Abstract
The period gene is a key regulator of biological rhythmicity in Drosophila melanogaster. The central part of the gene encodes a dipeptide Thr-Gly repeat that has been implicated in the evolution of both circadian and ultradian rhythms. We have previously observed that length variation in the repeat follows a latitudinal cline in Europe and North Africa, so we have sought to extend this observation to the southern hemisphere. We observe a parallel cline in Australia for one of the two major length variants and find higher levels of some Thr-Gly length variants, particularly at the tropical latitudes, that are extremely rare in Europe. In addition we examined >40 haplotypes from sub-Saharan Africa and find a very different and far more variable profile of Thr-Gly sequences. Statistical analysis of the periodicity and codon content of the repeat from all three continents reveals a possible mechanism that may explain how the repeat initially arose in the ancestors of the D. melanogaster subgroup of species. Our results further reinforce the view that thermal selection may have contributed to shaping the continental patterns of Thr-Gly variability.
THE period (per) gene in Drosophila controls a number of biological rhythms, the most prominent and best studied being the circadian cycle of behavior (Hall 2003). The PERIOD protein is an autoregulator of its own gene as well as of timeless, and the PER and TIM proteins form the negative limb of the feedback loop that is central to the generation of circadian rhythmicity (Hall 2003). Within PER lies a repetitive region that is composed of alternating dipeptide pairs of threonine and glycine residues (Yu et al. 1987). The Thr-Gly repeat has provided a useful model system for studying the mutational properties of the coding minisatellite that underlies this region, and as observed with repetitive regions in general, the repeat is highly polymorphic, both in sequence and length, within Drosophila melanogaster (Costa et al. 1991). The length mutation rate of the repeat within D. melanogaster has been estimated experimentally and as expected it is several orders of magnitude higher than the nucleotide substitution rate (Rosato et al. 1996, 1997). Not surprisingly, there is enormous interspecific length variation in this sequence between different dipteran species, but only within the drosophilid lineage has a dramatic expansion occurred, with D. pseudoobscura showing a modified pentapeptide repeat motif that has >30 copies compared to D. virilis, which has only a few pairs of the dipeptide (Peixoto et al. 1992, 1993; Nielsen et al. 1994). D. melanogaster is intermediate and encodes between 14 and 24 pairs of uninterrupted Thr-Gly dipeptides (Sawyer et al. 1997).
Functional studies of the Thr-Gly region have revealed that the repeat and its surrounding ∼700 bp encode species-specific patterns of rhythmic behavior between D. melanogaster and its sibling species D. simulans in some elements of circadian locomotor activity patterns and, more dramatically, in the courtship song cycle of the male (Wheeler et al. 1991; Rogers et al. 2004). Furthermore, repeat length variation in natural populations within Europe follows a latitudinal cline, so that high frequencies of the (Thr-Gly)20 and (Thr-Gly)17 alleles are found in northern and southern regions, respectively (Costa et al. 1992). Together these two length alleles make up ∼90% of the natural variation found on this continent, with variants carrying 14 (1%) and 23 (8%) Thr-Gly pairs and very rare variants with 18, 21, and 24 pairs (together accounting for 1%) making up the rest. Linkage disequilibrium pattern analysis of this region has suggested that balancing selection may be operating and this would seem to fit in nicely with the observed clinal distribution (Rosato et al. 1997).
In support of this idea, the temperature compensation of the clock, i.e., how well the ∼24-hr period is buffered against temperature changes, differs among the Thr-Gly variants (Sawyer et al. 1997). (Thr-Gly)17 variants on average show a 24-hr cycle at higher temperatures, but the period becomes shorter as the temperature is reduced. The (Thr-Gly)20 variants, on the other hand, show a period that is not sensitive to temperature change and is on average slightly shorter than 24 hr (Sawyer et al. 1997). Thus the two main variants appear to be adapted to the thermal environments in which they predominate, (Thr-Gly)17 in southern and (Thr-Gly)20 in northern Europe. Furthermore, per transgenes generated with different Thr-Gly encoding lengths also show similar thermal phenotypes to their natural counterparts, indicating that these effects are Thr-Gly length specific and are not caused by genetic background (Sawyer et al. 1997). Finally the 14–17–20–23 Thr-Gly series shows a linear pattern of temperature compensation, whereas the rare variants that are out of phase with this (Thr-Gly)3 interval (i.e., those variants with 15, 18, 21, or 24 Thr-Gly) do not lie on this temperature compensation gradient and generally show poorer compensation (Sawyer et al. 1997). As the conformational monomer from NMR studies is (Thr-Gly)3, which forms a β-turn, this may explain why the (Thr-Gly)3 interval seems to play such a prominent role in the population and behavioral biology of the per variants (Castiglione-Morelli et al. 1995).
The case for balancing selection maintaining the Thr-Gly polymorphism in Europe therefore encompasses many different levels, from the behavioral to the conformational (Costa and Kyriacou 1998). One further classic way of differentiating between natural selection and drift in the generation of a latitudinal cline is to study the polymorphism in a different continent (Oakeshott et al. 1981). D. melanogaster was probably introduced into Australia ∼100 years ago (Bock and Parsons 1981). Nevertheless, D. melanogaster shows a number of latitudinal clines in morphological characteristics such as body size (James et al. 1997) or in frequencies of various metabolic genes (Oakeshott et al. 1981, 1984) on this continent, suggesting that selection, in the face of considerable drift and migration, has already made its mark. While we might expect selection to be stronger on such morphological and biochemical phenotypes compared to more ephemeral behavioral characters, we have nevertheless examined the Thr-Gly length polymorphism on this continent. A positive result would further support and extend the adaptive scenario we have previously described for the European variants.
In addition we have sampled the Thr-Gly region in sub-Saharan African populations, from which D. melanogaster is believed to have evolved (David and Capy 1988), and these results also offer further insights into possible selective pressures that have molded the variability of this repetitive motif. Furthermore, the extensive Thr-Gly sequence variation we encounter in Africa, together with our European and Australian alleles, also allows us to investigate the possible mutational mechanisms that generate these functionally important polymorphisms.
MATERIALS AND METHODS
Australian populations:
Samples from 20 natural populations of D. melanogaster were collected at sites on a 2600-km transect along the eastern coast of Australia during February 1993 and kindly donated by Avis James and Linda Partridge. The isofemale lines from each collection were sampled from 13 latitudes spanning 2600 km. Seven of these latitudes had two independent collections made at different sites (Table 1) (see James et al. 1995 for map of collection sites). The nearest weather stations were noted and used for both latitude and longitudinal coordinates of the fly populations and the relevant recorded temperature data were referenced (Gentilli 1971). The computations for spatial autocorrelation were carried out using the SAAP program developed by David Wartenberg (Version 4.3, October 1989) and the correlations by Statistica (Statsoft).
TABLE 1.
Thr-Gly length variant frequencies in 20 natural Australian populations of D. melanogaster
| Population | Code | °Latitude | °Longitude | N | (Thr-Gly)14 | (Thr-Gly)17 | (Thr-Gly)18 | (Thr-Gly)20 | (Thr-Gly)21 | (Thr-Gly)22 | (Thr-Gly)23 | (Thr-Gly)24 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cairns | MO | 16.88 | 145.75 | 42 | 0.05 | 0.43 | 0.05 | 0.33 | 0.07 | 0 | 0.05 | 0.024 |
| Cairns | INN | 16.88 | 145.75 | 39 | 0.03 | 0.54 | 0 | 0.18 | 0.08 | 0 | 0.18 | 0 |
| Bowen | BRFS | 20.02 | 148.25 | 32 | 0 | 0.38 | 0 | 0.4 | 0 | 0.03 | 0.19 | 0 |
| Bowen | EP | 20.02 | 148.25 | 26 | 0 | 0.48 | 0 | 0.52 | 0 | 0 | 0.04 | 0 |
| Yeppoon | K | 23.13 | 150.75 | 27 | 0.07 | 0.52 | 0.04 | 0.33 | 0 | 0 | 0.04 | 0 |
| Yeppoon | LHF | 23.13 | 150.75 | 29 | 0.03 | 0.28 | 0.1 | 0.28 | 0.14 | 0 | 0.17 | 0 |
| Hervey Bay | AG | 25.55 | 152.68 | 36 | 0 | 0.56 | 0.06 | 0.17 | 0.06 | 0 | 0.17 | 0 |
| Hervey Bay | GL | 25.55 | 152.68 | 36 | 0 | 0.39 | 0 | 0.28 | 0 | 0.03 | 0.31 | 0 |
| S. Brisbane | BH | 27.95 | 153.4 | 30 | 0 | 0.47 | 0.1 | 0.40 | 0 | 0.06 | 0.03 | 0 |
| S. Brisbane | DG | 27.95 | 153.4 | 32 | 0 | 0.41 | 0.06 | 0.28 | 0 | 0 | 0.03 | 0.16 |
| Coff's Harbour | CH | 30.32 | 153.12 | 29 | 0 | 0.52 | 0 | 0.31 | 0.04 | 0 | 0.10 | 0.04 |
| Coff's Harbour | CD | 30.32 | 153.12 | 19 | 0 | 0.58 | 0.05 | 0.32 | 0 | 0 | 0.05 | 0 |
| Taree | CO | 31.9 | 152.48 | 29 | 0.03 | 0.55 | 0.07 | 0.24 | 0 | 0 | 0.10 | 0 |
| Taree | TY | 32.7 | 151.47 | 14 | 0 | 0.29 | 0.07 | 0.57 | 0 | 0 | 0.07 | 0 |
| Cobram | C | 35.82 | 145.57 | 30 | 0.03 | 0.4 | 0.03 | 0.53 | 0 | 0 | 0 | 0 |
| Cobram | PF | 35.82 | 145.57 | 35 | 0 | 0.57 | 0 | 0.37 | 0 | 0 | 0.06 | 0 |
| Melbourne | M | 37.68 | 145.53 | 23 | 0 | 0.35 | 0 | 0.61 | 0 | 0 | 0.04 | 0 |
| Melbourne | H | 38.23 | 145.03 | 33 | 0.09 | 0.42 | 0 | 0.45 | 0.03 | 0 | 0 | 0 |
| Tazmania | F | 41.18 | 146.37 | 21 | 0 | 0.38 | 0.05 | 0.57 | 0 | 0 | 0 | 0 |
| Tazmania | R | 42.88 | 147.33 | 38 | 0 | 0.66 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| Total (N) | — | — | — | 600 | 0.018 (11) | 0.47 (279) | 0.032 (19) | 0.36 (214) | 0.023 (14) | 0.007 (4) | 0.087 (52) | 0.012 (7) |
N is the number of Thr-Gly alleles determined from two males per isofemale line (odd numbers mean that the odd PCR did not work). Population codes were assigned as follows: High Falls Farm (MO), Innisfal Banana Farm (INN), Big Red Fruit Stand (BRFS), El Pedro Car Park (EP), Koppel Farm (K), Lazy Harry's Farm (LHF), Agnus Farm (AG), Goodlife Pools (GL), Brunswick Heads (BH), Dead Goose Farm (DG), Coff's Harbour (CH), Corindi (CD), Coopernook (CO), Tyrells (TY), Cornish Farm (C), Pullars Farm (PF), Chappies Farm (M), Hastings Farm (H), Forth (F), and Ranelagh (R). These populations have been previously described (James et al. 1995), where INN is IN, BRFS is BS, K is KL, LHF is LH, CD is CI, C is CS, M is ME, H is HS, F is FT, and R is RN. For those populations sampled at the same latitude but at different sites, an approximate single longitude value is given for both sites.
African populations:
The D. melanogaster populations used in this study were collected in four different geographical locations within Kenya (Kericho, Matuga, Nairobi, and Nguruman) during October and November 1995 by Stefan Escher and Kerstin Eriksson (European Drosophila Stock Centre, Umea, Sweden) and subdivided into 25 isofemale lines immediately after sampling. We obtained these lines in 1996 and four single males from each isofemale line were crossed to D. melanogaster females carrying an attached-X chromosome. Male progeny from these crosses were frozen and their Thr-Gly regions studied (see below).
European populations:
Flies were also collected from various European localities between 1989 and 1996 and isofemale lines set up immediately after collection. We thank Peter Corish for the flies from North Wooton (Somerset, UK).
Genotyping:
For the Australian populations, gene frequencies were required for the sex-linked per Thr-Gly repeat, so two males from each available isofemale line were selected at random and frozen immediately after receiving the flies in 1994–1995. DNA preparation, PCR, and gel electrophoresis were carried out as described in Costa et al. (1992). Two males were used to sample each isofemale line rather than a single female, because this facilitates Thr-Gly length identification on agarose gels. Markers composed of other known Thr-Gly length variants were used and any band that was not exactly in line with the markers was sequenced directly. The length variants defined were further characterized in a second reaction by coamplification of each DNA, with a previously sequenced DNA from an isolength Thr-Gly allele. Coamplification was made using a 1:1 ratio of the two DNAs. If coamplification produced a heteroduplex this indicated a difference in the DNA sequence of the variant vs. the isolength standard. For amplification, the following primers were used: 5′ primer, 5′-ATACACATGAGCAGTTGTGAC-3′ (5066–5085 in the per sequence published by Citri et al. 1987); 3′ primer, 5′-TTCTCCATCTCGTCGTCGTTGTG-3′ (5336–5355). For the African populations, DNA was prepared from a number of males from each attached-X line, and PCR was used with the 5′ primer, 5′-AACTATAACGAGAACCTGCT-3′ (4874–4893), and 3′ primer, 5′-CCGCGCGACTCCCGGTGCTTCTTC-3′ (5364–5387). All PCR fragments were sequenced directly, as for these populations we were interested in the sequences themselves rather than their frequencies.
For the Thr-Gly sequences described, the codon usage data were calculated using the CountCodon Program in the Codon Usage Database (http://www.kazusa.or.jp/codon/) and converted to the relative synonymous codon usage (RSCU) values (Shields et al. 1988). These sequences were also analyzed for periodicity using the autocorrelation procedure in the STATISTICA 5.0 package (Statsoft).
RESULTS
Australian populations:
Table 1 shows the frequencies of the different Thr-Gly length variants in 20 populations. The most frequent alleles are represented in order by the (Thr-Gly)17, (Thr-Gly)20, and (Thr-Gly)23 variants, making up ∼47, 36, and 9% of the Australian population, respectively. In addition at least one of the other rarer alleles—(Thr-Gly)14 (2%), (Thr-Gly)18 (3%), (Thr-Gly)21 (2%), (Thr-Gly)22 (0.7%), and (Thr-Gly)24 (1%)—was also present in most of the populations. If we consider the two Tasmanian samples, we also observe the greatest intersite variation in both (Thr-Gly)17 and (Thr-Gly)20 frequencies compared to the other samples collected at the same latitude. Island populations are particularly prone to the effects of drift, so this variance in allele frequencies is perhaps not unexpected. Table 2 shows the correlations with latitude for each of the Thr-Gly variant frequencies and reveals that only for (Thr-Gly)20 and (Thr-Gly)23 are they significant. The positive correlation for the (Thr-Gly)20 allele frequency mirrors that found in Europe, so the higher the latitude, the higher the frequency for this variant (Figure 1). The (Thr-Gly)21 frequencies were also significantly correlated with latitude, but as the majority of localities had zero frequencies for this length allele (Table 1) this result is largely meaningless. The (Thr-Gly)17 allele does not show any significant correlations; however, when we averaged the frequencies from the lines collected from the same latitude and removed the two island populations from Tasmania we obtained a correlation of −0.195, which is not significant but is the largest correlation we observed for this allele (Table 2). A much larger, significant negative correlation is observed between gene frequency and latitude for this allele in Europe.
TABLE 2.
Correlations of the frequency of the Thr-Gly variants with respect to latitude and mean yearly wet bulb temperature
| Latitude
|
Temperature
|
|||
|---|---|---|---|---|
| Allele | r | P | r | P |
| (Thr-Gly)14 | −0.10 | 0.670 | 0.10 | 0.673 |
| (Thr-Gly)17 | 0.09 | 0.718 | −0.12 | 0.626 |
| (Thr-Gly)18 | −0.10 | 0.671 | 0.14 | 0.548 |
| (Thr-Gly)20 | 0.47 | 0.036* | −0.54 | 0.014* |
| (Thr-Gly)21 | −0.45 | 0.046* | 0.41 | 0.073 |
| (Thr-Gly)22 | −0.19 | 0.422 | 0.17 | 0.473 |
| (Thr-Gly)23 | −0.54 | 0.015* | −0.44 | 0.053 |
| (Thr-Gly)24 | −0.08 | 0.724 | −0.05 | 0.844 |
r, Pearson product moment correlation. *P < 0.05.
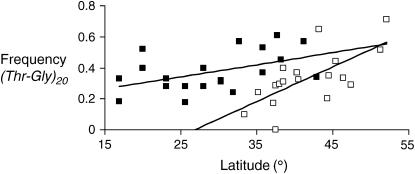
Figure 1.—
A cline in Thr-Gly frequencies in Australia. The frequency of (Thr-Gly)20 vs. latitude is shown for Australia (solid squares) compared to Europe (open squares). The data from Europe are redrawn from the results of Costa et al. (1992).
To further dissect these initial correlations, the frequencies of the (Thr-Gly)17 and the (Thr-Gly)20 allele were compared with the cumulative frequencies of all other alleles across localities using a G-test (Sokal and Rohlf 1995). The (Thr-Gly)23 could not be included in this analysis as a separate allele but was pooled with the rest of the rarer variants as it was not present in all localities. The populations that had been collected at the same latitudes were pooled but the Tasmanian populations excluded because only one fly in the two Tasmanian populations represented an allele other than the two common classes (see Table 1). This pooling and exclusion yielded 33 overall frequency classes that were then compared. The G-test was highly significant (G = 46.18, d.f. 10, P < 0.001), showing that the observed gene frequencies do not represent random samples drawn from a unique, panmictic, or genetically uniform population.
Allele frequency patterns were summarized using a spatial autocorrelation statistic, Moran's I (Sokal and Oden 1978), which represents the degree of similarity between populations as a function of their distance apart. The data were subdivided into six equally informative distance classes, each one containing from 30 to 34 comparisons; class limits are shown in Table 3. Moran's I was then calculated for the (Thr-Gly)17, (Thr-Gly)20, and (Thr-Gly)23 length alleles for each distance class. A cline is present when Moran's I values decrease continuously from significantly positive to significantly negative with an increase in distance between populations (Sokal 1979). A steady decline of Moran's I values was observed for the (Thr-Gly)20 allele from highly significantly positive within ∼350 km to significantly negative at large distances (Table 3). Moran's I for the (Thr-Gly)17 was insignificant for all distance classes (Table 3). The (Thr-Gly)23 variant was observed to have a significantly negative Moran's I for the largest distance class although there was no complementary significant positive value. Additional significance testing of the overall trend for each spatial correlogram was carried out using the highly conservative Bonferroni procedure (Oden 1984). This test, however, was not significant for (Thr-Gly)20 (P = 0.093) or for either the (Thr-Gly)17 or the (Thr-Gly)23 allele (Table 3).
TABLE 3.
Spatial autocorrelation statistics
| Distance classes (km)
|
(Thr-Gly)17
|
(Thr-Gly)20
|
(Thr-Gly)23
|
|||||
|---|---|---|---|---|---|---|---|---|
| Min | Max | N | I | P | I | P | I | P |
| 0.0 | 347.9 | 31 | −0.258 | 0.099 | 0.292 | 0.015* | 0.132 | 0.119 |
| 347.9 | 765.2 | 34 | 0.008 | 0.346 | 0.194 | 0.053 | −0.040 | 0.465 |
| 765.2 | 1065.8 | 30 | 0.032 | 0.297 | −0.128 | 0.319 | 0.153 | 0.093 |
| 1065.8 | 1388.2 | 31 | −0.238 | 0.124 | −0.276 | 0.082 | −0.049 | 0.491 |
| 1388.2 | 1778.3 | 32 | 0.166 | 0.086 | −0.117 | 0.345 | −0.151 | 0.266 |
| 1778.3 | 2898.3 | 32 | −0.037 | 0.455 | −0.298 | 0.039* | −0.344 | 0.017* |
Distance classes used in kilometers, the number of comparisons contained within each class, Moran's I, and its relative significance for the (Thr-Gly)17 (Thr-Gly)20, and (Thr-Gly)23 length alleles, respectively, are shown. *P < 0.05. Bonferroni approximations for correlograms: (Thr-Gly)17, 0.516; (Thr-Gly)20, 0.093; (Thr-Gly)23, 0.099.
The Royaltey–Astrachan–Sokal nonparametric test of departure from random geographic variation (Royaltey et al. 1975) was also applied. The 20 localities were connected by means of a Delaunay graph (Brassel and Reif 1979), which is a triangulation network connecting all neighboring localities together. The frequencies of the different Thr-Gly alleles were ranked among localities. The rank differences between the frequencies of pairs of adjacent localities, called edge lengths, were calculated, as was the expected edge length with a correction for continuity. The deviations from the observed mean edge length from the expected mean were then examined with a t-test. A Student's t of 0.93, with infinite degrees of freedom, was observed for the (Thr-Gly)17 allele frequencies, which agreed with the null hypothesis of random variation of this allele. The (Thr-Gly)20 allele frequencies, when calculated in the same way, gave a t = −2.55, which is significant beyond the 0.02 level with infinite degrees of freedom. The negative t-value shows that departure from randomness results from an underlying clinal pattern of allele frequencies (Royaltey et al. 1975). The frequencies of the (Thr-Gly)23 length allele were also treated in the same way and gave a Student's t of −2.18, which is significant beyond the 0.05 level, again with infinite degrees of freedom.
We next examined the relationship between the expected heterozygosity (based on length allele frequencies) and number of Thr-Gly alleles with latitude. In both cases significant negative correlations were observed (heterozygosity, r = −0.60, P = 0.0026; number of alleles, r = −0.66, P = 0.0008). A significant correlation was also observed between (Thr-Gly)20 frequency and yearly mean wet bulb temperature taken at 3 pm from the nearest field station (Gentilli 1971) (Table 2). While this result should be treated with caution because of multiple testing, the fact that statistical significance with temperature was associated specifically with (Thr-Gly)20 would seem to be more than just coincidence.
African populations:
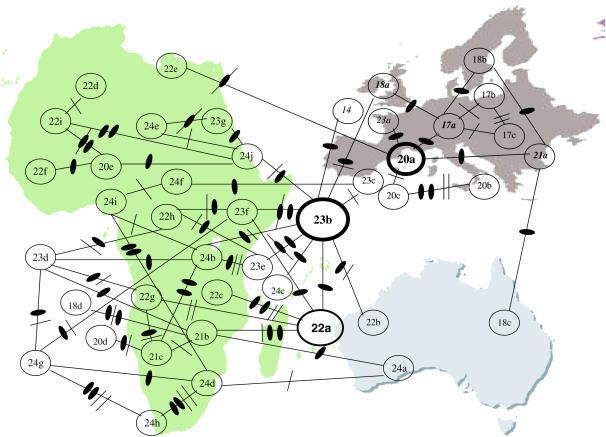
A large number of new length alleles were discovered. Surprisingly, one of the two major length variants in Europe and Australia, (Thr-Gly)17, was not found among the 41 sequences. The shortest length variant observed was the (Thr-Gly)18, with the rest ranging from (Thr-Gly)20 to (Thr-Gly)24. Of particular note is the observation of large numbers of variants that are found very rarely or not at all in Europe, including (Thr-Gly)21, (Thr-Gly)22, and (Thr-Gly)24. Table 4 shows all the Thr-Gly length variants from Europe, Australia, and Africa, using a single letter code that describes each Thr-Gly encoding cassette. From these we generated a Thr-Gly network that is illustrated in Figure 2 and that describes the derivation of each length allele from its closest sequence neighbor (Costa et al. 1991; Peixoto et al. 1992; Rosato et al. 1996). All of the variants can be easily derived from each other by single deletions/duplications of Thr-Gly cassettes, plus the odd nucleotide substitution. This provides an unrooted parsimony network with two alleles, (Thr-Gly)23b and (Thr-Gly)20a, found on all three continents and one allele, (Thr-Gly)22a, found in sub-Saharan Africa and Australia but not in Europe/North Africa. These observations would suggest that these three “cosmopolitan” alleles are ancestral. About half the European/North African alleles (6/13) are also found in Australia, but none of these haplotypes other than the cosmopolitan (Thr-Gly)23b and (Thr-Gly)20a variants (see above) are present in sub-Saharan Africa.
TABLE 4.
Structure of the Thr-Gly repeat
| Repeat | Sequence |
|---|---|
| TG24a | a b c c - - - d d d d d e f d2 - - - - - - - -g c - h d g h d h h h - d1 h |
| TG24b | a b c c c - - - d d d d e f d4 - - - - - - - -g c - h d g h d h h h - d1 h |
| TG24c | a b c c c c - - - d d d e f d - - - - - - - -g c - h d g h d h h h - d1 h |
| TG24d | a b c c - - - d d d d d e f d - - - - - - - -g c - h d g h d h h h - d1 h |
| TG24e | a b c c c2 - - - - d d d e f d - - - - - - - - g c h1 h - g h d h h h - d1 h |
| TG24f | a b c c c - - - d d d d e f d - - - - - - - -g c - h d g h d h h h - d1 h1 |
| TG24g | a b c c - - - - d d d d e f d - - - - - - - - g c - h d g h d h h h h d1 h |
| TG24h | a b c - c2 - - d d d d d e f d4 - - - - - - - g c - h d g - d h h h h - h |
| TG24i | a b c c c - - - d d d d e f d - - - - - - - g c - h d g h d h h h - d1 h |
| TG24j | a b c c c - - - - - d d d e f d - - - - - - - g c h1 h d g h d h h h - d1 h |
| TG23a | a b c c - - - - - - d d d e f d e f d e f d g c - - - - - - - - h h h - d h |
| TG23b | a b c c c - - - - - d d d e f d - - - - - - - g c - h d g h d h h h - d1 h |
| TG23c | a b c c c - - - - d d d e f d - - - - - - - - g c - h d g h d h h h - d1 h1 |
| TG23d | a b c c - - - - d d d d e f d4 - - - - - - - - g c - h d g h d h h h - d1 h |
| TG23e | a b c c c - - -d d d d e f d - - - - - - - - g c - h d g h d h h2 - - d1 h |
| TG23f | a b c c - - - -d d d d3 e f d - - - - - - - - g c - h d g h d h h h - d1 h |
| TG23g | a b c c - - - - - d d d e f d - - - - - - - - g c h1 h d g h d h h h - d1 h |
| TG22a | a b c c - - - - - d d d e f d - - - - - - - - - g c - h d g h d h h h - d1 h |
| TG22b | a b c c c - - - - d d d e f d - - - - - - - - -g c - h d g h d h h2 h - - h |
| TG22c | a b c c - - - - - d d d e f d e f - - - - - - - - - - h d g h d3 h h h - d h |
| TG22d | a b c c c - - - - - d d e f d - - - - - - - - - g c c h d g h3 d h h - d1 h |
| TG22e | a b c c - - - - - d d d e f d e f d - - - - - g c h h - - h d h- - - d1 h |
| TG22f | a b c c c c c - d d d e f d - - - - - - - - - g c h1h d g h d h - - - - - - |
| TG22g | a b c c - - - - - d d d e f d4 - - - - - - - - g c - h d4 g h d h h h - d1 h |
| TG22h | a b c c - - - - d d d d e f d - - - - - - - - g c - h d g h d h h - - d1 h |
| TG22i | a b c c c - - - - - d d e f d - - - - - - - - g c h1h d g h d h h - - -d1h |
| TG21a | a b c c - - - - - d d d e f d e f d - - - - - g c - - - - - - - - -h h h h d h |
| TG21b | a b c c - - - - - - d d e f d2 - - - - - - - - -g c - h d g h d h h - h d1 h |
| TG21c | a b c c - - - - - - d d e f d4 - - - - - - - - -g c – h d g h d h h h - d1 h |
| TG20a | a b c c - - - - - d d d e f d e f d - - - - - g c - - - - - - - - - -h h h d h |
| TG20b | a b c1 c - - - - -d d d e1f d e - d - - - g g c - - - - - - - - - -h h h d1h |
| TG20c | a b c c - - - - - d d d e f d e f d - - - - - g c - - - - - - - - - -h h h d1 h |
| TG20d | a b c c - - - - - - d d e f d4 - - d - - - - - - -c – h d g h d h h - - - d1h |
| TG20e | a b c c c - - - - d d d e f d - - - - - - - - g c h1h d g h - - - - - - - d h |
| TG18a | a b c c c - - - - d d d e f d - - - - - - - - g c - - - - - - - - - - h h h d h |
| TG18b | a b c c - - - - - d d d e f d - - - - - - - - g c - - - - - - - - - h h h h d h |
| TG18c | a b c c - - - - - d d d e f d e f d - - - - -g c - - - - - - - - - - - - - h d h |
| TG18d | a b c c - - - - - d d d e f d4 - - - - - - - -g c - h d - h d h - - - - h |
| TG17a | a b c c - - - - - d d d e f d - - - - - - - - g c - - - - - - - - - - - h h h d h |
| TG17b | a b c c - - - - - d d d e1 f d - - - - - - - -g c - - - - - - - - - - - h h h d h |
| TG17c | a b c c - - - - - d d d1 e f d - - - - - - - -g c - - - - - - - - - - - h h h d h |
| TG14 | a b c c c - - - - - - - - - - - - - - - - - - - - - - - - - d g h d - - h h h d1 h |
Each letter represents a Thr-Gly encoding hexameric cassette. If any of the cassettes have a synonymous substitution they are given a subscript indicating their possible common mode of decent. a, ACGGGC; b, ACTGGT; c, ACAGGT; c1, ACAGGA; c2, ACTGGT; d, ACTGGA; d1, ACTGGC; d2, ACAGGC; d3, ACTGGT; d4, ACTGGG; e, ACCGGG; e1, ACAGGG; f, ACAGGA; g, ACGGGA; h, ACAGGC; h1, ACAGGT; h2, ACGGGC; h3, ACAGGG.
Figure 2.—
Global Thr-Gly allelic network. Thr-Gly alleles are connected by deletion/duplication (solid ovals) and single base changes (slashes). A single nucleotide change within a duplicated/deleted cassette is shown as a slash through an oval. Heavy circles around (Thr-Gly)20a and (Thr-Gly)23b represent alleles that are found on all three continents. The circle (Thr-Gly)22a is found in Australia and Africa. Otherwise all alleles are placed around the continent in which they are found. European alleles also found in Australia are shown in italic type.
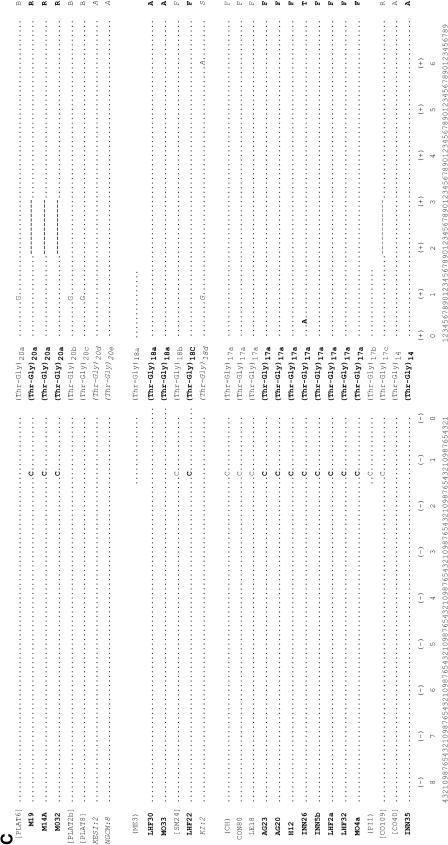
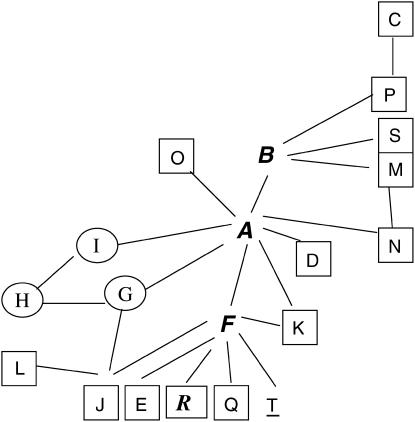
In addition we examined the ∼150 bp flanking each type of Thr-Gly repeat from the three continents and these are shown in Figure 3. There are 10 polymorphic sites (4 replacement, 6 silent), plus one small deletion surrounding these sequences, that give rise to 20 different haplotypes (A to T), of which 5 (G, H, I, R, and T) are found outside Africa. We observe that the (Thr-Gly)22 allelic series is flanked by the highest number of haplotypes, 9, followed by the (Thr-Gly)23, which has 8. However, the (Thr-Gly)23b, (Thr-Gly)22a, and (Thr-Gly)20a repeats represent the specific alleles that are each associated with the highest number of flanking haplotypes, namely 4. Formal phylogenetic analyses of these haplotypes were not informative, given the paucity of informative sites, but we did generate a network for the flanking haplotypes in which we connected all those that differ by a single mutational event (Figure 4). It is clear to see that most haplotypes are African in origin and can be connected to either the African haplotype A or F via a single mutation. In turn, A and F are also connected together by a single event. There are some private European haplotypes, G, H, I, and one exclusive Australian, T.
Figure 3.—
Thr-Gly flanking haplotypes from Europe, Australia, and Africa. DNA sequence of the regions flanking the uninterrupted Thr-Gly repeat of European and Australian (boldface type) and Kenyan (italic type) flies. Dots indicate conserved positions; dashes indicate deletions. The amino acid sequence is shown at the top and the only two amino acid substitutions (S/F and G/R) are indicated. The numbers at the bottom label each nucleotide position. Published sequences are reported in brackets. Note that the BX1, ME3, and PI1 sequences shown here are 3 bp longer, both in the 5′ and the 3′ regions flanking the main repeat, than in Costa et al. (1991); however, the sequences for the entire flanking regions were not available. Unless otherwise stated, the sequences described by Rosato et al. (1996) are shown in parentheses. The sequence prefixed with # is a personal communication (C. Aquadro). AG, Yeppoon (Queensland, Australia); AL, Albaida (Spain); BRFS, Bowen (Queensland, Australia); BX, Bordeaux (France); CH, Chieti (Italy) (Yu et al. 1987); CO, Cognac (France); CON, Conselve (Italy); CS, Canton-S (Jackson et al. 1986); DG, South Brisbane (New South Wales, Australia); GL, Hervey Bay (Queensland, Australia); H, Melbourne (Victoria, Australia); ICIPE, K, L, P, Nairobi (Kenya); INN, Cairns (Queensland, Australia); KESI, Kericho (Kenya); LE, Leiden (Netherlands); LEC, Lecce (Italy); LHF, Yeppoon (Queensland, Australia); M, Melbourne (Victoria, Australia); MA, Matuga (Kenya); ME, Merano (Italy); MO, Cairns (Queensland, Australia); NGMI and NGCM, Nguruman (Kenya); NW, North Wooton (United Kingdom); OR, Oregon-R (Citri et al., 1987); PI, Pietrastornina (Italy); PLAT, Platanistassa (Cyprus); SM, San Mateo (Spain).
Figure 4.—
A network of Thr-Gly allele flanking haplotypes from three continents. The haplotypes from Figure 3 are shown and can be connected in a network by single base changes. African haplotypes are boxed but if also found in Europe and/or Australia they are italic boldface and not boxed. Private European haplotypes are circled and the single private Australian haplotype T is underlined. The boxed Australian haplotype R is also found in Europe.
Mutational mechanisms:
The RSCU (Sharp et al. 1988) values for ACN Thr and GGN Gly codons within eight per Thr-Gly repeat length categories (14, 17, 18, 20, 21, 22, 23, 24) were compared with those of the full-length per cDNA sequence of Oregon-R per (Citri et al. 1987), both with and without the corresponding Oregon (Thr-Gly)20 fragment (Table 5). The ACN Thr and GGN Gly RSCU indices showed elevations of about twofold for ACT and ACA, and GGT, respectively, compared with Oregon-R, although the favored codons within the repeat are ACA for Thr and GGA for Gly. We also compared the codon distributions between two 5′ and 3′ subregions of equal length spanning 16 codons in each sequence—except for (Thr-Gly)14—that was subdivided into two equal portions of 14 codons (Table 6). Along each repeat analyzed, we noted a characteristic distribution of the codon species, with significant higher frequencies of ACT Thr and GGT Gly codons in the 5′ subregion (ANOVA F1,84 = 208, P ≪ 0.001 for ACT Thr; F1,84 = 453, P ≪ 0.001 for GGT Gly) and significant higher frequencies of ACA Thr and GGC Gly codons in the 3′ subregion (F1,84 = 291, P ≪ 0.001 for ACA Thr; F1,84 = 611, P ≪ 0.001 for GGC Gly). Furthermore, alignment of the repeat shows that the first 24 bp in the 5′ region encodes four Thr-Gly cassettes that are the most highly conserved, being identical with the exception of two cases, (Thr-Gly)20b, which shows a T → A transversion at the 18th nucleotide, and (Thr-Gly)24h, which carried an A → T transversion at the 21st nucleotide. In contrast, the 24-bp 3′ subregion reveals one or more modifications for every Thr-Gly encoding cassette.
TABLE 5.
RSCU index for per Thr-Gly alleles
| RSCU ± SD
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Sequence | ACT | ACC | ACA | ACG | GGT | GGC | GGA | GGG |
| TG14 (1) | 1.14 | 0 | 2.28 | 0.57 | 1.14 | 2.00 | 0.85 | 0 |
| TG17 (3) | 1.41 ± 0.00 | 0.15 ± 0.13 | 1.96 ± 0.13 | 0.47 ± 0.00 | 0.94 ± 0.00 | 1.25 ± 0.14 | 1.56 ± 0.13 | 0.23 ± 0.00 |
| TG18 (4) | 1.43 ± 0.11 | 0.28 ± 0.11 | 1.83 ± 0.22 | 0.44 ± 0.00 | 0.94 ± 0.12 | 1.05 ± 0.28 | 1.66 ± 0.22 | 0.33 ± 0.13 |
| TG20 (5) | 1.44 ± 0.09 | 0.28 ± 0.11 | 1.80 ± 0.00 | 0.48 ± 0.11 | 0.84 ± 0.22 | 1.12 ± 0.23 | 1.68 ± 0.18 | 0.36 ± 0.09 |
| TG21 (3) | 1.27 ± 0.11 | 0.26 ± 0.11 | 1.96 ± 0.11 | 0.51 ± 0.11 | 0.76 ± 0.00 | 1.46 ± 0.29 | 1.46 ± 0.22 | 0.32 ± 0.11 |
| TG22 (10) | 1.40 ± 0.12 | 0.22 ± 0.08 | 1.85 ± 0.11 | 0.53 ± 0.10 | 0.89 ± 0.25 | 1.27 ± 0.24 | 1.56 ± 0.15 | 0.27 ± 0.13 |
| TG23 (7) | 1.46 ± 0.09 | 0.23 ± 0.13 | 1.79 ± 0.13 | 0.52 ± 0.10 | 0.84 ± 0.12 | 1.27 ± 0.19 | 1.64 ± 0.14 | 0.25 ± 0.13 |
| TG24 (10) | 1.50 ± 0.11 | 0.16 ± 0.00 | 1.83 ± 0.11 | 0.50 ± 0.00 | 0.83 ± 0.16 | 1.35 ± 0.09 | 1.62 ± 0.11 | 0.19 ± 0.07 |
| pera | 0.54 [15] | 1.24 [34] | 0.84 [23] | 1.38 [38] | 0.46 [17] | 1.94 [71] | 1.12 [41] | 0.46 [17] |
| per ΔTG20b | 0.35 [8] | 1.42 [32] | 0.62 [14] | 1.6 [36] | 0.41[13] | 2.09 [66] | 1.02 [32] | 0.48 [15] |
D. melanogaster Oregon-R per full-length cDNA characterized by a (Thr-Gly)20 (TG20) repeat (Citri et al. 1987) GenBank sequence no. M30114.
The same per cDNA sequence without the TG20 repeat. The number of codons is given in brackets and the number of Thr-Gly haplotypes is given in parentheses.
TABLE 6.
Thr-Gly repeat codon distribution
| % of codons (mean ± SD)
|
||
|---|---|---|
| Codons | 5′ region | 3′ region |
| ACT | 49 ± 9.4 | 25 ± 6.1** |
| ACC | 6 ± 6.3 | 0.3 ± 1.9** |
| ACA | 31 ± 8.9 | 62 ± 7.8** |
| ACG | 13 ± 2.4 | 13 ± 4.7 |
| GGT | 43 ± 8.9 | 5 ± 7.3** |
| GGC | 13 ± 1.9 | 65 ± 13.6** |
| GGA | 38 ± 7.5 | 29 ± 8.0** |
| GGG | 6.5 ± 6.4 | 0.6 ± 2.6** |
**P ≪ 0.0001.
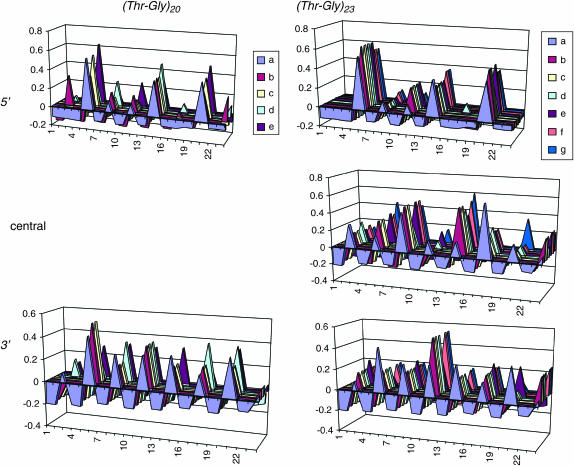
To evaluate the duplication process we examined each Thr-Gly sequence for periodicity, using a modification of a binary code method (Nielsen et al. 1994). For each codon, we gave the value of 0 to the invariant nucleotides in the first and second positions and considered the nucleotide in third position and scored the presence of adenine (the most frequent nucleotide in this position) as 1 and its absence as 0. Each repeat sequence was subdivided into three portions: a 5′ and 3′ subregion of 16 codons each, except for Thr-Gly14, which was subdivided into two subregions of 14 codons each and a variable length central region (from 2 to 16 codons). We then performed an autocorrelation, searching for periodicity in any sequence ≥36 bp long. Figure 5 shows the graphical representations of the autocorrelation coefficients for one short allelic series, (Thr-Gly)20, and one long one, (Thr-Gly)23. For the majority of the shorter repeats (Thr-Gly < 21 pairs), the hexanucleotide subunit gave the largest significant correlation coefficient in the 5′ and also usually in the 3′ subregion. The only exception was the (Thr-Gly)14, in which the 3′ regions showed a strongly significant periodicity corresponding to the dodecanucleotide subunit. In the longer alleles (Thr-Gly > 20), the same 5′ periodicity was observed, but the most significant 3′ periodicity was always the 12-bp subunit, whereas the central region was more heterogeneous in its repeatability.
Figure 5.—
Periodic structure of Thr-Gly repeats. Autocorrelograms for the adenine nucleotide are shown for the 5′ and 3′ region of the (Thr-Gly)20 series of alleles and for the 5′, central, and 3′ regions for the (Thr-Gly)23 allelic series. In the 5′ regions both allelic series show a predominant periodicity corresponding to 6 bp. In the 3′ region of the repeat, the periodicity also peaks at 6 bp for the (Thr-Gly)20 series (P < 0.05), whereas for the (Thr-Gly)23 the periodicity 3′ is 12 bp (all P < 0.05). This difference in pattern is generally observed between the shorter alleles [Thr-Gly < 21, except for (Thr-Gly)14, see discussion] and longer alleles (Thr-Gly > 20). In the central regions of the longer alleles, the autocorrelogram reveals a more heterogeneous set of periodicities. A supplemental file showing these correlograms for all allelic series can be downloaded from http://www.genetics.org/supplemental/.
DISCUSSION
Latitudinal clines are not observed on all continents at allozyme loci thought to be under climatic selection (Bubliy et al. 1999). In Australia, however, we have observed a spatial pattern of distribution for the frequency of the (Thr-Gly)20 allele that is reminiscent of the similar cline observed in Europe with this variant (Costa et al. 1992). This observation provides additional support for the hypothesis that these clines in Thr-Gly repeat length may have been shaped by selection. The decline of spatial autocorrelation with distance for the (Thr-Gly)20 allele and the result of the Royaltey–Astrachan–Sokal test agree in suggesting that this variant is geographically structured along an approximately north–south axis of Australia. Also, the correlations and spatial structure for the (Thr-Gly)23 allele suggest a higher prevalence in the north compared to the south of Australia. Although the (Thr-Gly)17 allele did not show any evidence for a cline, an inverse relationship between allele frequency and latitude was observed when steps were taken to reduce the variance of the data by pooling samples from the same latitude and removing the Tasmanian populations, which seemed particularly prone to drift. Such an inverse but statistically significant relationship is observed in Europe, so perhaps the data for this allele are not so dissimilar between the two continents (Costa et al. 1992). Weeks et al. (2006) have recently published findings suggesting that neither the (Thr-Gly)20 nor (Thr-Gly)17 per alleles show the expected relationship with latitude in samples from eastern Australia collected between 2000 and 2004. While there is some uncertainty associated with the data presented by these authors, it is interesting that a reanalysis of their data reveals a weak positive correlation between latitude and (Thr-Gly)20 frequency and the inverse relationship for the (Thr-Gly)17, as expected from the European studies (C. P. Kyriacou, A. A. Peixoto and R. Costa, unpublished data). Thus the conclusion that the Thr-Gly repeat cline is less robust than in Europe (see below) and that its strength may be modulated by interyearly environmental factors seems to be merited.
In this regard, Figure 1 compares the spatial distributions of the (Thr-Gly)20 allele from our studies on the two continents. We can see clearly that the European (Thr-Gly)20 cline is much steeper than that of Australia. A similar Old/New World relationship has been observed in the clines of wing size in male (but not female) D. subobscura, where again the cline is less steep in the Americas. As with D. melanogaster in Australia, D. subobscura is a species that was recently introduced to North and South America, in this case almost 30 years ago (Gilchrist et al. 2001, 2004; Huey et al. 2000). Australia's recent colonization by D. melanogaster means that perhaps only 90–100 years have elapsed since the introduction of this species (Bock and Parsons 1981). If weak selection is acting on the Thr-Gly repeat, as suggested by a study of linkage disequilibrium patterns around this repetitive region (Rosato et al. 1997), then longer periods of time would be required for the stabilization of any spatial pattern. The Australian colonization is considered to have occurred from Africa, Asia, and Europe (David and Capy 1988), and along the eastern coast the colonization and subsequent migration by humans and their fly commensals contributed to a constant admixture of the founder Thr-Gly variants. It might therefore appear surprising that any cline in Thr-Gly length at all was discovered on this continent, given the relatively short chronological period available for the allelic spatial patterns to establish themselves. However, given the effective population size of D. melanogaster of ∼106 (Aquadro 1992), even a relatively weak selection coefficient could be effective and generate these geographical patterns.
As in northern Europe, we see a higher frequency of (Thr-Gly)20 alleles in the more thermally variable southern regions of Australia, with a hint of the opposite relationship for the (Thr-Gly)17. Furthermore, variants that are out of phase with (Thr-Gly)3 intervals observed for the most common alleles in Europe, the 14–17–20–23 series, are observed at much higher frequencies in Australia: 7.3% compared to <1% in Europe (Table 1). The circadian temperature compensation of these European out-of-phase variants does not follow the linear trend seen for the in-phase variants and they are generally poorer in maintaining 24-hr cycles at varying temperatures (Sawyer et al. 1997). Taken as a whole, the rarer out-of-phase alleles are observed more frequently in the northern tropical Australian latitudes (Table 1). In fact, there are significant negative correlations between Thr-Gly expected heterozygosity and latitude and between number of alleles and latitude. These observations suggest that the polymorphism might tend toward neutrality in the lower tropical latitudes. This might be because selection related to circadian temperature compensation may not be as important in the tropics given the warmer, more constant thermal environments, and therefore more Thr-Gly alleles could be tolerated. Thus even the data from these rare Australian variants provide some further support for Thr-Gly variation playing a role in thermal adaptation (Costa et al. 1992; Ewer et al. 1992; Peixoto et al. 1993, 1998; Sawyer et al. 1997). To this can also be added the significant correlation between mean annual temperature with the frequency of the (Thr-Gly)20 allele across Australia (Table 2). In addition, recent geographical analyses of Thr-Gly length variation from flies in Evolution Canyon in Israel reveal not only that, as expected, the (Thr-Gly)17 is the most frequent allele in southern Europe/Middle East but also that the (Thr-Gly)20 variant is nearly three times as frequent on the colder north-facing slope compared to the south, whereas the opposite is found for the (Thr-Gly)17 variant (Zamorzaeva et al. 2005). These results further cement the emerging relationship between Thr-Gly length variation and temperature.
One important question that needs to be resolved is whether the cline in Thr-Gly variation, even though it is seen in two continents for at least one of the major alleles, is actually driven by selection at another nearby locus. This is a thorny issue that has received some attention, particularly with studies of polymorphic sites around the Adh locus (Berry and Kreitman 1993). If linkage disequilibrium is strong and spans many tens of kilobases, then it is conceivable that clines could be generated in many genes simply by their spatial relationships to the site under selection (Duvernell et al. 2003). With the Thr-Gly region this potential problem is less pervasive because of the relatively rapid length mutation rate that has been observed in the Thr-Gly array and that is at least three orders of magnitude greater than the point mutation rate (Rosato et al. 1997). Consequently, linkage disequilibrium, although present, would be expected to break down much more rapidly with respect to Thr-Gly length alleles (Rosato et al. 1997). In fact this is exactly what we see in that repeatedly the same Thr-Gly allele is found on a variety of different flanking haplotypes.
The African alleles have been extremely informative, and we observe that two Thr-Gly repeats, the (Thr-Gly)23b and the (Thr-Gly)20a, are found on all continents, with (Thr-Gly)22a present in Africa and Australia. It is these three repeats that also have the highest number of flanking haplotypes, suggesting that they might represent the oldest alleles. The (Thr-Gly)23b is particularly important because it connects the vast majority of the African repeats to the European network as seen in Figure 2. Indeed, within Europe, each European and North African repeat can be derived from the (Thr-Gly)23b by simple deletions and duplications (Costa et al. 1991). As the (Thr-Gly)23b and (Thr-Gly)22a differ by only a single deletion/insertion event, we can see how these two variants could be considered to be the ancestral alleles from which all the others are derived. However, one puzzle is that the (Thr-Gly)20a is derived from the (Thr-Gly)23b most parsimoniously via a single deletion involving five Thr-Gly pairs from the (Thr-Gly)23b and a base change to create the (Thr-Gly)18a, then by deletion of a single Thr-Gly encoding hexamer to generate the (Thr-Gly)17a, followed by a duplication of the efd cassettes to create the (Thr-Gly)20a (see Costa et al. 1991 and Table 4). Thus if (Thr-Gly)20a is an older allele, why are not the likely older intermediates by which it arose, (Thr-Gly)18a and (Thr-Gly)17a, also found in Africa, particularly given that the (Thr-Gly)17 series is found at an average frequency of 45% in Europe and North Africa? One could be tempted to speculate that the (Thr-Gly)17 length alleles represent a relatively new series of mutations that arose after the last ice age, when flies colonized Europe. This view would be supported by the observation that (Thr-Gly)17a has two flanking haplotypes, F and T, compared to (Thr-Gly)20a, which has four, B, F, Q, and R. Thus our tentative conclusion is that the (Thr-Gly)20a was derived in Africa via the 23b-18a-17a-20a mutational pathway. The (Thr-Gly)17a was subsequently lost in Africa, but the (Thr-Gly)20a migrated to Europe, from which a back mutation to (Thr-Gly)17a was readily generated by a single deletion involving efd, from which a single base change independently generated the (Thr-Gly)17b/c repeats. This scenario might explain the smaller number of flanking haplotypes for the latter alleles. Thermal selection on the circadian clock would then act to favor the newly derived (Thr-Gly)17a allele, particularly in southern Europe (Sawyer et al. 1997; Peixoto et al. 1998).
The F flanking sequence predominates in both Europe and Australia for the two major Thr-Gly length variants (20 and 17) but in Africa is only observed in association with a (Thr-Gly)22 repeats, suggesting a founder-flush event during the colonization of Europe and the New World. The other major European/Australian flanking region is G, which is associated with the (Thr-Gly)23b in Europe and Australia but is not observed in Africa, but this is most likely derived by a single base difference from the flanking African haplotypes A and J. The general lack of variability in both the repeat itself and its flanking regions in Europe and the New World is to be expected, as ancestral population should retain more variability. However, this effect will be amplified in the seasonal environments of Europe and mid- to southern Australia, which will further impose thermal selection on the repeat and it associated regions by reducing variation (Sawyer et al. 1997; Peixoto et al. 1998). Such thermal selection would be expected to be weak at best in sub-Saharan Africa and tropical Australia, where Thr-Gly length variability should be, and indeed appears to be, more easily tolerated.
We also took the opportunity to use the large sample of repeat sequences to study the mutational mechanisms that generate the length variability. Duplications of the Thr-Gly encoding hexanucleotide appear to generate the 5′ part of the repeat, but the dodecanucleotide encoding two repeat cassettes appears to be the unit of duplication in the 3′ region in the longer Thr-Gly allelic series. This also included the shortest allele, (Thr-Gly)14, but as this is most likely derived by a simple nine-cassette deletion from the central region of the (Thr-Gly)23b we can appreciate why its repeatability profile is similar to that of the longer alleles. The shorter alleles generally have a different pattern of repeatability in the 3′ region because compared to the (Thr-Gly)23b, a large five-cassette deletion has probably occurred in the 3′ region to generate the (Thr-Gly)18a from which all the other shorter European alleles are derived [save for the African (Thr-Gly)18d] (see Table 4, Figure 2). Thus from the ancestral (Thr-Gly)23b allele we can account, although somewhat speculatively, for the types of deletions that may have originally given rise to the repeat (see below).
A rather different pattern of duplication has been observed in the more extensive pentapeptide repeat of D. pseudoobscura per, where 30-bp duplications in the 5′ region are replaced by 15-bp duplications in the central and 3′ region (Nielsen et al. 1994). The 5′ region of the D. melanogaster repeat is the most highly conserved, with the 5′ cassettes abcc almost invariant within D. melanogaster and also observed in nearly all per alleles examined so far within the melanogaster complex of species, which also includes D. simulans, D. mauritiana, and D. sechellia (Peixoto et al. 1992; Rosato et al. 1996). While it would be tempting to conclude that these cassettes may represent the oldest repeats, with newer duplications of these cassettes with their single nucleotide variations being generated in the 3′ direction, the fact that these are largely invariant could also suggest that they are the newest repeats and not enough time has elapsed since species divergence for them to accrue any mutations. On the basis of this argument, the 3′ variable repeats would represent the oldest sequences, as suggested for the per repeat in D. pseudoobscura (Nielsen et al. 1994), so that in the common ancestors of the melanogaster complex the length of the repeat extended from the 3′ to 5′ direction. Comparison with the species of the melanogaster subgroup—D. yakuba, D. orena, D. erecta, and D. teissieri, which are more distantly related to the melanogaster complex (Ko et al. 2003)—reveals that the dgh-type repeats are also generally found in the 3′ region but that the 5′ region has only the a cassette in D. erecta, D. teissieri, and D. orena, whereas D. yakuba contains no 5′ abc-type cassettes at all (Peixoto et al. 1992). Thus these data would also support the view that abc cassettes may be the most recent and that in the ancestors of the subgroup the repeat extended in a 3′ to 5′ direction. Under this scenario, repeat expansion may have originally relied more on two-cassette, 12-bp duplications, but with the more recent sequences added by one-cassette duplications in the 5′ region. The central regions that show more evidence of turnover (see Table 4) would also represent the more unpredictable double/single cassette duplications/deletions. Similar scrambling has been observed with the D. pseudoobscura pentapeptide cassettes in the central part of the repeat (Nielsen et al. 1994).
In spite of the rapid evolution of the repeat the preferred use of Thr ACT and ACA codons within this region matched the most abundant isoacceptor tRNA types in D. melanogaster (8 AGT and 6 TGT tRNA isoacceptors; genomic tRNA database, http://lowelab.ucsc.edu/GtRNAdb/)(Lowe and Eddy 1997), yet was very different from the codon usage for the rest of the per gene. Similarly for Gly the most commonly used codons were consistent with the relative abundance of the tRNA species in D. melanogaster: the major tRNA class is GCC (14 GCC tRNA isoacceptors that optimally bind both GGC and GGT) (Moriyama and Powell 1997) followed by TCC (6 tRNA isoacceptor types), which binds GGA (Lowe and Eddy 1997). Given the stochastic nature of duplications/deletions in the repetitive region, it might be argued that no such coherent overall pattern of optimal codon use would be expected. If duplication did indeed initiate from ancestral 3′ cassettes that happened to be dh-like (ACTGGA, ACAGGC), then the derived codons would be similar, thereby explaining the Thr codon preferences in the repeat. This scenario would also explain why GGA and GGC are the most frequent codons in the 3′ and central region of the repeat. The fact that these and the Thr codons are consistent with the major tRNA isoacceptors would be accidental. In support of this view, the ACN and GGN RSCU values were not high enough to resemble those of typical highly expressed genes in D. melanogaster (Sharp et al. 1988) so it is difficult to argue that any kind of selection might be acting on codon identity. Thus the Thr-Gly codon profile may reflect the ancestral expansion of the repeat 3′ to 5′.
In conclusion, finding that the frequency of a major Thr-Gly variant shows a latitudinal cline in Australia that is parallel to that found in Europe provides further evidence that natural selection is operating to maintain this polymorphism. Although allele frequency gradients on such large geographical scales can be generated by other processes, such as range expansion, a classical explanation for a latitudinal cline is an adaptive response to climatic variation. Climate-related selection is thought to be responsible for latitudinal clines at other loci in D. melanogaster (Oakeshott et al. 1981, 1984; Berry and Kreitman 1993) as well as in phenotypic traits such as thorax length and wing area (James et al. 1995, 1997). The correlation observed with the frequency of (Thr-Gly)20 alleles in Australia and temperature suggests that this cline is also temperature related and that selection, in addition to historical factors in Thr-Gly allele evolution (Rosato et al. 1996), has produced these continental patterns of polymorphism.
Acknowledgments
C.P.K. and R.C. acknowledge grants from EC-Biotechnology programme ERB-B104-CT960096 and the 6th Framework Project EUCLOCK (No. 018741) and from the British Council-Ministero dell'Università e della Ricerca Scientifica e Tecnologica. C.P.K. also acknowledges a grant from the National Environmental Research Council and a Royal Society Wolfson Research Merit Fellowship. R.C. also acknowledges grants from the Agenzia Spaziale Italiana and Ministero dell'Università e della Ricerca. A.A.P. was funded by a Brazilian Conselho Nacional de Desenvolvimento Cientifico e Tecnològico fellowship and the Howard Hughes Medical Institute and CP by an Erasmus studentship
References
- Aquadro, C. F., 1992. Why is the genome variable? Insights from Drosophila. Trends Genet. 8: 355–362. [DOI] [PubMed] [Google Scholar]
- Berry, A., and M. Kreitman, 1993. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134: 869–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, I. R., and P. A. Parsons, 1981. Species of Australia and New Zealand, pp. 291–308 in Genetics and Biology of Drosophila, edited by M. Ashburner, H. L. Carsons and J. N. Thompson. Academic Press, London.
- Brassel, K., and D. Reif, 1979. A procedure to generate Theissen polygons. Geogr. Anal. 11: 289–303. [Google Scholar]
- Bubliy, O. A., B. A. Kalabushkin and A. G. Imasheva, 1999. Geographic variation of six allozyme loci in Drosophila melanogaster: an analysis of data from different continents. Hereditas 130: 25–32. [DOI] [PubMed] [Google Scholar]
- Castiglione-Morelli, M. A., V. Guantieri, V. Villani, C. P. Kyriacou, R. Costa et al., 1995. Conformational study of the Thr-Gly repeat in the Drosophila clock protein, PERIOD. Proc. R. Soc. Lond. B Biol. Sci. 260: 155–163. [DOI] [PubMed] [Google Scholar]
- Citri, Y., H. V. Colot, A. C. Jacquier, Q. Yu, J. C. Hall et al., 1987. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326: 42–47. [DOI] [PubMed] [Google Scholar]
- Costa, R., and C. P. Kyriacou, 1998. Functional and evolutionary implications of natural variation in clock genes. Curr. Opin. Neurobiol. 8: 659–664. [DOI] [PubMed] [Google Scholar]
- Costa, R., A. A. Peixoto, J. R. Thackeray, R. Dalgleish and C. P. Kyriacou, 1991. Length polymorphism in the threonine-glycine-encoding repeat region of the period gene in Drosophila. J. Mol. Evol. 32: 238–246. [DOI] [PubMed] [Google Scholar]
- Costa, R., A. A. Peixoto, G. Barbujani and C. P. Kyriacou, 1992. A latitudinal cline in a Drosophila clock gene. Proc. R. Soc. Lond. B Biol. Sci. 250: 43–49. [DOI] [PubMed] [Google Scholar]
- David, J. R., and P. Capy, 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Duvernell, D., P. Schmidt and W. Eanes, 2003. Clines and adaptive evolution in the methuselah gene region in Drosophila melanogaster. Mol. Ecol. 12: 1277–1285. [DOI] [PubMed] [Google Scholar]
- Ewer, J., B. Frisch, M. J. Hamblen-Coyle, M. Rosbash and J. C. Hall, 1992. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J. Neurosci. 12: 3321–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentilli, J., 1971. Climates of Australia and New Zealand, in World Survey in Climatology, Vol. 13, edited by J. Gentilli. Elsevier, Amsterdam.
- Gilchrist, G. W., R. B. Huey and L. Serra, 2001. Rapid evolution of wing size clines in Drosophila subobscura. Genetica 112–113: 273–286. [PubMed] [Google Scholar]
- Gilchrist, G. W., R. B. Huey, J. Balanya, M. Pascual and L. Serra, 2004. A time series of evolution in action: a latitudinal cline in wing size in South American Drosophila subobscura. Evolution Int. J. Org. Evolution 58: 768–780. [DOI] [PubMed] [Google Scholar]
- Hall, J. C., 2003. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv. Genet. 48: 1–280. [DOI] [PubMed] [Google Scholar]
- Huey, R. B., G. W. Gilchrist, M. L. Carlson, D. Berrigan and L. Serra, 2000. Rapid evolution of a geographic cline in size in an introduced fly. Science 287: 308–309. [DOI] [PubMed] [Google Scholar]
- Jackson, F. R., T. A. Bargiello, S. H. Yun and M. W. Young, 1986. Product of per locus of Drosophila shares homology with proteoglycans. Nature 320: 185–188. [DOI] [PubMed] [Google Scholar]
- James, A. C., R. B. Azevedo and L. Partridge, 1995. Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics 140: 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, A. C., R. B. R. Azevedo and L. Partridge, 1997. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics 146: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W. Y., R. M. David and H. Akashi, 2003. Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol. 57: 562–573. [DOI] [PubMed] [Google Scholar]
- Lowe, T. M., and S. R. Eddy, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1997. Synonymous substitution rates in Drosophila: mitochondrial versus nuclear genes. J. Mol. Evol. 45: 378–391. [DOI] [PubMed] [Google Scholar]
- Nielsen, J., A. A. Peixoto, A. Piccin, R. Costa, C. P. Kyriacou et al., 1994. Big flies, small repeats: the “Thr-Gly” region of the period gene in Diptera. Mol. Biol. Evol. 11: 839–853. [DOI] [PubMed] [Google Scholar]
- Oakeshott, J. G., G. K. Chambers, J. B. Gibson and D. A. Willcocks, 1981. Latitudinal relationships of esterase-6 and phosphoglucomutase gene frequencies in Drosophila melanogaster. Heredity 47: 385–396. [DOI] [PubMed] [Google Scholar]
- Oakeshott, J. G., S. W. Mckechnie and G. K. Chambers, 1984. Population-genetics of the metabolically related Adh, Gpdh and Tpi polymorphisms in Drosophila-melanogaster.1. Geographic-variation in Gpdh and Tpi allele frequencies in different continents. Genetica 63: 21–29. [Google Scholar]
- Oden, N., 1984. Assessing the significance of a spatial autocorrelogram. Geogr. Anal. 16: 1–16. [Google Scholar]
- Peixoto, A. A., R. Costa, D. A. Wheeler, J. C. Hall and C. P. Kyriacou, 1992. Evolution of the threonine-glycine repeat region of the period gene in the melanogaster species subgroup of Drosophila. J. Mol. Evol. 35: 411–419. [DOI] [PubMed] [Google Scholar]
- Peixoto, A. A., S. Campesan, R. Costa and C. P. Kyriacou, 1993. Molecular evolution of a repetitive region within the per gene of Drosophila. Mol. Biol. Evol. 10: 127–139. [DOI] [PubMed] [Google Scholar]
- Peixoto, A. A., J. M. Hennessy, I. Townson, G. Hasan, M. Rosbash et al., 1998. Molecular coevolution within a Drosophila clock gene. Proc. Natl. Acad. Sci. USA 95: 4475–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, A. S., E. Rosato, R. Costa and C. P. Kyriacou, 2004. Molecular analysis of circadian clocks in Drosophila simulans. Genetica 120: 213–222. [DOI] [PubMed] [Google Scholar]
- Rosato, E., A. A. Peixoto, A. Gallippi, C. P. Kyriacou and R. Costa, 1996. Mutational mechanisms, phylogeny, and evolution of a repetitive region within a clock gene of Drosophila melanogaster. J. Mol. Evol. 42: 392–408. [DOI] [PubMed] [Google Scholar]
- Rosato, E., A. A. Peixoto, R. Costa and C. P. Kyriacou, 1997. Linkage disequilibrium, mutational analysis and natural selection in the repetitive region of the clock gene, period, in Drosophila melanogaster. Genet. Res. 69: 89–99. [DOI] [PubMed] [Google Scholar]
- Royaltey, H., E. Astrachan and R. Sokal, 1975. Tests for patterns of geographical variation. Geogr. Anal. 8: 369–395. [Google Scholar]
- Sawyer, L. A., J. M. Hennessy, A. A. Peixoto, E. Rosato, H. Parkinson et al., 1997. Natural variation in a Drosophila clock gene and temperature compensation. Science 278: 2117–2120. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., E. Cowe, D. G. Higgins, D. C. Shields, K. H. Wolfe et al., 1988. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens: a review of the considerable within-species diversity. Nucleic Acids Res. 16: 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, D. C., P. M. Sharp, D. G. Higgins and F. Wright, 1988. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol. Biol. Evol. 5: 704–716. [DOI] [PubMed] [Google Scholar]
- Sokal, R., 1979. Ecological parameters inferred from spatial autocorrelograms, pp. 167–196 in Contemporary Quantitative Ecology and Related Econometrics, edited by G. Patel and M. Rosenzweig. International Co-operative Publishing House, Fairland, MD.
- Sokal, R., and N. Oden, 1978. Spatial autocorrelation in biology. Biol. J. Linn. Soc. 10: 199–228. [Google Scholar]
- Sokal, R., and F. Rohlf, 1995. Biometry: The Principles and Practice of Statistics in Biological Research. W. H. Freeman, New York.
- Weeks, A. R., S. W. Mckechnie and A. A. Hoffman, 2006. In search of clinal variation in the period and clock timing genes in Australian Drosophila melanogaster populations. J. Evol. Biol. 19: 551–557. [DOI] [PubMed] [Google Scholar]
- Wheeler, D. A., C. P. Kyriacou, M. L. Greenacre, Q. Yu, J. E. Rutila et al., 1991. Molecular transfer of a species-specific behavior from Drosophila simulans to Drosophila melanogaster. Science 251: 1082–1085. [DOI] [PubMed] [Google Scholar]
- Yu, Q., H. V. Colot, C. P. Kyriacou, J. C. Hall and M. Rosbash, 1987. Behavior modification by in vitro mutagenesis of a variable region within the period gene of Drosophila. Nature 326: 765–769. [DOI] [PubMed] [Google Scholar]
- Zamorzaeva, I., E. Rashkovetsky, E. Nevo and A. Korol, 2005. Sequence polymorphism of candidate behavioral genes in Drosophila melanogaster flies from ‘Evolution canyon’. Mol. Ecol. 14: 3235–3245. [DOI] [PubMed] [Google Scholar]