Abstract
The meiotic drive system on maize abnormal chromosome 10 (Ab10) is contained within a terminal domain of chromatin that extends the long arm of Ab10 to ∼1.3 times the size of normal chromosome 10L. Ab10 type I (Ab10-I) does not recombine with normal chromosome 10 (N10) over an ∼32-cM terminal region of the long arm. Comparative RFLP mapping demonstrates that multiple independent rearrangements are responsible for the current organization of Ab10-I, including a set of nested inversions and at least one long supernumerary segment at the end of the chromosome. Four major meiotic drive functions, i.e., the recombination effect, smd3, 180-bp neocentromere activity, and the distal tip function, all map to the distal supernumerary segment. TR-1-mediated neocentromere activity (the fifth known drive function) is nonessential in the type II variant of Ab10 and maps to a central region that may include a second supernumerary insertion. Both neocentromere activity and the recombination effect behave as dominant gain-of-function mutations, consistent with the view that meiotic drive involves new or alien gene products. These and other data suggest that the Ab10 meiotic drive system was initially acquired from a related species and that a complex haplotype evolved around it.
MEIOTIC drive describes a variety of phenomena that “beat Mendel's rules” and cause the preferential segregation of alleles or haplotypes to the next generation (Lyttle 1991). Unlike gamete competition, where segregation is distorted by alleles that are unfit for gametogenesis or fertilization (Bernasconi et al. 2004), meiotic drive is a dominant, active phenomenon. Meiotic drive genes are better described as ultrafit for gametogenesis—genetic “hackers” that have evolved to manipulate and exploit the mechanics of chromosome transmission. The most complex drive systems tend to affect autosomal chromosomes and have several consistent features: (1) a selfish drive locus, which encodes a trans-acting product that activates segregation distortion; (2) a target locus, which is a cis-acting chromosomal domain that is susceptible to the activity of the drive product; and (3) one to several modifiers that increase the efficiency of meiotic drive. In addition, the genes of a meiotic drive system tend to be held in tight linkage by inversions or structural polymorphisms. If the elements of the system are separated by recombination, drive becomes ineffective or even self-destructive. Linkage disequilibrium is viewed as a necessity for both the origin and maintenance of successful meiotic drive systems (Lyttle 1991).
Most well-studied meiotic drive systems induce gametes that contain wild-type chromosomes to malfunction during some stage of gametogenesis or fertilization (e.g., Segregation Distorter of Drosophila and t-haplotype of mouse; Lyttle 1991; Lyon 2003). Maize abnormal chromosome 10 (Ab10), on the other hand, modifies female meiosis to favor its own transmission (Birchler et al. 2003). The targets of the Ab10 meiotic drive system are condensed heterochromatic regions called knobs (Rhoades and Vilkomerson 1942). Knobs may be found at any of 22 different loci (Kato 1976), but the largest known knob is within the drive system on Ab10. According to Rhoades (1942), Ab10 is preferentially transmitted in several steps. First, recombination must occur between knobbed and non-knobbed chromosomes, producing heteromorphic dyads (Kikudome 1959; Rhoades and Dempsey 1966). Next, Ab10 causes the transformation of knobs into neocentromeres, which move rapidly poleward (Yu et al. 1997) and ultimately pull the knobs to the upper and lower daughter cells of the linear tetrad, excluding the middle two daughter cells. Because the lower megaspore is the only daughter cell that develops into an egg, knobs and the linked drive loci are preferentially transmitted. An interesting aspect of the Ab10 system is that most, if not all, of the other 21 knobs are subject to meiotic drive as well, as long as Ab10 is present. These knobs are thought to have evolved after Ab10 to take advantage of the trans-acting factors supplied by Ab10. Since nearly every chromosome arm has a knob-forming site, Ab10 has presumably had a major impact on Zea evolution (Buckler et al. 1999).
There are three different types of maize knobs: those composed primarily of 180-bp repeats, those composed primarily of TR-1 repeats, and those containing both repeats in separate parts of the knob (Ananiev et al. 1998; Hiatt et al. 2002). Both 180-bp-containing and TR-1-containing knobs behave as neocentromeres, although they are controlled by different trans-acting factors on Ab10 (Hiatt et al. 2002). A mutant of meiotic drive (Smd1) with reduced neocentromere activity has been described (Dawe and Cande 1996). There are also at least three other functions required for efficient meiotic drive that map to Ab10 (Dawe and Hiatt 2004). One of these is the recombination effect, which increases recombination between centromeres and knobs, thus increasing the efficiency of drive. Two other functions are defined only by their phenotypes and map positions. These are the distal tip function (a deletion) and an apparent point mutation known as smd3 (Hiatt and Dawe 2003a). Both mutants abolish the meiotic drive of Ab10 as well as the capacity of Ab10 to mediate meiotic drive at other knobbed chromosomes.
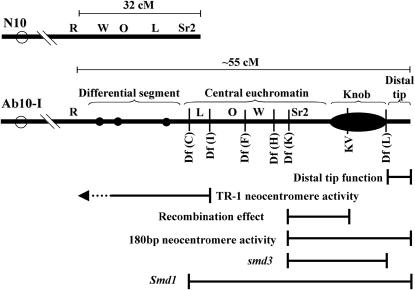
Cytologically, Ab10 type I (Ab10-I) can be distinguished from N10 by a large segment of unique chromatin that extends the long arm of the chromosome by ∼1.3-fold. It has been speculated that at least a portion of this extra chromatin was originally transposed from another genome (Rhoades 1981), although the location of the alien chromatin relative to segments derived from N10 remains unknown. As shown in Figure 1, the unique domain is composed of a differential segment with three small chromomeres, a central euchromatin region containing an inverted portion of N10, a large heterochromatic knob, and a small euchromatic distal tip (Rhoades and Vilkomerson 1942; Rhoades and Dempsey 1985). Each of the five functions involved in meiotic drive—180-bp neocentromere activity, TR-1 neocentromere activity, recombination effect, distal tip function, and smd3—has been mapped to the Ab10-I haplotype with various degrees of precision (Hiatt and Dawe 2003b; Figure 1).
Figure 1.—
Graphical representations of the N10 and Ab10-I chromosomes. The illustration shows the breakpoints of the deficiency chromosomes used or described in this study as well as the mapped meiotic functions (see Hiatt and Dawe 2003b for a more complete description). The genetic lengths are indicated for comparison, but more detail on map distances is provided in Figures 2 and 8. The differential segment (with three chromomeres), central euchromatin, knob, and distal tip are indicated. Letters represent known genes. The segment including the W (W2), O (O7), and L (L13) genes is inverted on Ab10-I.
In an effort to better understand the structure and origin of the Ab10 meiotic drive system, we integrated the molecular and genetic maps of N10 and developed an RFLP map of Ab10-I. We found that Ab10-I contains a series of rearrangements that extend over much of the 32 cM shared with N10. Ab10-I also contains a long supernumerary segment, apparently translocated to the end of N10 several million years ago, which contains the bulk of the meiotic drive system. Neocentromere activity and the recombination effect both behave as dominant gain-of-function mutations, consistent with the view that meiotic drive is an introduced feature of the maize genome.
MATERIALS AND METHODS
Integration of Sr2 with the molecular map:
Two strains, one homozygous for R-Isr (Colored1-Inhibitor of striate1) and Sr2 (striate leaves2; in the B73 background) and another homozygous for r-isr and sr2 (probably in the W22 background), were obtained from the Maize Genetics Cooperation Stock Center (Urbana, IL). The lines were used to produce a segregating F2 population. Only F2 r-isr/r-isr seeds were planted because the Isr locus inhibits the striate leaves phenotype. Tissue was collected from 32 striate (sr2/sr2) and 50 green (Sr2/Sr2 or Sr2/sr2) plants for Southern blot analysis. Linkage analysis was performed using Mapmaker version 3.0 (Lander et al. 1987) using an LOD score of 3.0 and a recombination fraction of 35 Kosambi centimorgans.
RFLP mapping of Ab10-I:
Ab10-I, Df(C), Df(I), Df(F), Df(H), and Df(K) were obtained from Marcus Rhoades (Rhoades and Dempsey 1985) and deficiency Df(L) was identified in the Dawe laboratory (Hiatt and Dawe 2003a). Ab10-I and all deficiencies were backcrossed into a modified (ACr) W23 background at least five times and maintained in the heterozygous condition. For preliminary screening, DNA from homozygous N10 (W23 ACr), Ab10-I/N10, and Df(C)/N10 were analyzed for polymorphisms using six different restriction enzymes and 23 RFLP markers from bins 10.06 and 10.07. Southern analysis was performed using standard protocols (Chomet et al. 1987). Eleven RFLPs showed no detectable polymorphism between N10 and Ab10-I (csu300b, uaz294, asg50d, bnl7.02, csu844, csu615b, isu53b, asg19b, php20568a, bcd135b, and bcd1092a).
PCR and sequencing:
The strains used for PCR were B73 and Mo17 (standard inbreds), lines confirmed to be homozygous for Ab10-I and Ab10-II, and three lines from the North Central Regional Plant Introduction Station in Ames, Iowa: Papago Flour corn (PI 217410), Gourdseed (PI 217405), and Tom Thumb (PI 217412). Primers for amplifying the 3′ UTRs from AY109829 (F-TGACATAGCTCGTCCAACTGTAGC, R-ATGCTGCTATTGGTCTTCTTCTGG) were obtained from http://www.maizegdb.org (see also Wright et al. 2005). We recovered two different AY110016 homologs when the primers designed by Wright et al. (listed on http://maizegdb.org) were used: the original gene that maps to chromosome 10 (AY110016_a) and a second, AY110016_b, which can be found on GenBank as BZ680382 (the map position for AY110016_b is unknown). To simplify the analysis, we used new primers that are specific to AY110016_a (F-CGCTCAATGTGCAGAAAGCCATAGA, R-CCTCCAGCTCCAAATGCTTGCGT). Primers for csu48 (F-CAGTGCATGCATGCGACTCTAA, R-CCTTGATCTGAGGAATTTAGTGCA) were designed to amplify the 3′ UTR. The three markers were amplified and sequenced from all seven maize lines, with the exception of AY110016 from Tom Thumb, which we were unable to recover. PCR products were cloned into pCR4-TOPO (Invitrogen), at least two different clones from each PCR product sequenced, and the consensus sequences determined. The resulting 20 sequences can be viewed in the STS division of GenBank (BV682871–BV682890).
Bridge and fragment assays:
Strains heterozygous for a knobbed form of Inv3a and Ab10-I (marked by the recessive r allele) were crossed as a female to a TB-10L32 strain provided by the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/). The B centromere on TB-10L32 is linked to Rnj, an R allele that expresses pigment in the cap of the endosperm (aluerone) and embryo. The dosage of TB-10L32 was monitored using Rnj. If Rnj was present in both endosperm and embryo, the seed (and resulting plant) was scored as euploid; if Rnj was observed in the endosperm but not the embryo, it was scored as hypoploid; and if the embryo was pigmented but not the endosperm, then the seed was scored as hyperploid. Plants containing Inv3a were processed for the bridge and fragment assay as described previously (Hiatt and Dawe 2003b). Data from euploid, hypoploid, and hyperploid plants were compared pairwise using a Z-test.
Cytological analysis and FISH:
Stocks homozygous for Ab10-I and Ab10-II were imaged in three-dimensional (3D) form at pachytene using a DeltaVision 3D light microscope workstation. The Ab10 haplotype domains were computationally extracted and straightened (Dawe et al. 1994). For visual assays of neocentromere activity, cells were processed for fluorescent in situ hybridization (FISH) to reveal the TR-1 and 180-bp repeats as previously described (Hiatt et al. 2002).
RESULTS
An integrated map of distal 10L:
Given that Ab10L is ∼1.3 times longer than N10L, it is likely that additional chromatin from N10, from another location in the maize genome, or from another species has been added to the chromosome at some time during its evolution. It is not clear, however, whether meiotic drive was introduced with (or evolved from) novel chromatin, since the location of the meiotic drive system relative to DNA derived from N10 has not been established. We set out to answer this question using a set of 12 RFLPs and the genetic markers R and Sr2.
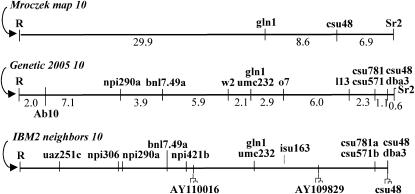
Since the molecular and genetic maps of N10 are poorly integrated, our first step was to map the distal-most genetic marker (Sr2) to two of the most distal RFLP markers (gln1 and csu48). From a mapping population segregating for r and sr2, 82 progeny were scored for RFLPs at gln1 and csu48. As shown in Figure 2, the results placed the four markers in the following order (proximal to distal): R-(29.9 cM)–gln1-(8.6 cM)–csu48-(6.9 cM)–Sr2.
Figure 2.—
Three maps of the region distal to R on N10. Mroczek 10 is from data presented here, Genetic 2005 10 is a consensus map that combines several genetic and RFLP maps, and IBM2 neighbors 10 is the most recently computed genetic map from a population of inbred lines (http://www.maizegdb.org). Actual genetic distances between loci (in centimorgans) are indicated below the Mroczek and Genetic 2005 maps. The position where N10 and Ab10 diverge (Rhoades 1942) is marked 2 cM from R on the Genetic 2005 map; most, if not all, of the 32 cM distal to this position are present on Ab10 in rearranged form. The isu163 locus was added to the IBM2 map on the basis of its tight linkage to rz569a on the CU 99 10 map. The three loci used for sample sequencing are indicated beneath the IBM2 neighbors 10 map (and marked with tree-like symbols). The maps have been normalized to the same length and include only those markers used in the Ab10-I mapping effort.
An integrated map of distal Ab10-I:
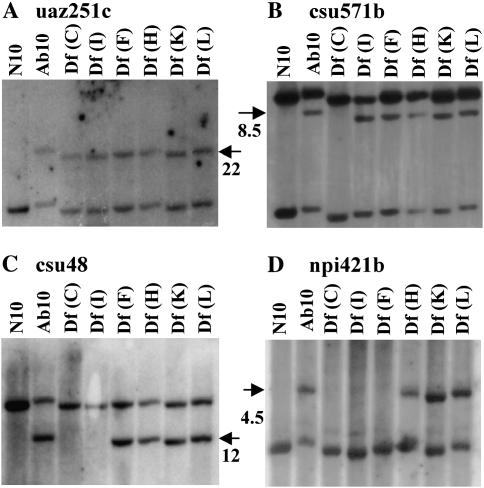
A molecular map of the Ab10-I chromosome was prepared using a series of terminal deficiencies. The breakpoints of Df(C), Df(I), Df(F), Df(H), Df(K), and Df(L) are distributed across the distal approximately one-sixth of the long arm of Ab10 separating genetic markers and dividing functional domains of the meiotic drive system. A subset of the deficiencies was previously used to show that White2 (W2), Opaque7 (O7), and Luteus13 (L13) are inverted on Ab10-I relative to N10 (Rhoades and Dempsey 1985; Figure 1). To orient the same deficiency breakpoints with the molecular map, we chose 12 markers with restriction site polymorphism between N10 and Ab10-I. The RFLPs were then used to determine whether the marker was present or absent on each of the deficiencies (Figure 3). For example, the csu48 probe (Figure 3C) revealed a unique band in deficiencies Df(F), Df(H), Df(K), and Df(L), but not in Df(C) and Df(I), placing it between the Df(I) and Df(F) breakpoints. The map positions of csu48 and the 11 other RFLPs mapped in the same manner are shown in Figure 4.
Figure 3.—
RFLP mapping of Ab10-I. The first lane contains DNA from the (N10/N10) W23 inbred. The remaining lanes contain DNA from plants heterozygous for N10 (W23) and the chromosome indicated. DNA was digested with HindIII (uaz251, csu571b, csu48) or BglII (npi421b). RFLPs specific to Ab10-I and their approximate molecular weights are indicated with arrows. Only portions of the gels are shown. The uaz251, csu571, and npi421 probes hybridize to more than one gene; in these cases we do not know if the nonpolymorphic bands shown are from N10 or one of the homologs elsewhere. (A) The uaz251c probe shows a polymorphic band in Ab10-I and all of the deficiencies. The data indicate that uaz251c maps proximal to the Df(C) breakpoint. (B) An Ab10-I-specific csu571b fragment is absent in Df(C) but present in Df(I). The data show that csu571b maps between these two markers. (C) The csu48 marker maps between the Df(I) and Df(F) breakpoints. (D) The npi421b marker maps between Df(F) and Df(H) breakpoints.
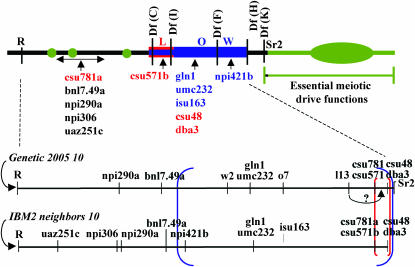
Figure 4.—
The Ab10-I molecular map. The RFLPs are positioned with respect to the deficiencies used in the mapping effort. The comparison of Ab10-I to N10 reveals two nested inversions, the larger in blue and the smaller in red and blue (to indicate that region has been inverted twice). Only a very small segment of the region distal to Df(K) (containing the Sr2 locus) can be confirmed as having been derived from N10; the remaining DNA is supernumerary. Supernumerary DNA is indicated in green. Below the Ab10-I map are the N10 RFLP maps from Figure 2. The boundaries of the inversions are indicated with brackets of the appropriate color. As noted, the position of L13 appears to be incorrect on the Genetic 2005 map, since both L13 and csu571b lie in the Df(C)–Df(I) interval.
At least four conclusions can be drawn from these data. First, we detected no evidence of duplicated or extra bands with the markers used, suggesting that the extended chromatin is not derived from a terminal duplication event(s).
Second, the fact that uaz251c, npi306, npi290a, and bnl7.49a map proximal to the Df(C) breakpoint suggests that the 18- to 24-cM segment they encompass lies within the differential segment. The data do not allow us to determine the order of these markers. However, the fact that recombination between R and the Ab10-I haplotype is limited to 2 cM (Rhoades 1942) strongly suggests the presence of one or more structural polymorphisms.
Third, a comparison of the newly created N10 and Ab10-I molecular maps indicates that the relative order of markers in the central euchromatin cannot be explained by a single inversion event. As shown in Figure 4, the most parsimonious model invokes two inversion events, a large inversion that includes the interval between npi421b and csu48, and a smaller inversion that includes the interval between csu781a and csu48 (such that the csu781a-csu48 region was inverted twice). It is unlikely that the order of csu781a and csu48 is incorrect on the standard genetic map, since csu781a mapped proximal to csu48 in each of the three comprehensive maps where both markers were included (UMC 98, Pioneer composite 1999, and IBM2). Our data do not allow us to determine whether the small or large inversion occurred first.
Fourth, the data reveal that all 12 RFLPs map proximal to the Df(H) breakpoint. The genetic marker Sr2 is the only marker that maps distal to this position and is the distal-most known marker on the long arms of both N10 and Ab10-I (Figures 1 and 2). The portion of Ab10-I distal to Sr2 appears to be supernumerary, i.e., a duplicated region from another portion of the maize genome or an alien segment of DNA derived from an interspecific cross.
Meiotic drive is a dominant gain of function:
If meiotic drive originated from an ancient interspecific cross, we would expect at least some of the genes in the system to behave as gain-of-function mutations, i.e., as neomorphs (“an effect not produced, or at least not produced to an appreciable extent, by the original normal gene”; Muller 1932, p. 246). The number of normal genes or chromosomes present does not generally affect the expressivity of a neomorphic mutation. The phenotype is relatively constant whether the neomorph is heterozygous with a deletion, a single copy of the normal gene/chromosome, or two copies of the normal gene/chromosome. To determine whether Ab10-mediated meiotic drive behaves as a neomorph, we tested the effects of increasing and decreasing the dosage of N10 on neocentromere activity and the recombination effect.
N10 dosage was varied using TB-10L32, a reciprocal translocation that links the B chromosome centromere to the distal half of chromosome 10L. The translocation breakpoint is 3.0-cM proximal to the R locus. The B centromere is known for its propensity to nondisjoin at the second pollen mitosis, causing the two sperm nuclei to differ in their genetic constitution (Carlson 1986). In a cross where a male TB-10L32 heterozygote is crossed to an Ab10-I female, the progeny will be N10/Ab10-I (normal, euploid), N10/N10/Ab10-I (hyperploid), and (−)/Ab10-I (hypoploid).
We developed an assay for quantifying the recombination effect and neocentromere activity in a previous study (Hiatt and Dawe 2003b). When a strain carrying paracentric inversion Inv3a is crossed to strain with a normal (but knobbed) chromosome 3, recombination within the inversion creates a dicentric bridge and an acentric fragment. The recombination effect gene(s) on Ab10-I tightens pairing and increases recombination within the inversion. In addition, although acentric fragments are generally immobile, neocentromere activity draws the knobbed acentric fragment poleward. The knob on Inv3a contains long arrays of both the 180-bp and TR-1 repeats (Hiatt et al. 2002), and neocentromere activity of either alone is probably sufficient to move the fragment.
The data were quantified by scoring the frequency of bridges (i.e., recombination) and the absence of the knobbed acentric fragments at the spindle midzone (i.e., neocentromere activity). The resulting data are shown in Table 1. In the key comparison between hypoploids and hyperploids, there was no statistical difference in the overall levels of recombination and neocentromere activity (P < 0.01). The values from euploids (Ab10-I/N10) were similar to the values obtained in previous experiments with Ab10-I/N10 heterozygotes; recombination effect was slightly higher than expected (52% compared to a previous average of 50%) and neocentromere activity was slightly less than expected (46% compared to a previous average of 49%) (Hiatt and Dawe 2003b). Wild-type (N10 homozygous) plants generally show 40% recombination within Inv3a and a loss of the acentric fragment in <15% of the cells (Hiatt and Dawe 2003b).
TABLE 1.
Effect of Ab10-I dosage on neocentromere activity and recombination effect
| Genotype | Neocentromere activity (%) | Recombination effect (%) |
|---|---|---|
| Hypoploid (−/Ab10-I) | 58.5a ± 7.6, 217b (4) | 44.5c ± 4.6, 593b (4) |
| Euploid (N10/Ab10-I) | 46.4 ± 5.1, 392 (4) | 51.9 ± 3.3, 876 (4) |
| Hyperploid (N10/N10/Ab10-I) | 56.0 ± 7.3, 225 (3) | 47.3 ± 2.4, 582 (3) |
There were no statistically significant differences between hypoploid and hyperploid plants for neocentromere activity or recombination effect. The data from euploid plants were significantly different from both hypoploid and hyperploid plants (P ≤ 0.01).
Percentage of anaphase I cells (± standard deviation) with a bridge but no fragment present, indicating neocentromere activity had moved the knobbed fragment to a pole.
Total number of anaphase I cells. Number in parenthesis indicates number of plants from which meiocytes were harvested.
Percentage of anaphase I cells (± standard deviation) with a bridge and/or fragment, indicating recombination within Inv3a.
Since increasing and decreasing the dosage of N10 had little or no effect on the phenotypes measured, we conclude that the Ab10-I recombination effect and neocentromere activities behave as gain-of-function mutations. We note that our data do not allow us to determine whether 180-bp arrays, TR-1 arrays, or a combination of both were responsible for the neocentromere activity measured in these experiments.
Ab10-II lacks TR-1-mediated neocentromere activity:
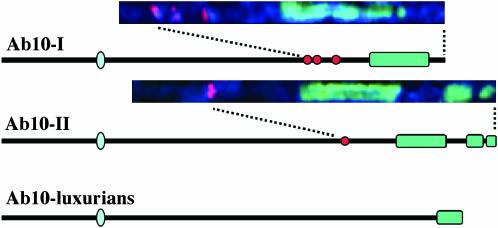
Whereas there is only one known form of N10, there are at least three known variants of Ab10 (Figure 5). Aside from Ab10-I, the best studied of these variants is Ab10-II (Longley 1937; Rhoades and Dempsey 1985). Preliminary cytogenetic analyses suggest that Ab10-II has a similar organization to Ab10-I, at least with respect to the inversion of the L13-O7-W2 region (Rhoades and Dempsey 1988). However, Ab10-II is longer than Ab10-I, and instead of three chromomeres and one large knob, Ab10-II contains one chromomere and two large knobs.
Figure 5.—
Three known variants of Ab10. Line drawings of each chromosome were adapted from previous images (Longley 1937; Rhoades and Dempsey 1985; Kato and Lopez 1990). Actual chromosome images of the haplotype domains are shown in larger view above Ab10-I and Ab10-II. Chromosome images were computationally extracted (and straightened; Dawe et al. 1994) from in situ-hybridized pachytene spreads of strains homozygous for the respective chromosomes. TR-1 repeats are in red and 180-bp repeats in green.
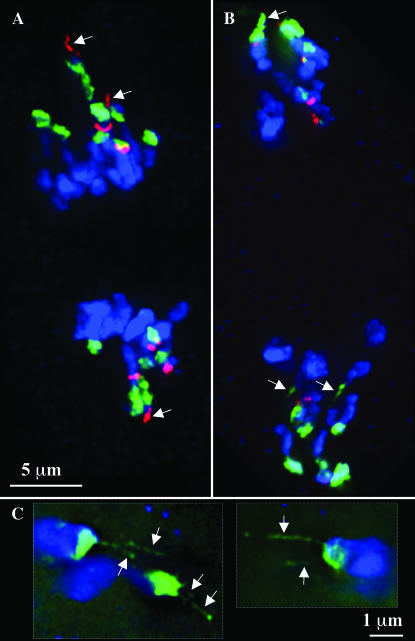
By analyzing pachytene preparations we were able to confirm the early descriptions of Ab10-II and demonstrate that the single chromomere of the differential segment corresponds to a single TR-1-positive domain (Figure 5). Since the differential segment also contains the genes required for TR-1-mediated neocentromere activity (Hiatt et al. 2002), we went on to test whether the reduction in TR-1 corresponds to changes in the neocentromere activity of TR-1 repeats. In Ab10-I strains, TR-1 neocentromere activity is visible as long “leaders” that precede the 180-bp-containing neocentromeres (Figure 6A). In homozygous Ab10-II strains, however, there was no evidence of TR-1-mediated leader activity; no TR-1 neocentromeres were observed at all, despite the fact that TR-1 knobs were present in the strains used (Figure 6B). Numerous leaders were observed in Ab10-II strains but they were always composed of 180-bp arrays. Multiple 180-bp-containing leaders were seen emanating from single knobs when Ab10-II was present (Figure 6C), which is a phenomenon we never observed in Ab10-I lines (but which has been observed in rye; Manzanero and Puertas 2003).
Figure 6.—
Neocentromere leaders in Ab10-I and Ab10-II lines. Each image represents a partial projection from a 3D data set. DNA is shown in blue, the 180-bp repeat in green, and the TR-1 repeat in red. Arrows point to leaders. (A) TR-1-mediated leader activity at anaphase II in a strain homozygous for Ab10-I. (B) 180-bp-mediated leader activity at anaphase II in a strain homozygous for Ab10-II. (C) 180-bp-mediated leaders at prometaphase II in a homozygous Ab10-II line. At this stage multiple leaders were observed emanating from single knobs. Two images from different data sets are shown (outlined with dotted lines).
In a direct test for phenotypic differences between Ab10-I and Ab10-II, we paired them together [Ab10-I (linked to r)/Ab10-II (linked to R)] and test crossed the resulting plants to r/r testers. In this head-to-head comparison the more efficient drive system should be preferentially recovered. In field crosses the R-Ab10-II chromosome was preferentially recovered in 59.8% of the progeny (n = 433 kernels from five ears). The same experiment was repeated in the greenhouse and gave similar results (61.2% R kernels; n = 521 from five ears). These are significant levels of meiotic drive (i.e., different from a 1:1 ratio, P < 0.05). However, the measured drive is also significantly less than what was observed when either Ab10-I/N10 or Ab10-II/N10 was test crossed alone (P < 0.05). We observed 78.9% preferential segregation in Ab10-I/N10 test crosses (n = 1039 kernels from nine ears) and 73.8% in Ab10-II/N10 test crosses (n = 503 kernels from four ears).
Ab10-II lacks visible manifestations of TR-1 motility and yet is more efficient at meiotic drive. These results suggest that TR-1-mediated neocentromere activity is a dispensable feature of the Ab10 meiotic drive system.
Sample sequencing from Ab10:
It is difficult to estimate the age and origin of Ab10 since the major meiotic drive functions lie in a region that lacks known markers [distal to Df(H)]. However, the proximal portion of the haplotype contains numerous known genes. The fact that the known markers lie within a set of inversions (which can limit the spread of alleles; Navarro et al. 1997) suggests that it might be possible to date the proximal half by sample sequencing. Toward this aim, we sequenced 3′ UTRs from three genes within the inversion-containing region of the haplotype: AY110016, AY109829, and csu48 (see Figure 2 for locations of markers). Sequence data from the three loci were obtained from Ab10-I and Ab10-II, the inbreds B73 and Mo17, and the more divergent races Gourdseed, Tom Thumb, and Papago Flour corn (for races see Goodman and Brown 1988). AY109829 and AY110016 had been used as markers previously (Wright et al. 2005, http://www.panzea.org), allowing us to compare our sequences to those from a larger collection of inbreds.
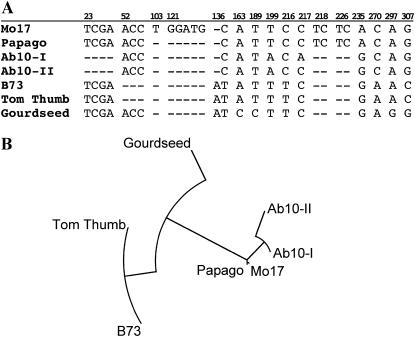
The analyses revealed no significant phylogenetic separation between the N10 and Ab10 chromosomes, at least for the three loci chosen. The terminal marker csu48 proved to have the largest number of polymorphisms, but as shown in Figure 7, existing variation among N10 alleles is sufficient to account for the polymorphisms observed on Ab10.
Figure 7.—
Results of sequencing the 3′ UTR from csu48. (A) Alignment of the sequences obtained here, showing only the polymorphisms. The numbers above indicate the bases relative to the appropriate GenBank entries (BV682883–BV682889). (B) A neighbor-joining tree showing the phylogenetic relationships among the seven sequences.
DISCUSSION
The goal of this study was to better understand the origin of the Ab10 meiotic drive system. Using comparative mapping we show that the meiotic drive system resides in a large haplotype and that the terminal section of the chromosome is supernumerary. Extensive new mapping data, a phenotypic description of the Ab10-II variant, and the results of a dosage series analysis all suggest that the essential features of the meiotic drive system are limited to the terminal supernumerary region.
The Ab10 meiotic drive system resides in a large haplotype:
The demonstration by Rhoades and Dempsey (1985) that the central euchromatin contains a transposed and inverted segment of N10 was an important turning point in our understanding of Ab10. Using the same set deficiencies used by Rhoades and Dempsey (1985), we sought to extend the description of Ab10-I using molecular markers. The intent was to delimit the breakpoints of the inversion and identify the location of the presumed supernumerary chromatin.
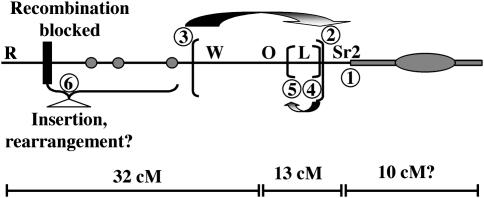
Our molecular analysis suggests that at least six chromosome breakage and reunion events occurred during the evolution of Ab10-I. These are illustrated in Figure 8, where each of the presumed breaks are numbered individually. The fact that the region distal to Df(H) does not hybridize to any of 12 RFLP markers suggests that there is a translocation breakpoint close to Sr2 (labeled 1). We also confirmed the presence of an ∼20-cM inversion extending from npi421b to csu48/dba3, and identified a second nested inversion that includes csu781a and csu48/dba3 (labeled 2–5). The differential segment is largely derived from N10 but it probably contains at least one substantial insertion (labeled 6), since Df(I), which lacks the W2-Sr2 region, is roughly the same length as N10 (Figure 1). Recombination between N10 and Ab10 is effectively blocked 2 cM from R (Rhoades 1942). There may be other rearrangements in the differential segment as well.
Figure 8.—
The positions of six putative chromosome break points that separate Ab10 and N10. The addition of alien chromatin—and the major meiotic drive functions—to the end of chromosome 10L was a key event in Ab10-I evolution (labeled 1). There are at least two separate inversions in the central euchromatin, requiring four chromosomal breakage events (labeled 2–5). The length and polymorphism (with Ab10-II) of the differential segment suggests that it contains an insertion and perhaps other rearrangements (labeled 6). Estimated genetic distances are shown below the cartoon. The distance between R and Df(F) was estimated by Rhoades and Dempsey (1985) to be 32 cM; the Genetic 2005 10 map shows the distance between O7 and Sr2 to be 13 cM, and the supernumerary region cannot be measured, but by cytological comparison appears to be at least 10 cM.
As in other drive systems, we believe that the complex chromosomal rearrangements on Ab10 serve to maintain linkage among the various components required for meiotic drive (Wu and Hammer 1990; Hammer et al. 1991). A crossover that separated the 180-bp neocentromere-activating gene from the Ab10 knob would render both loci useless, since neither can preferentially segregate themselves. Thus, emerging meiotic drive systems benefit from polymorphisms that reduce recombination among the component genes (Lyttle 1991), and inversions are particularly effective in this regard (e.g., Rieseberg 2003). Nested inversions are more effective than single inversions at preventing recombination (Beadle and Sturtevant 1935), and nested inversions with intermingled regions of nonhomology will further deepen the linkage disequilibrium. Given the apparent barriers between the two chromosomes, it is not surprising that no instance of recombination between Ab10 and N10 has been reported.
Visual inspection of the Ab10 haplotype suggests it extends over a remarkably long distance. Estimates of its genetic length are confounded by the facts that Ab10 does not recombine with N10 and that large knobs, such as those on Ab10, tend to impair recombination (Rhoades 1978). Rhoades and Dempsey (1985) were able to minimize these effects by measuring recombination between two deficiency chromosomes [Df(F) and Df(H)], both of which lack the knob. The results suggested that the portion of the haplotype that lies proximal to Df(F) is ∼32 cM (not to be confused with the 32-cM segment of N10 that does not recombine with Ab10, Figure 2). Since the size of the analogous region on N10 is only 24 cM, these data support the idea that there is an insertion within the differential segment. To estimate the length of the segment distal to Df(F), we can use the standard Genetic 2005 map of N10, where O7 and Sr2 are separated by 13 cM (Figure 2). There is no way of measuring the length of the supernumerary region, although visual comparisons to other segments of the chromosome indicate it is at least 10 cM. The available information suggests that 55 cM would be a conservative estimate of the overall size of the Ab10-I haplotype.
The key elements of the drive system are novel:
The results of a mapping strategy based on first integrating the genetic and RFLP maps (Figure 2) and then placing RFLP markers relative to known Ab10-I deficiency breakpoints (Figure 4) indicate that most of the homology to N10 markers is restricted to the proximal side of the Df(H) breakpoint (one exception is the Sr2 gene). Within this proximal region, there is substantial structural polymorphism but apparently little sequence polymorphism. Nearly half of the RFLP markers initially targeted for Ab10 mapping were abandoned due to a lack of polymorphism relative to N10 (see materials and methods for list). In addition, we assayed three loci by sequence analysis and found no evidence that the Df(H)-proximal portion of Ab10 is phylogenetically distinct from N10 (Figure 7). Such sequence similarity may indicate that the inversions on Ab10 occurred recently. A second, perhaps more likely, scenario is that the inversions are relatively old and sufficient time has elapsed that many of the alleles from N10 have been transferred to Ab10 by gene conversion and double crossing over (Chovnick 1973; Navarro et al. 1997).
In contrast, the large region distal to the Df(H) breakpoint has no known homology to N10, is not required for plant growth (Hiatt and Dawe 2003a), and contains all of the primary features of the meiotic drive system. The cis-acting knob smd3 and the genes that control 180-bp neocentromere activity, distal tip function, and recombination effect all map to the distal supernumerary region. We also show that the neocentromere and recombination effect phenotypes are not enhanced or diluted by subtracting or adding doses of N10; i.e., they behave as dominant gain-of-function mutations (Table 1). Taken together, the available data suggest that the key elements of the Ab10-I drive system did not originate in maize, but rather evolved in a different species and were introduced into maize by an interspecific cross.
Classical maize literature provides ample support for the idea that the additional chromatin on Ab10-I lacks homology to the normal maize complement (Rhoades 1952; Snope 1967). In haploids, there is no evidence that Ab10-I shows preferential pairing with any other chromosome: the bivalent frequency in haploid Ab10-I plants is indistinguishable from the bivalent frequency in haploids with normal chromosome 10 (Snope 1967). In trisomic N10/N10/Ab10-I plants the Ab10-I chromosome is present as a univalent 86% of the time and never shows a consistent association with any other maize chromosome (Rhoades 1952). If there were substantial regions of homology between the Ab10-I haplotype and any other portion of the maize genome, at least occasional pairing would be observed. Reciprocal translocations containing much smaller segments of homology than the presumed novel segment of Ab10-I result in cross-shaped pairing figures at pachytene and rings of four at diakinesis (Burnham 1962).
Rhoades (1981) suggested that the terminal segment of Ab10 may have been acquired from a rare cross with Tripsacum. Indeed, Tripsacum is the only extant non-Zea genus that hybridizes with maize (Mangelsdorf and Reeves 1939) and it contains 180-bp knob repeats that are remarkably similar to those in maize (Dennis and Peacock 1984). However, in situ hybridization data suggest that at least three maize-specific retroelement families (Huck, Cinful, and Prem/Ji) are just as abundant on Ab10-I as they are in any other part of the genome (Mroczek and Dawe 2003). These results suggest the system is at least ∼4–6 million years old (SanMiguel et al. 1998), which is prior to the time that maize diverged from Tripsacum (Hilton and Gaut 1998).
Ab10 is structurally and functionally polymorphic:
While meiotic drive evolves selfishly and without regard to host fitness, the host is under selection to maintain normal Mendelian segregation (Ardlie 1998). The result can be an “arms-race”-like conflict that causes remarkable rates of evolution (Pennisi 2003). In strains carrying the Drosophila Segregation Distorter (SD) haplotype, meiotic drive is frequently counterbalanced by host-encoded suppressors of drive, which in turn are subverted by new haplotypes that evade suppression (Hartl 1975; Hartl and Hiraizumi 1976). Numerous structural and functional variants of the SD haplotype have been described (e.g., Palopoli and Wu 1996), most of which are presumed to represent adaptations to different environments and genetic backgrounds. It is very likely that similar forces have shaped the Ab10 meiotic drive system.
Evidence for at least two variants of Ab10 was published in the first comprehensive survey of chromosomal variation in the genus Zea (Longley 1937) and was confirmed in a much larger effort by Kato (1976). The data suggest that Ab10-I exists in ∼2% of maize throughout the Americas as well as ∼10% of the natural populations of annual teosinte (Kato 1976). Ab10-II has never been found in maize naturally (although it has been introduced into maize) and is restricted to annual teosintes in central Mexico (Kato 1976). Kato also described a third form of abnormal 10 in Zea luxurians, a more distant relative of maize (Kato 1976; Kato and Lopez 1990; Buckler and Holtsford 1996). The third form has an arm ratio similar to that of Ab10-I and Ab10-II (i.e., an extra-long 10L), but it has a single large terminal knob (Figure 5). It remains to be seen whether this chromosome demonstrates meiotic drive. It is likely, however, that Ab10–luxurians is a either a progenitor or variant of Ab10-I and Ab10-II, since all other chromosomes in Z. luxurians have arm ratios that very nearly match those in maize.
Our data demonstrate that at least some of the structural variation among Ab10 chromosomes correlates with phenotypic variation. Ab10-II, which has only one of the three TR-1-rich chromomeres found on Ab10-I (Figure 5), also lacks the trans-acting factors that allow TR-1 repeats to become neocentromeres (Figure 6). Nevertheless, genetic tests indicate Ab10-II is a more efficient meiotic drive system than Ab10-I. These results indicate that TR-1 activity is a dispensable feature of the Ab10 haplotype and suggest that the entire TR-1 cassette on Ab10-I (Hiatt et al. 2002) is more accurately described as a modifier, i.e., a gene(s) that facilitates but is not required for meiotic drive. TR-1 may represent an innovation that allowed Ab10-I to out-compete other Ab10 haplotypes or a mechanism to evade a host-encoded suppressor of meiotic drive (Hiatt et al. 2002; Dawe and Hiatt 2004). The data provide an intriguing first glance at the genetic diversity on maize chromosome 10L and highlight the potential for using natural variation to better understand the mechanism of Ab10-mediated meiotic drive.
Acknowledgments
We thank Phil Stinard for advice on the sr2 Isr strain, Amy Bouck for explaining Mapmaker, Andy Paterson for advice on RFLP analysis, and Jon Willis, Chris Topp, and Mike Scanlon for help with Southerns. This research was completed at the University of Georgia under a grant from the National Science Foundation (9513556) to R.K.D. R.J.M. was also supported by an interdisciplinary training grant (BIR9220329).
References
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: Are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95: 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie, K. G., 1998. Putting the brake on drive: meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 14: 189–193. [DOI] [PubMed] [Google Scholar]
- Beadle, G. W., and A. H. Sturtevant, 1935. X chromosome inversions and meiosis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 21: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi, G., T. L. Ashman, T. R. Birkhead, J. D. Bishop, U. Grossniklaus et al., 2004. Evolutionary ecology of the prezygotic stage. Science 303: 971–975. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., R. K. Dawe and J. F. Doebley, 2003. Marcus Rhoades, preferential segregation and meiotic drive. Genetics 164: 835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S. I., T. L. Phelps-Durr, C. S. K. Buckler, R. K. Dawe, J. F. Doebley et al., 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S. T., and T. P. Holtsford, 1996. Zea systematics: ribosomal ITS evidence. Mol. Biol. Evol. 13: 612–622. [DOI] [PubMed] [Google Scholar]
- Burnham, C., 1962. Discussions in Cytogenetics. Burgess Publishing, Minneapolis.
- Carlson, W. R., 1986. The B-chromosome of maize. CRC Crit. Rev. Plant Sci. 3: 201–226. [Google Scholar]
- Chomet, P. S., S. Wessler and S. L. Dellaporta, 1987. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick, A., 1973. Gene conversion and transfer of genetic information within the inverted region of inversion heterozygotes. Genetics 75: 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and W. Z. Cande, 1996. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc. Natl. Acad. Sci. USA 93: 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and E. N. Hiatt, 2004. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 12: 655–669. [DOI] [PubMed] [Google Scholar]
- Dawe, R. K., J. W. Sedat, D. A. Agard and W. Z. Cande, 1994. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76: 901–912. [DOI] [PubMed] [Google Scholar]
- Dennis, E. S., and W. J. Peacock, 1984. Knob heterochromatin homology in maize and its relatives. J. Mol. Evol. 20: 341–350. [DOI] [PubMed] [Google Scholar]
- Goodman, M. M., and W. L. Brown, 1988. Races of corn, pp. 33–75 in Corn and Corn Improvement, edited by G. F. Sprague and J. W. Dudley. American Society of Agronomy, Madison, WI.
- Hammer, M. F., S. Bliss and L. M. Silver, 1991. Genetic exchange across a paracentric inversion of the mouse T-complex. Genetics 128: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. K., 1975. Segregation distortion in natural and artificial populations of Drosophila melanogaster, pp. 83–91 in Gamete Competition in Plants and Animals, edited by D. L. Mulcahy. North-Holland Publishing, Amsterdam.
- Hartl, D. L., and Y. Hiraizumi, 1976. Segregation distortion, pp. 615–666 in The Genetics and Biology of Drosophila, edited by E. Novitski and M. Ashburner. Academic Press, New York.
- Hiatt, E. N., and R. K. Dawe, 2003. a The meiotic drive system on maize abnormal chromosome 10 contains few essential genes. Genetica 117: 67–76. [DOI] [PubMed] [Google Scholar]
- Hiatt, E. N., and R. K. Dawe, 2003. b Four loci on Abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt, E. N., E. K. Kentner and R. K. Dawe, 2002. Independently-regulated neocentromere activity of two classes of satellite sequences in maize. Plant Cell 14: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, H., and B. S. Gaut, 1998. Speciation and domestication in maize and its wild relatives: evidence from the globulin-1 gene. Genetics 150: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y. T. A., 1976. Cytological studies of maize (Zea mays L.) and teosinte (Zea mexicana Shrader Kuntze) in relation to their origin and evolution. Mass. Agric. Exp. Stn. Bull. 635: 1–58. [Google Scholar]
- Kato, Y. T. A., and A. R. Lopez, 1990. Chromosome knobs of the perennial teosintes. Maydica 35: 125–141. [Google Scholar]
- Kikudome, G. Y., 1959. Studies on the phenomenon of preferential segregation in maize. Genetics 44: 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Longley, A. E., 1937. Morphological characters of teosinte chromosomes. J. Agric. Res. 54: 835–862. [Google Scholar]
- Lyon, M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. [DOI] [PubMed] [Google Scholar]
- Lyttle, T., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf, P. C., and R. G. Reeves, 1939. The origin of Indian corn and its relatives. Texas Agric. Exp. Stn. Bull. 574: 1–80. [Google Scholar]
- Manzanero, S., and M. J. Puertas, 2003. Rye terminal neocentromeres: characterization of the underlying DNA and chromatin structure. Chromosoma 111: 408–415. [DOI] [PubMed] [Google Scholar]
- Mroczek, R. J., and R. K. Dawe, 2003. Distribution of retroelements in centromeres and neocentromeres of maize. Genetics 165: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Further studies on the nature and causes of gene mutations, edited by D. F. Jones. Proceedings of the 6th International Congress of Genetics, Ithaca, NY, Vol. I, pp. 213–255.
- Navarro, A., E. Betran, A. Barbadilla and A. Ruiz, 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli, M. F., and C. I. Wu, 1996. Rapid evolution of a coadapted gene complex: evidence from the Segregation Distorter (SD) system of meiotic drive in Drosophila melanogaster. Genetics 143: 1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E., 2003. Meiotic drive—bickering genes shape evolution. Science 301: 1837–1839. [DOI] [PubMed] [Google Scholar]
- Rhoades, M. M., 1942. Preferential segregation in maize. Genetics 27: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M. M., 1952. Preferential segregation in maize, pp. 66–80 in Heterosis, edited by J. W. Gowen. Iowa State College Press, Ames, IA.
- Rhoades, M. M., 1978. Genetic effects of heterochromatin in maize, pp. 642–671 in Maize Breeding and Genetics, edited by D. B. Walden. John Wiley & Sons, New York.
- Rhoades, M. M., 1981. Did abnormal chromosome 10 arise from a translocation between Tripsacum and maize chromosomes? Maize Genet. Coop. News Lett. 55: 13. [Google Scholar]
- Rhoades, M. M., and E. Dempsey, 1966. The effect of abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics 53: 989–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M. M., and E. Dempsey, 1985. Structural heterogeneity of chromosome 10 in races of maize and teosinte, pp. 1–18 in Plant Genetics, edited by M. Freeling. Alan R. Liss, New York.
- Rhoades, M. M., and E. Dempsey, 1988. Structure of K10-II chromosome and comparison with K10-I. Maize Genet. Coop. News Lett. 62: 33. [Google Scholar]
- Rhoades, M. M., and H. Vilkomerson, 1942. On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA 28: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., 2003. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. Gaut, A. Tikhonov, Y. Nakajima and J. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45. [DOI] [PubMed] [Google Scholar]
- Snope, A. J., 1967. On the origin of abnormal 10. Maize Genet. Coop. News Lett. 41: 114–116. [Google Scholar]
- Wright, S. I., I. Vroh Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I., and M. F. Hammer, 1990. Molecular evolution of ultraselfish genes of meiotic drive systems, pp. 177–203 in Evolution at the Molecular Level, edited by R. K. Sclander, T. Whittam and A. Clark. Sinauer, Sunderland, MA.
- Yu, H.-G., E. N. Hiatt, A. Chan, M. Sweeney and R. K. Dawe, 1997. Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]