplemental Figure 1 at http://www.genetics.org/supplemental/). We first tested whether this effect is specific to these genes or can be obtained by reducing the levels of any of the anterior PAR proteins. We found that disruption by RNAi (Fire et al. 1998) of par-3, par-6, pkc-3, or cdc-42 can effectively suppress the lethal phenotype of the temperature-sensitive allele par-2(it5ts) at a semirestrictive temperature (Table 1). This indicates that genes encoding proteins that regulate anterior PAR function can be identified on the basis of their ability to suppress par-2(it5ts) embryonic lethality.
TABLE 1.
Suppressors of par-2(it5ts) lethality
| RNAi clone | Gene | Embryonic viability (%)a | Domains and description |
|---|---|---|---|
| Vector | Negative control | 5.5 ± 2.6 | |
| C32E12.2 | Negative control | 8.4 ± 4.5 | |
| F54E7.3 | par-3 | 75.1 ± 5.6 | PDZ domains, component of the anterior PARs |
| T26E3.3 | par-6 | 29.2 ± 16.1 | PDZ and CRIB domains, component of the anterior PARs |
| F09E5.1 | pkc-3 | 38.3 ± 19.8 | Kinase domain, component of the anterior PARs |
| R07G3.1 | cdc-42 | 48.2 ± 27.5 | GTPase domain, activator of the anterior PARs |
| Y53C12B.3 | nos-3 | 10.0 ± 6.3b | Q/N-rich domain, zf-nanos-type zinc-finger domain, Drosophila Nanos and Xenopus Xcat-2 homolog |
| F57C2.6 | spat-1 | 15.4 ± 5.5 | Conserved in C. briggsae only |
| F54F2.5 | ztf-1 | 16.3 ± 5.2 | C2H2-type zinc fingers |
| Y48A6B.13 | spat-2 | 32.0 ± 8.8 | Conserved in C. briggsae only |
| Y43E12A.1c | cyb-2.1 cyb-2.2 | 59.3 ± 6.1 | Cyclin B homologs |
| W02A2.1 | fat-2 | 22.7 ± 9.6 | Fatty acid desaturase |
| ZC64.3 | ceh-18 | 20.9 ± 7.0 | POU-class homeodomain transcription factor |
| T13H2.5 | spat-3 | 50.9 ± 12.1 | C3HC4-type RING-finger domain and Q/N-rich domain |
| ZK20.5 | rpn-12 | 48.0 ± 18.5 | Component of the 26S proteasome non-ATPase regulatory particle |
| T06D8.8 | rpn-9 | 22.9 ± 6.6 | Component of the 26S proteasome non-ATPase regulatory particle |
| B0205.3 | rpn-10 | 25.4 ± 10.3 | Component of the 26S proteasome non-ATPase regulatory particle |
Suppressors of par-2 (it5ts) lethality were determined by feeding RNAi clones to animals at the L1 stage. All of these clones were not identified in the genomewide screen; see text and materials and methods for details.
The value corresponds to the average percentage of embryos that hatched over the total number of embryos ± standard deviation over eight assays.
Although nos-3 was not found to suppress par-2 in this particular assay, it was included as a suppressor of par-2 since it suppressed all of the par-2 phenotypes in other assays.
Clone Y43E12A.1 might disrupt the function of both cyb-2.1 and cyb-2.2 since these two genes are 95% identical at the nucleic acid level.
We therefore devised a method, based on RNAi, to screen for genes that suppress the lethal phenotype of par-2(it5ts) (see materials and methods). We used this method to assay whether individual bacterial clones from an available collection (Kamath et al. 2003), each expressing a unique dsRNA molecule, can restore viability to the progeny of par-2(it5ts) animals at a semirestrictive temperature. Using an RNAi assay in liquid, we tested 16,458 bacterial clones in two independent screens and found 136 clones that qualitatively showed partial suppression of par-2(it5ts) embryonic lethality in both assays (supplemental Figure 1 and supplemental Table 1 at http://www.genetics.org/supplemental/). We then quantitatively tested each of the 136 clones on solid assay and found that embryonic viability following disruption by RNAi of 8 of them was significantly higher than the viability of par-2(it5ts) embryos when tested multiple times at a semirestrictive temperature (supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). The description of these 8 suppressors of par-2(it5ts) embryonic lethality can be found in Table 1. We collectively refer to the genes identified as suppressor of par-two (spat) genes.
In an independent assay, we tested whether available alleles for the genes disrupted by the 136 RNAi clones identified in the primary screen could suppress the lethal phenotype of par-2 when the par-2 gene itself is disrupted by RNAi. Among the available strains mutant for the 136 clones identified, we found that lethality resulting from par-2(RNAi) was efficiently suppressed by nos-3(q650) and by spat-3(gk22) (supplemental Table 3 at http://www.genetics.org/supplemental/). Interestingly, while nos-3 was recovered in the primary screen, we found that nos-3(RNAi) did not significantly suppress par-2(it5ts) lethality in subsequent assays (Table 1). This suggests that our solid assay for RNAi was not exhaustive and that other suppressors of par-2(it5ts) lethality might be present among the RNAi clones that were negative in the subsequent validation steps (see materials and methods).
Finally, we scored embryonic viability in animals mutant for both par-2(it5ts) and either nos-3(q650) or spat-3(gk22). At semirestrictive temperature, the progeny of all of the double-mutant strains has a significantly increased embryonic viability compared to par-2(it5ts) (Table 2). Taken together, these results suggest that the spat genes that we identified are bona fide suppressors of par-2.
TABLE 2.
Mutations in nos-3, spat-3, or rpn-10 can suppress par-2 lethality
| Genotype | Embryonic viability at 20° (%)a | Embryonic viability at 25° (%)a |
|---|---|---|
| Wild type | 99.8 ± 0.0 | 99.3 ± 0.5 |
| par-2(it5ts) | 16.9 ± 3.7 | 0.8 ± 0.8 |
| par-2(lw32) | 1.7 ± 1.0b | ND |
| nos-3(q650) | 99.4 ± 0.3 | 75.2 ± 3.9 |
| spat-3(gk22) | 99.8 ± 0.0 | 98.7 ± 0.1 |
| rpn-10(tm1180) | 99.0 ± 0.1 | 99.2 ± 0.3 |
| rpn-10(tm1349) | 99.0 ± 0.2 | 97.3 ± 0.8 |
| nos-3(q650); par-2(it5ts) | 86.5 ± 2.0 | 53.6 ± 1.6 |
| nos-3(q650); par-2(lw32) | 54.6 ± 4.7b | ND |
| par-2(it5ts); spat-3(gk22) | 86.6 ± 2.2 | 22.9 ± 5.9 |
| par-2(lw32); spat-3(gk22) | 43.1 ± 6.8b | ND |
| rpn-10(tm1180); par-2(it5ts) | 62.2 ± 2.9 | 10.3 ± 1.8 |
| rpn-10(tm1349); par-2(it5ts) | 73.5 ± 2.7 | 8.8 ± 0.8 |
The value corresponds to the average percentage of embryos that hatched over the total number of embryos ± standard error of the mean over three independent assays; see materials and methods for details.
Viability for these particular assays was determined at 15° instead of 20° because par-2(lw32) animals also contain the mutation unc-45(e286ts), which confers an egg-laying-defective phenotype at higher temperatures.
Sequence analysis of the spat genes suggests that they are quite diverse in their function. The most common domain identified consists of zinc-binding motifs, which are found in three of our suppressor proteins (NOS-3, ZTF-1, and SPAT-3). Two of these proteins (NOS-3 and SPAT-3) also contain Gln/Asn (Q/N)-rich domains, which have been found in prions and implicated in neurodegenerative disorders (Michelitsch and Weissman 2000; Forman et al. 2004). We also identified RNAi clone Y43E12A.1, which is predicted to disrupt the function of the two cyclin B homologs cyb-2.1 and cyb-2.2 (together referred to as cyb-2). Since these two genes are >95% identical at the nucleic acid level, we could not assess whether RNAi disruption of only one of them is sufficient to suppress par-2(it5ts) embryonic lethality. One of the suppressors, ceh-18, encodes a homeodomain-containing transcription factor that was previously shown to function during oocyte maturation (Greenstein et al. 1994; Miller et al. 2003). Another suppressor, fat-2, encodes a fatty-acid desaturase that is required to maintain normal levels and composition of polyunsaturated fatty acids in the animal (Watts and Browse 2002). Two of the suppressors encode proteins that do not have well-defined homologs in other species (SPAT-1 and SPAT-2).
One of the suppressors identified consists of the gene encoding the non-ATPase proteasome subunit RPN-12. In various systems, RPN-12 has been shown to function in the regulatory particle of the proteasome and was proposed to share some activities with the subunit RPN-10 (Wilkinson et al. 2000; Glickman and Ciechanover 2002; Takahashi et al. 2002). In C. elegans, only 3 of the ∼30 genes shown so far to encode proteasome subunits (Davy et al. 2001) do not result in embryonic lethality upon disruption by RNAi: rpn-9, rpn-10, and rpn-12 (Takahashi et al. 2002; Kamath et al. 2003; Sonnichsen et al. 2005). While we did not identify rpn-9 or rpn-10 in the genomewide screen, subsequent analysis revealed that RNAi from either of these two genes can also suppress par-2(it5ts) embryonic lethality (Table 1), albeit less efficiently than RNAi from rpn-12. Furthermore, we found that embryos double mutant for par-2(it5ts) and either allele of rpn-10 (tm1180 or 1349) are viable at a semipermissive temperature (Table 2), indicating that rpn-10 is a suppressor of par-2. This demonstrates that additional suppressors of par-2(it5ts) can be found by looking for genes that are related in sequence or function to the identified suppressors.
Taken together, our results describe several suppressors of par-2 lethality that, on the basis of their protein sequence, are likely to play different roles in polarity.
Some of the par-2 suppressors could function independently of par-2:
The spat genes identified could conceivably suppress the phenotypes associated with a loss of function of par-2 by at least two distinct but nonexclusive mechanisms: (1) by downregulating the function of the anterior PARs (as in the case of RNAi to par-3, par-6, pkc-3, or cdc-42) or (2) by upregulating residual PAR-2 function in par-2(it5ts) mutants. To distinguish between these two possibilities, we first tested whether some of our suppressors can suppress the lethal phenotype of par-2(it5ts) at a fully restrictive temperature. Except for par-3 and par-6, none of the RNAi clones listed in Table 1 could significantly suppress par-2(it5ts) lethality at a fully restrictive temperature in our assay (data not shown). However, we found that the progeny of animals double mutant for par-2(it5ts) and either nos-3(q650) or spat-3(gk22) have a much higher viability at a fully restrictive temperature than the progeny of par-2(it5ts) single mutants (Table 2). On the contrary, viability of the progeny of animals double mutant for par-2(it5ts) and either allele of rpn-10 (tm1180 or tm1349) is only slightly higher than that of par-2(it5ts) in the same conditions (Table 2). Furthermore, the lethality of par-2(RNAi) is not suppressed in animals mutant for either allele of rpn-10 (supplemental Table 3 at http://www.genetics.org/supplemental/). This suggests that nos-3 and spat-3 function independently of PAR-2 and that rpn-10 requires some residual PAR-2 activity to suppress the embryonic lethal phenotype of par-2.
To assess whether nos-3 and spat-3 could exert their function independently of PAR-2, we tested whether nos-3 or spat-3 can suppress the embryonic lethality associated with par-2(lw32). The lw32 allele contains a stop codon in the middle of the par-2 coding sequence and was shown to behave as a strong loss of function (Levitan et al. 1994; Boyd et al. 1996). We found that mutations in both nos-3 and spat-3 can suppress par-2(lw32): the viability of embryos double mutant for par-2(lw32) and either nos-3(q650) or spat-3(gk22) was much higher compared with par-2(lw32) single-mutant embryos (Table 2). Furthermore, the viability of nos-3(q650); par-2(it5ts); par-2(RNAi) embryos at a restrictive temperature was also increased compared to par-2(RNAi) (supplemental Table 3 at http://www.genetics.org/supplemental/). Taken together, these results suggest that some of the spat genes, such as nos-3 and spat-3, could suppress par-2 embryonic lethality independently of PAR-2 activity, possibly by modulating the function of the anterior PARs.
Codisruption of both fbf-1 and fbf-2 suppresses par-2 lethality:
The gene nos-3 encodes a C. elegans homolog of the Drosophila protein Nanos. NOS-3 functions in the germline and is also found associated with P granules in the early embryo (Kraemer et al. 1999; Subramaniam and Seydoux 1999). There are two other paralogs of nos-3 in C. elegans, nos-1 and nos-2, and all three genes have been shown to have redundant functions in the germline (Kraemer et al. 1999; Subramaniam and Seydoux 1999). We therefore tested whether any of these Nanos orthologs can also suppress par-2(it5ts) lethality. Unlike nos-3, we found that individual disruption of nos-1 or nos-2 by RNAi does not suppress par-2(it5ts) lethality (data not shown). Furthermore, codisruption of nos-1 or nos-2 by RNAi in nos-3(q650); par-2(it5ts) animals did not result in any increase in embryonic viability (data not shown). These results suggest that, in contrast to nos-3, nos-1 and nos-2 do not modulate PAR protein function.
In Drosophila, Nanos functions as a negative regulator of translation for various mRNAs in conjunction with the ribonucleoprotein Pumilio (reviewed in Wickens et al. 2002; Wilhelm and Smibert 2005). There are 11 Pumilio homologs predicted in the C. elegans genome (Wickens et al. 2002). We found that RNAi disruption of none of these genes individually can suppress par-2(it5ts) lethality (Table 3 and data not shown). Furthermore, embryos double mutant for par-2(it5ts) and fbf-1(ok91), a presumptive null allele of the pumilio homolog fbf-1 (Crittenden et al. 2002), also largely fail to hatch (Table 3), indicating that disruption of fbf-1 alone is not sufficient to suppress par-2 lethality. However, fbf-1 has been shown to function redundantly with fbf-2 in the C. elegans germline (Crittenden et al. 2002). These two genes are required, along with nos-3, to regulate the synthesis of the sex-determining FEM-3 protein and the translational repressor GLD-1 protein (Zhang et al. 1997; Kraemer et al. 1999; Hansen et al. 2004). We therefore tested whether codisruption of fbf-1 and fbf-2 can suppress par-2(it5ts) lethality. We found that the viability of fbf-1(ok91); par-2(it5ts); fbf-2(RNAi) triply disrupted embryos is significantly higher than that of any double-disruption combination at a semirestrictive temperature (Table 3). This indicates that both FBF-1 and FBF-2 can modulate the function of the C. elegans PAR protein pathway.
TABLE 3.
Codisruption of fbf-1 and fbf-2 can suppress par-2(it5ts) lethality
| RNAi clone | Gene | Embryonic viability in par-2(it5ts) (%)a | Embryonic viability in fbf-1(ok91); par-2(it5ts) (%)a |
|---|---|---|---|
| Vector | Negative control | 4.6 ± 3.1 | 12.9 ± 7.3 |
| C32E12.2 | Negative control | 7.3 ± 7.5 | 19.8 ± 4.9 |
| ZK20.5 | rpn-12b | 50.8 ± 11.5 | 64.1 ± 8.5 |
| H12I13.a | fbf-1 | 7.5 ± 4.3 | 30.9 ± 5.8c |
| F21H12.5 | fbf-2 | 5.2 ± 5.2 | 45.5 ± 14.6 |
Suppression was determined by feeding RNAi clones to animals at the L4 stage; see materials and methods for details.
The value corresponds to the average percentage of embryos that hatched over the total number of embryos ± standard deviation over three assays.
RNAi to rpn-12 was used as a positive control in this assay.
This fbf-1 RNAi clone might partially disrupt the function of fbf-2 since these two genes are 93% identical at the nucleic acid level.
nos-3 suppresses the embryonic phenotypes associated with disruption of par-2:
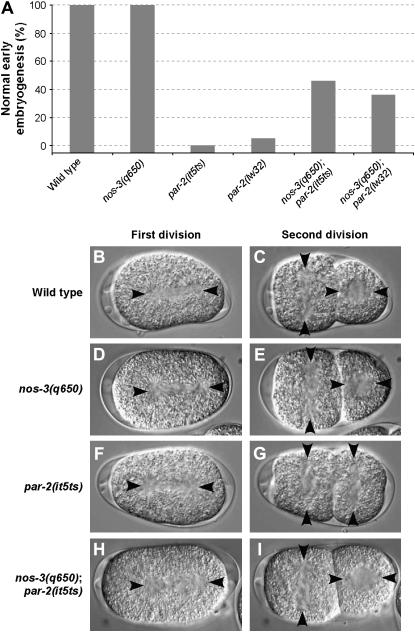
To further understand the mechanism of par-2 suppression, we characterized in more detail the par-2 phenotypes that are suppressed by nos-3, in addition to embryonic lethality. We first monitored early patterns of cell division by generating time-lapse images of early embryonic development and compared the phenotype of nos-3; par-2 double-mutant embryos to that of wild-type, nos-3, and par-2 mutant embryos (Figure 1). In nos-3 mutants, early embryonic development proceeds as in wild type and most embryos develop into adults at all temperatures (Figure 1 and Tables 2 and 4). In one-cell-stage par-2 mutant embryos, the normal posterior displacement of the first mitotic spindle does not occur, resulting in two cells of equal size after cytokinesis (Kemphues and Strome 1997). Furthermore, while blastomeres of wild-type two-cell-stage embryos have perpendicular spindle orientations and asynchronous cytokineses, the two spindles align parallel to each other and the division of both blastomeres is synchronous in par-2 mutant embryos (Kemphues and Strome 1997). We found that all of these par-2 phenotypes are suppressed in embryos double mutant for nos-3 and par-2 (Figure 1 and Table 4). This indicates that nos-3 can efficiently suppress the early cell division phenotypes of par-2 mutants and that the NOS-3 protein can regulate some early embryonic development processes.
Figure 1.—
nos-3(q650) suppresses the early embryonic phenotypes of par-2 mutants. (A) While nos-3(q650) embryos have a wild-type pattern of early development (n = 22), such a pattern is rarely observed in par-2(it5ts) (n = 24) or in par-2(lw32) (n = 19) mutant embryos. This phenotype is suppressed in nos-3(q650); par-2(it5ts) (n = 22) and nos-3(q650); par-2(lw32) (n = 31) double-mutant embryos. (B–I) Images from time-lapse movies of embryos undergoing first (B, D, F, and H) or second (C, E, G, and I) division. Phenotypes were scored in wild-type (B and C), nos-3(q650) (D and E), par-2(it5ts) (F and G), or nos-3(q650); par-2(it5ts) (H and I) embryos at 25°. In B–I, anterior is to the left and arrowheads indicate centrosome positions.
TABLE 4.
Mutations in nos-3 can suppress the early embryonic phenotypes associated with mutations in par-2
| Genotype | Posterior spindle displacement at first mitosis (%) | Perpendicular spindle orientation at second mitosis (%) | Asynchronous cytokinesis at second mitosis (%) | No. recorded |
|---|---|---|---|---|
| Wild type | 100 | 100 | 100 | 20 |
| nos-3(q650) | 100 | 100 | 100 | 22 |
| par-2(it5ts) | 17 | 0 | 4 | 24 |
| par-2(lw32) | 16 | 5 | 16 | 19 |
| nos-3(q650); par-2(it5ts) | 86 | 45 | 73 | 22 |
| nos-3(q650); par-2(lw32) | 48 | 39 | 45 | 31 |
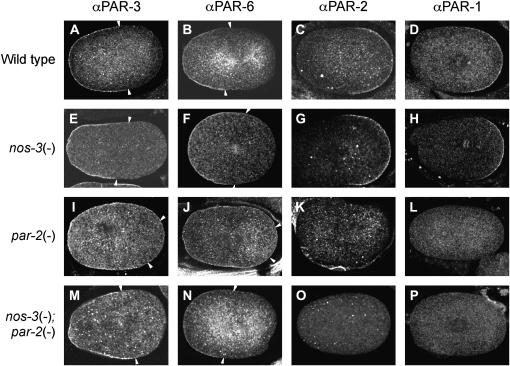
We then used immunofluorescence to monitor the localization of PAR proteins in early embryos. PAR-3 and PAR-6 localize to the anterior cortex in wild-type embryos (Etemad-Moghadam et al. 1995; Hung and Kemphues 1999). In par-2 mutants, polarity cannot be maintained and the anterior PARs expand toward the posterior (Cuenca et al. 2003). We observed that PAR-3 and PAR-6 proteins are restricted to the anterior cortex of most nos-3(q650); par-2(it5ts) double-mutant embryos (Figure 2). Therefore, disruption of nos-3 restores the asymmetric localization pattern of anterior PARs in embryos mutant for par-2.
Figure 2.—
PAR-3 and PAR-6, but not PAR-2 and PAR-1, localization is restored in nos-3(q650); par-2(it5ts) mutants. (A–D) In wild-type embryos, PAR-3 (A) and PAR-6 (B) localize at the anterior cortex while PAR-2 (C) and PAR-1 (D) localize at the posterior cortex. (E–H) In nos-3 mutants, PAR protein localization is as in wild type: 100% of embryos showed anterior PAR-3 (E, n = 32), anterior PAR-6 (F, n = 24), posterior PAR-2 (G, n = 20), and posterior PAR-1 (H, n = 27). (I–L) In par-2(it5ts) mutants, the localization of PAR-3 (I) and PAR-6 (J) is expanded toward the posterior pole (in 90% of embryos, n = 19 for PAR-3 and n = 29 for PAR-6) whereas PAR-2 (K) and PAR-1 (L) are absent from the cortex (in 100% of embryos, n = 15 each). (M–P) In nos-3(q650); par-2(it5ts) mutants, wild-type localization of PAR-3 (M) and PAR-6 (N) is restored [in 68% of embryos for PAR-3 (n = 22) and in 74% of embryos for PAR-6 (n = 19)] whereas PAR-2 (O) and PAR-1 (P) are absent from the cortex [in 100% of embryos for PAR-2 (n = 20) and in 91% of embryos for PAR-1 (n = 22)]. All animals were shifted to 25° at 24 hr before dissection and staining. In A–P, anterior is to the left.
The posterior localization of PAR-2 was not restored by disrupting NOS-3 function: nos-3(q650); par-2(it5ts) double mutants showed no detectable PAR-2 staining at the posterior embryonic cortex (Figure 2). This is consistent with the notion that nos-3 can suppress par-2 lethality independently of PAR-2 function. Taken together, these results indicate that nos-3(q650) can suppress most of the early phenotypes associated with loss of PAR-2 function.
PAR-1 is undetectable at the cortex of nos-3; par-2 mutant embryos:
A surprising result was obtained when we monitored the localization of the protein PAR-1. PAR-1 is a Ser/Thr kinase that is asymmetrically localized at the posterior cortex of wild-type one-cell embryos (Guo and Kemphues 1995). Upon disruption of par-2, PAR-1 cortical localization is lost and PAR-1 is found in the cytoplasm (Boyd et al. 1996). Interestingly, we observed that PAR-1 was undetectable at the cortex of 20 of 22 nos-3(q650); par-2(it5ts) double-mutant embryos at restrictive temperature (Figure 2). A very weak posterior cortical localization of PAR-1 was observed in 2 of 22 embryos. The 54% of nos-3(q650); par-2(it5ts) double-mutant embryos that hatch at restrictive temperature (Table 2) suggests that the asymmetric localization of PAR-1 at the posterior cortex is not essential in these double mutants.
PAR-1 has been shown to be important in stabilizing P granules in the early embryo (Cheeks et al. 2004). P granules consist of large ribonucleoprotein complexes that localize to the posterior end of the embryo and that have been proposed to play a role in germline specification (Kemphues and Strome 1997; Kawasaki et al. 2004). In par-2 mutant embryos, in which PAR-1 is cytoplasmic, P-granule localization is largely normal in one- and two-cell embryos, but becomes abnormal in four-cell-stage embryos (Boyd et al. 1996). Interestingly, we found that P granules are correctly localized in 12 of 18 nos-3(q650); par-2(it5ts) double-mutant, four-cell-stage embryos (Figure 3). Since P granules are required for germline development, we monitored maternal-effect sterility in single- and double-mutant animals at both permissive and restrictive temperatures for par-2(it5ts). As shown in Figure 3, nos-3(q650) can efficiently suppress the maternal-effect sterility of par-2(it5ts) animals at permissive and restrictive temperatures. This suggests that the P granules that are localized at the posterior of nos-3(q650); par-2(it5ts) embryos are functional, despite the observation that PAR-1 is not detectably enriched at the posterior cortex in these double-mutant embryos at the restrictive temperature.
Figure 3.—
Fertility and P-granule localization are restored in nos-3(q650); par-2(it5ts) mutants. (A) Proportion of sterile animals determined for the indicated genotype at 15° and 25°. Maternal-effect sterility was scored in all cases except in par-2 mutant animals at 25° because of embryonic lethality. However, escapers were previously shown to have a fully penetrant sterile phenotype (Watts et al. 1996). (B–E) P-granule localization in wild-type (B), nos-3(q650) (C), par-2(it5ts) (D), and nos-3(q650); par-2(it5ts) (E) four-cell-stage embryos. In every wild-type and nos-3 embryo observed, P granules were restricted to the posterior-most cell (n = 10) whereas they were localized in several cells in all embryos mutant for par-2(it5ts) (n = 10). P granules were restricted to the posterior-most cell in 66% of nos-3(q650); par-2(it5ts) embryos (n = 18). All animals were shifted to 25° at 24 hr before dissection and staining. In B–E, anterior is to the left.
One possibility was that PAR-1 was not functional in these embryos and that its function was compensated by a secondary, redundant activity. To test this, we disrupted par-1 by RNAi in embryos double mutant for nos-3(q650); par-2(it5ts) and found that none of these triply disrupted embryos were viable at restrictive temperature (0% viability, n = 200 embryos counted in three assays). This indicates that PAR-1 is active in nos-3(q650); par-2(it5ts) double mutants and that posterior cortical enrichment of PAR-1 is not required for its essential function(s).
DISCUSSION
In this work, we have described a screen for suppressors of par-2(it5ts) lethality in which we uncovered 13 genes that modulate the function of the PAR polarity pathway. Of these, the three suppressors that we characterized in more detail fall into distinct classes: those that require residual PAR-2 activity to suppress par-2 lethality (rpn-10) and those that can suppress a strong loss-of-function allele of par-2 and could therefore exert their activity independently of PAR-2, perhaps by modulating the function of the anterior PARs (nos-3 and spat-3). We also found that while nos-3 can suppress most of the early embryonic phenotypes of par-2(it5ts), it does not restore cortical asymmetry of PAR-1. PAR-1 is active in these embryos, suggesting that its essential function is independent of its asymmetric enrichment at the posterior cortex. Taken together, these results identify several putative regulators of PAR protein function and further implicate NOS-3 in PAR protein-dependent cell polarity.
Some of the suppressors that we identified could function upstream of anterior PAR signaling. Upstream components could include genes involved in regulating the levels of the anterior PARs or in the activation or the anchoring of anterior PARs at the cell cortex. Both nos-3 and spat-3 could fall in this class: NOS-3 is a translational regulator and SPAT-3 contains a RING-finger domain, which has been implicated in ubiquitin-dependent degradation in other systems (Petroski and Deshaies 2005). Since both suppressors function independently of par-2, they might be involved in regulating protein levels of components of the anterior PARs. Some other suppressors could also function downstream of PAR proteins. For example, disruption of a gene required for transducing the signal of the anterior PARs could result in weaker signaling, and thus in suppression of lethality. It is unclear at the moment whether any of the candidates that we identified fall into this class. Such candidates could conceivably be potential targets for phosphorylation by PKC-3 and might further cooperate with each other to suppress the various phenotypes associated with disruption of par-2. It will be interesting in the future to determine the functional and biochemical relationships between the PAR proteins and the suppressors identified in this work.
Some of the suppressors could also function via par-2 itself. For instance, our results demonstrated that rpn-10 can suppress par-2(it5ts) lethality in double-mutant animals at a semirestrictive temperature, indicating that rpn-10 is a suppressor of par-2. However, embryonic lethality in double mutants was not suppressed at a fully restrictive temperature or when par-2 dsRNA was injected into rpn-10 mutant animals. This suggests that rpn-10 can suppress par-2 lethality only in embryos that have residual PAR-2 levels or activity. RPN-10 is a subunit of the 26S proteasome and could directly regulate the levels of PAR-2 protein. PAR-2 contains a RING-finger domain, which has often been shown to function in E3 ubiquitin ligase complexes (Petroski and Deshaies 2005). Interestingly, this RING-finger domain was recently proposed to function in regulating PAR-2 protein levels in the embryo (Hao et al. 2006). However, while the levels of PAR-2 are decreased in par-2(it5ts) embryos, we could not observe a detectable increase in PAR-2 levels in either rpn-10 mutants or rpn-10; par-2(it5ts) double mutants (data not shown). It is possible that an undetectable increase in PAR-2 levels is sufficient to suppress lethality. Alternatively, RPN-10 may regulate the levels of an upstream or downstream component of the PAR-2 pathway that has yet to be identified.
We find that disruption of nos-3 or fbf-1/fbf-2 suppresses par-2 lethality. Nanos homologs, together with Pumilio homologs, have been best characterized for their role in repressing mRNA translation (Wickens et al. 2002; Wilhelm and Smibert 2005). In C. elegans, for instance, nos-3, fbf-1, and fbf-2 were shown to repress the translation of the fem-3 mRNA at the sperm-to-oocyte transition during gamete production (Zhang et al. 1997; Kraemer et al. 1999). These genes also participate in the regulation of GLD-1 function during meiotic progression (Hansen et al. 2004). While all three C. elegans orthologs of Drosophila Nanos have redundant roles in the germline, we found that nos-1 or nos-2 do not seem to have a role in cell polarity. The protein NOS-3 contains an N-terminal Q/N-rich domain that is not present in the other two C. elegans Nanos proteins. It is possible that this domain confers different functions on NOS-3, perhaps allowing it to modulate the polarity pathway. Many proteins in C. elegans contain a Q/N-rich domain and preliminary analysis indicated that mutations in many of these are unable to suppress par-2(RNAi) lethality. This indicates that proteins bearing this domain do not systematically play a role in cell polarity. Nanos and Pumilio were previously shown to repress the translation of cyclin B mRNA in both Drosophila and Xenopus (Sonoda and Wharton 2001; Nakahata et al. 2003). Interestingly, we found that disruption of cyb-2 can also suppress par-2 lethality. However, given that both nos-3 and cyb-2 suppress par-2 lethality, it is unlikely that NOS-3 acts as a translational repressor of cyb-2 in this pathway.
Strikingly, we observed that PAR-1 enrichment at the posterior cortex is not restored in nos-3(q650); par-2(it5ts) mutant embryos at restrictive temperature. Since the anterior localization of the anterior PARs is normal in these embryos, this indicates that PAR-2 is required to recruit PAR-1 at high levels at the cortex. A high proportion of nos-3(q650); par-2(it5ts) mutant embryos hatch and P-granule localization is largely normal in these embryos. The activity of PAR-1 is required in these double mutants because disrupting PAR-1 function results in embryonic lethality. This indicates that PAR-1 can exert its functions independently of its asymmetric localization at the posterior cortex and that PAR-2 is not required for PAR-1 activity. A possible explanation is that cytoplasmic PAR-1 activity could be asymmetrically regulated, for instance, by the activity of the anterior PARs. In support of this latter hypothesis, previous reports demonstrated that phosphorylation of mammalian PAR-1 by a PKC-3 homolog negatively regulates its activity in vivo (Hurov et al. 2004). Consistent with our findings, Boyd et al. (1996) have shown that P granules are enriched at the posterior of one-cell par-2 mutant embryos where PAR-1 is not cortical, also suggesting that PAR-1 can function independently of its cortical localization.
Finally, it is interesting to note that many of the par-2 suppressors that we have identified have homologs in other species. Since the function of PAR proteins is conserved in many polarized cell types, it will be of interest to investigate whether some of the suppressors identified also participate in cell polarity in other organisms. While Nanos has already been implicated in establishing polarity of the Drosophila embryo, its role in cell polarity had not been described in other systems. Our results suggest that the conservation of Nanos function in cell polarity might be more general than previously appreciated.
Acknowledgments
We are indebted to Matthias Peter for providing us with access to robotic equipment and Jan Riemer for help with Northern blot analysis. We are also grateful to Yves Barral, Damien D'Amours, Patrick Meraldi, and Matthias Peter for comments on the manuscript; Ulrike Margelisch, Astrid Volmer, and Anton Lehmann for technical assistance; as well as the Gotta and Peter labs for helpful advice and stimulating discussions. We also thank the Light Microscopy Center of the Eidgenössische Technische Hochschule for providing state-of-the-art microscopes and Gabor Csucs for technical help with image acquisition. We thank Pierre Gönczy, Ken Kemphues, Shohei Mitani (Japanese National Bioresource Project for C. elegans), the C. elegans Gene Knockout Consortium, and the Caenorhabditis Genetics Center (which is funded by the National Center for Research Resources of the National Institutes of Health) for strains and reagents. This work was supported by postdoctoral fellowships from the European Molecular Biology Organization to J.-C.L., the Human Frontier Science Program to T.M., and the Fondation Pour la Recherche Medicale to A.P., as well as grants from the Swiss National Foundation and from Novartis to M.G.
References
- Betschinger, J., and J. A. Knoblich, 2004. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14: R674–R685. [DOI] [PubMed] [Google Scholar]
- Boyd, L., S. Guo, D. Levitan, D. T. Stinchcomb and K. J. Kemphues, 1996. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122: 3075–3084. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks, R. J., J. C. Canman, W. N. Gabriel, N. Meyer, S. Strome et al., 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14: 851–862. [DOI] [PubMed] [Google Scholar]
- Cox, D. N., S. A. Seyfried, L. Y. Jan and Y. N. Jan, 2001. Bazooka and atypical protein kinase C are required to regulate oocyte differentiation in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 98: 14475–14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden, S. L., D. S. Bernstein, J. L. Bachorik, B. E. Thompson, M. Gallegos et al., 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. [DOI] [PubMed] [Google Scholar]
- Cuenca, A. A., A. Schetter, D. Aceto, K. Kemphues and G. Seydoux, 2003. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development 130: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, A., P. Bello, N. Thierry-Mieg, P. Vaglio, J. Hitti et al., 2001. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, L. G., 1995. Blastomere culture and analysis. Methods Cell Biol. 48: 303–321. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam, B., S. Guo and K. J. Kemphues, 1995. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83: 743–752. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and A. Hall, 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell 106: 489–498. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and A. Hall, 2003. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr. Opin. Cell Biol. 15: 67–72. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Forman, M. S., J. Q. Trojanowski and V. M. Lee, 2004. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 10: 1055–1063. [DOI] [PubMed] [Google Scholar]
- Glickman, M. H., and A. Ciechanover, 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., J. M. Bellanger, M. Kirkham, A. Pozniakowski, K. Baumer et al., 2001. zyg-8, a gene required for spindle positioning in C. elegans, encodes a doublecortin-related kinase that promotes microtubule assembly. Dev. Cell 1: 363–375. [DOI] [PubMed] [Google Scholar]
- Gotta, M., M. C. Abraham and J. Ahringer, 2001. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11: 482–488. [DOI] [PubMed] [Google Scholar]
- Greenstein, D., S. Hird, R. H. Plasterk, Y. Andachi, Y. Kohara et al., 1994. Targeted mutations in the Caenorhabditis elegans POU homeo box gene ceh-18 cause defects in oocyte cell cycle arrest, gonad migration, and epidermal differentiation. Genes Dev. 8: 1935–1948. [DOI] [PubMed] [Google Scholar]
- Guo, S., and K. J. Kemphues, 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. [DOI] [PubMed] [Google Scholar]
- Hansen, D., L. Wilson-Berry, T. Dang and T. Schedl, 2004. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104. [DOI] [PubMed] [Google Scholar]
- Hao, Y., L. Boyd and G. Seydoux, 2006. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell 10: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp, M., C. Schroder, C. Pfeifle, H. Jackle and D. Tautz, 1989. Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature 338: 629–632. [DOI] [PubMed] [Google Scholar]
- Hung, T. J., and K. J. Kemphues, 1999. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126: 127–135. [DOI] [PubMed] [Google Scholar]
- Hurd, T. W., L. Gao, M. H. Roh, I. G. Macara and B. Margolis, 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5: 137–142. [DOI] [PubMed] [Google Scholar]
- Hurov, J. B., J. L. Watkins and H. Piwnica-Worms, 2004. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14: 736–741. [DOI] [PubMed] [Google Scholar]
- Huynh, J. R., M. Petronczki, J. A. Knoblich and D. St Johnston, 2001. Bazooka and PAR-6 are required with PAR-1 for the maintenance of oocyte fate in Drosophila. Curr. Biol. 11: 901–906. [DOI] [PubMed] [Google Scholar]
- Irish, V., R. Lehmann and M. Akam, 1989. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338: 646–648. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kawasaki, I., A. Amiri, Y. Fan, N. Meyer, S. Dunkelbarger et al., 2004. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167: 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues, K., and S. Strome, 1997. Fertilization and establishment of polarity in the embryo, pp. 335–359 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Kraemer, B., S. Crittenden, M. Gallegos, G. Moulder, R. Barstead et al., 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Levitan, D. J., L. Boyd, C. C. Mello, K. J. Kemphues and D. T. Stinchcomb, 1994. par-2, a gene required for blastomere asymmetry in Caenorhabditis elegans, encodes zinc-finger and ATP-binding motifs. Proc. Natl. Acad. Sci. USA 91: 6108–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D., A. S. Edwards, J. P. Fawcett, G. Mbamalu, J. D. Scott et al., 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2: 540–547. [DOI] [PubMed] [Google Scholar]
- Macara, I. G., 2004. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 5: 220–231. [DOI] [PubMed] [Google Scholar]
- Michelitsch, M. D., and J. S. Weissman, 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 97: 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. A., P. J. Ruest, M. Kosinski, S. K. Hanks and D. Greenstein, 2003. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 17: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, S., T. Kotani, K. Mita, T. Kawasaki, Y. Katsu et al., 2003. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120: 865–880. [DOI] [PubMed] [Google Scholar]
- Nance, J., 2005. PAR proteins and the establishment of cell polarity during C. elegans development. BioEssays 27: 126–135. [DOI] [PubMed] [Google Scholar]
- Petroski, M. D., and R. J. Deshaies, 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- Qiu, R. G., A. Abo and G. Steven Martin, 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10: 697–707. [DOI] [PubMed] [Google Scholar]
- Roegiers, F., and Y. N. Jan, 2004. Asymmetric cell division. Curr. Opin. Cell Biol. 16: 195–205. [DOI] [PubMed] [Google Scholar]
- Shi, S. H., L. Y. Jan and Y. N. Jan, 2003. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112: 63–75. [DOI] [PubMed] [Google Scholar]
- Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Sonoda, J., and R. P. Wharton, 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev. 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, K., and G. Seydoux, 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871. [DOI] [PubMed] [Google Scholar]
- Tabuse, Y., Y. Izumi, F. Piano, K. J. Kemphues, J. Miwa et al., 1998. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125: 3607–3614. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., H. Iwasaki, H. Inoue and K. Takahashi, 2002. Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol. Chem. 383: 1263–1266. [DOI] [PubMed] [Google Scholar]
- Watts, J. L., and J. Browse, 2002. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99: 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, J. L., B. Etemad-Moghadam, S. Guo, L. Boyd, B. W. Draper et al., 1996. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122: 3133–3140. [DOI] [PubMed] [Google Scholar]
- Wickens, M., D. S. Bernstein, J. Kimble and R. Parker, 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J. E., and C. A. Smibert, 2005. Mechanisms of translational regulation in Drosophila. Biol. Cell. 97: 235–252. [DOI] [PubMed] [Google Scholar]
- Wilkinson, C. R., K. Ferrell, M. Penney, M. Wallace, W. Dubiel et al., 2000. Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J. Biol. Chem. 275: 15182–15192. [DOI] [PubMed] [Google Scholar]
- Zhang, B., M. Gallegos, A. Puoti, E. Durkin, S. Fields et al., 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]