Abstract
Activation of the Chk1 protein kinase by DNA damage enforces a checkpoint that maintains Cdc2 in its inactive, tyrosine-15 (Y15) phosphorylated state. Chk1 downregulates the Cdc25 phosphatases and concomitantly upregulates the Wee1 kinases that control the phosphorylation of Cdc2. Overproduction of Chk1 causes G2 arrest/delay independently of DNA damage and upstream checkpoint genes. We utilized this to screen fission yeast for mutations that alter sensitivity to Chk1 signaling. We describe three dominant-negative alleles of cdr1, which render cells supersensitive to Chk1 levels, and suppress the checkpoint defects of chk1Δ cells. Cdr1 encodes a protein kinase previously identified as a negative regulator of Wee1 activity in response to limited nutrition, but Cdr1 has not previously been linked to checkpoint signaling. Overproduction of Cdr1 promotes checkpoint defects and exacerbates the defective response to DNA damage of cells lacking Chk1. We conclude that regulation of Wee1 by Cdr1 and possibly by related kinases is an important antagonist of Chk1 signaling and represents a novel negative regulation of cell cycle arrest promoted by this checkpoint.
GENOME integrity is essential for normal tissue homeostasis. DNA damage, either from extrinsic sources such as radiation and genotoxic drugs or from intrinsic sources from metabolism and DNA replication, poses a threat to the stable inheritance of the genome. Not surprisingly, therefore, cells have evolved a plethora of pathways to sense and repair DNA damage and also to alter cell cycle progression to allow time for DNA repair to complete. The latter, known as DNA damage checkpoints, respond to many different lesions throughout the cell cycle and ultimately regulate the cyclin-dependent kinases that drive the cell cycle transitions.
The transition from G2 into mitosis is controlled by the cyclin B/Cdc2 kinase. Cdc2 is the catalytic subunit of this complex, although its kinase activity requires its binding to the cyclin partner. While Cdc2 levels remain constant throughout the cell cycle, cyclin B accumulates during interphase, forms complexes with Cdc2, and is rapidly destroyed by ubiquitination at the metaphase-to-anaphase transition. As cyclin B/Cdc2 complexes form, they are subjected to an inhibitory tyrosine phosphorylation on residue 15 (Y15) by specific tyrosine kinases, generally known as Wee1. Such inactive complexes accumulate during G2, and when conditions are appropriate for mitotic entry, Y15 is dephosphorylated by the Cdc25 phosphatases, and cells rapidly enter mitosis (Nurse 1990; Dunphy 1994).
DNA damage in S-phase and G2 activates a checkpoint pathway that is largely conserved from the fission yeast Schizosaccharomyces pombe through to humans (O'Connell et al. 2000). The effector of this signaling cascade is a serine/threonine protein kinase, Chk1. Activation of Chk1 by DNA damage requires several protein complexes, including the PCNA-related 9-1-1 complex, its loader Rad17-Rfc2-5, and the PI-3K-related kinases ATM and ATR, which phosphorylate Chk1 on C-terminal residues, an event required for its activation. Several mediator proteins are also required for Chk1 activation, including the BRCT-domain proteins Crb2 and Cut5 in S. pombe (O'Connell et al. 2000) and BRCA1, Mdc1, and TopBP1 in metazoans (Garcia et al. 2005). When Chk1 is overexpressed, it can signal a G2 delay/arrest independently of DNA damage and these upstream regulators (Walworth et al. 1993).
Activated Chk1 ensures a delay in G2 until completion of DNA repair by enforcing the maintenance of Cdc2 Y15 phosphorylation. This is achieved through regulation of both Wee1 kinases and Cdc25 phosphatases (O'Connell et al. 2000). In S. pombe, phosphorylation of Wee1 by Chk1 leads to a transient stabilization of this short-lived protein and hence to increased cellular levels of Wee1 kinase (O'Connell et al. 1997; Raleigh and O'Connell 2000). Similarly, Chk1 has been implicated in the regulation of Wee1 homologs in Xenopus (Lee et al. 2001; Stanford and Ruderman 2005), Drosophila (Price et al. 2000; Stumpff et al. 2004), and humans (Li et al. 2002). For Cdc25, its phosphorylation by Chk1 in S. pombe leads to its nuclear exclusion and catalytic inactivation (Furnari et al. 1999; Lopez-Girona et al. 1999, 2001). In mammals, there are three isoforms of Cdc25, Cdc25A, -B, and -C. Each can be phosphorylated by Chk1, although only Cdc25A is essential for cell cycle progression in the mouse (Ferguson et al. 2005). Chk1 phosphorylation of Cdc25A leads to its degradation by ubiquitin-dependent proteolysis (Sorensen et al. 2003; Xiao et al. 2003). Either mode of regulating Cdc2 Y15 phosphorylation is sufficient for a checkpoint delay; that is, cells lacking Wee1 or cycling independently of Cdc25 are largely checkpoint proficient. When both are absent, and the cell cycle is driven only by cyclin accumulation, the checkpoint fails, and cells become hypersensitive to DNA damage (Raleigh and O'Connell 2000).
Inactivation of Chk1 is both necessary and sufficient to signal resumption of the cell cycle through activation of Cdc2 (Latif et al. 2004). This is achieved via dephosphorylation of the activating phospho-residues by Dis2, a protein phosphatase 1 (PP1) homolog, in S. pombe (den Elzen and O'Connell 2004), and by PP1 and the p53-target PPMID (Wip1) in humans (Lu et al. 2005). How this is spatially and temporally regulated is not clear at this stage, and it remains likely that additional mechanisms to antagonize Chk1 signaling exist. Other mechanisms of regulating Cdc2 activity have been described. These include regulation of Wee1 activity by the Cdr kinases in S. pombe under conditions of limited nutrition, especially nitrogen starvation, where downregulation of Wee1 is an important event in advancing mitotic entry to achieve G1 cell cycle arrest (Young and Fantes 1987; Feilotter et al. 1991; Coleman et al. 1993; Parker et al. 1993; Wu and Russell 1993). Further, Polo kinases play an important role in upregulating Cdc25 activity at mitotic entry, and regulation of Polo kinases has been implicated in DNA damage checkpoint proficiency (Barr et al. 2004). Although cells lacking Chk1 fail to delay in response to DNA damage, it is not known whether other pathways converging on Cdc2 activation contribute to the delay in wild-type cells.
We have used genetic screens in S. pombe to attempt to identify additional regulators and effectors of the G2 DNA damage checkpoint. Here we describe a screen in which we have isolated mutations that alter the cellular sensitivity to ectopic Chk1 expression. We isolated three alleles of cdr1 that render cells supersensitive to Chk1. These mutations suppress the DNA damage sensitivity of mutants in the checkpoint pathway. We show that the cdr1 mutations are dominant-negative alleles, linking Cdr1 for the first time to the DNA damage response, in addition to its role in nutritional signaling. Upon limited nutrition, particularly nitrogen deprivation, Cdr1 phosphorylates and negatively regulates the kinase activity of Wee1, thus activating Cdc2, and cells enter into mitosis at a smaller cell size (Feilotter et al. 1991; Coleman et al. 1993; Wu and Russell 1993). As cdr1Δ does not suppress the checkpoint mutations, this dominant-negative effect may extend to other related protein kinases. Finally, upregulation of Cdr1 leads to a checkpoint defect, DNA damage hypersensitivity, and exacerbation of the defects of chk1Δ cells. We conclude that Cdr1 and related kinases antagonize Chk1 in the regulation of Wee1 in the context of the DNA damage checkpoint.
MATERIALS AND METHODS
Fission yeast methods:
All strains used were derivatives of 972 h− and 975 h+, and detailed genotypes are listed in the supplemental material at http://www.genetics.org/supplemental/. Standard procedures and media were used for growth and genetic manipulation (Moreno et al. 1991). Methods for transformation, microscopy, and DAPI staining have been described previously (O'Connell et al. 1997). For nmt1 promoter induction experiments, exponentially growing cells were washed three times in medium lacking thiamine and then inoculated into defined medium lacking thiamine and collected after 15–18 hr. For testing of dominance without background sporulation, h−/h− diploids were constructed by protoplast fusion and selected by cross complementation of leu1-32 and his3-D1 markers.
Screening to identify the location of the D8 mutation:
On the basis of the recessive lethal phenotype of the D8 strain, suppressed by the tRNA sup3-5, we designed a plasmid shuffle screen. For this we cloned an adh1-driven herpes simplex virus 1 thymidine kinase (TK) gene (Padh1∷TK) (Kiely et al. 2000) into the SacI site of pREP5 (sup3-5, nmt1). ade6-704 cells were transformed with this plasmid, conferring adenine prototrophy (sup3-5 suppression of ade6-704) and sensitivity to the thymidine analog (+)-5-fluoro-2′-deoxyuridine (FUdR), which, when phosphorylated by Padh1-driven TK, inhibits thymidylate synthetase and blocks dTTP synthesis, killing the cells. This strain was crossed to nmt1∷chk1(int)sup3-5 D8 ade6-704 ura4-D18 to derive D8 (pREP5-sup3-5-Padh1∷TK) ade6-704 ura4-D18 progeny (i.e., no longer containing sup3-5 nmt1∷chk1). The presence of the D8 mutation, now suppressed by the sup3-5 gene within the Padh1∷TK construct, was confirmed by transformation with pREP41X∷chk1. This strain was then transformed with a ura4-based genomic library (Barbet et al. 1992), and transformants were selected on minimal medium with adenine to allow loss of the sup3-5 marker (within pREP5-Padh∷TK plasmid) with D8's recessive lethality, presumably now rescued by a library plasmid. Such events were screened for by replica plating to minimal medium with adenine (to allow ade6-704 cells to grow in the absence of sup3-5) and 50 μm FUdR to select for the absence of Padh1∷TK. Surviving colonies, now adenine auxotrophs, had library plasmid-borne rescue of the D8-associated lethality. From this, a total of five independent clones of cdc5 were obtained. However, further analysis (see results) showed that the cdc5 mutation in D8 was not the cause of the supersensivity to Chk1, but rather due to the closely linked cdr1 gene, and that the D8 strain contained these two linked mutations.
DNA damage survival assays:
For ultraviolet-C (UV-C) assays, 100, 1000, and 10,000 cells were plated in triplicate on yeast extract plus supplements (YES) agar (90-mm plate) or on EMM2 agar without thiamine for the overexpression of Cdr1 protein and irradiated in a Stratalinker. Colonies were counted and the percentage of survival was expressed as a proportion of unirradiated controls after 4 days at 30°. For methyl methanesulfonate (MMS) sensitivity assays, cultures were diluted to an OD595 of 0.1 (2 × 106 cells/ml) and 5 μl of 10-fold serial dilutions were spotted onto YES, or EMM2 agar for the overexpression of Cdr1 protein, containing the indicated concentrations of MMS. Plates were incubated for 4 days at 30°.
Construction of cdr1-Flag3-His6 strains:
A Flag3-His6 tag was integrated into the 3′-end of the cdr1 ORF using PCR products derived from pFA6a-Flag3-His6-kanMX6 (Morikawa et al. 2004). G418-resistant colonies were selected and checked for correct integration in the genome by PCR with primers inside and outside the ORF of the cdr1 gene and by Western blotting. We confirmed that the tagged protein was functional by proficiency for G1 arrest upon nitrogen depletion. A PCR product from this strain that spanned 300 bp on each side of the integration was derived and used to tag the dominant cdr1 alleles by homologous recombination.
Protein methods:
For detection of Chk1, GST-Cdr1, Cdc2, or Wee1-His6HA3, frozen cells were disrupted with glass beads using a bead beater and extracted into lysis buffer 1 (8 m urea, 100 mm Na2HPO4, 10 mm Tris–HCl, pH 8.0). The extract was cleared by centrifugation at 13,000 × g for 15 min, and the supernatant was boiled in SDS sample buffer. Protein extracts were run on 7% (Wee1) or 10% (Chk1, GST-Cdr1, Cdc2) SDS–PAGE gels and transferred to nitrocellulose membrane in 10 mm 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11, and 10% methanol for 1 hr. Overexpressed Chk1 was detected by rabbit polyclonal anti-Chk1 (O'Connell et al. 1997), HA epitope with 12CA5 (Roche), GST with monoclonal anti-GST (Santa Cruz, B14), Y15 phosphorylated Cdc2 (Cell Signaling Technology), total Cdc2 (PSTAIRE, Santa Cruz), and tubulin as a loading control by monoclonal-α-tubulin (Sigma, St. Louis, B-5-1-2). For Cdr1-Flag3-His6, the extracts were prepared in lysis buffer 2 (10 mm NaPO4, pH 7.5, 150 mm NaCl, 1% NP-40, 10 mm EDTA, 50 mm NaF, 2 mm DTT) and 5× protease inhibitor cocktail (Sigma). Cdr1-Flag3-His6 was immunoprecipitated with anti-Flag M2 agarose (Sigma). Extracts were run on a 7% gel and transferred as above, and the Flag-Tag was detected with the anti-Flag M2 (Sigma). Immune complexes were detected with horseradish-peroxidase-linked secondary antibodies (Amersham, Buckinghamshire, UK) followed by chemiluminescence. Dephosphorylation of Wee1 was performed as described (O'Connell et al. 1997). For Cdr1 autophosphorylation assays, wild-type and mutant GST-Cdr1 expressed from the nmt1 promoter was recovered on glutathione sepharose from 500 μg of extract prepared in lysis buffer 2. After extensive washing in lysis buffer 2 and kinase buffer (50 mm Tris, pH 8, 10 mm MgCl2, 1 mm MnCl2, 1 mm DTT), cold ATP was added to 100 μm together with 10 μCi of [γ-32P]ATP (3000 Ci/mmol) and incubated at 30° for 30 min. Reactions were stopped by boiling in sample buffer and separated on SDS–PAGE gels, and the signal quantified with a Bio-Rad (Hercules, CA) FX phosphorimager.
Nitrogen deprivation:
Midlogarithmic asynchronous cultures were washed three times in EMM2 medium (Moreno et al. 1991) lacking ammonium chloride and then inoculated into minimal medium with and without ammonium chloride to an OD595 of 0.1. The cells were collected at 24 and 48 hr and fixed in 70% ethanol. Cells were resuspended in 50 mm sodium citrate, pH 6.5, 20 μg/ml propidium iodide, and 40 μg/ml RNaseA and incubated at 37° overnight. DNA content was determined using a FACSCalibur (Becton Dickinson) and CellQuest software.
RESULTS
Isolation of mutants with altered sensitivity to Chk1:
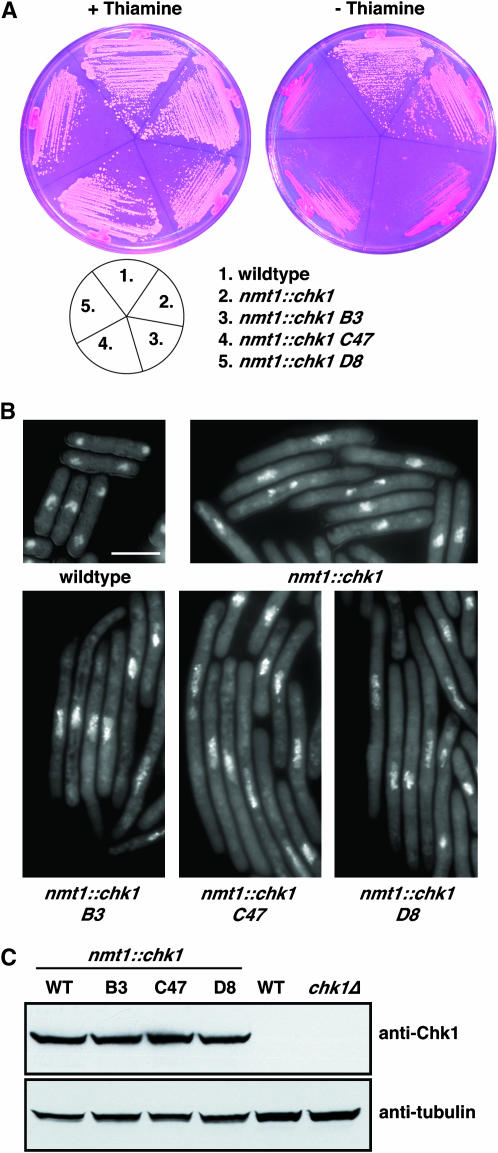
To screen for novel regulators and effectors of Chk1 signaling, we sought to identify genes that modulate the duration of a Chk1-mediated G2 delay. To this end, we constructed a strain of S. pombe in which Chk1 is expressed from the nmt1 promoter, integrated in single copy using the sup3-5 tRNA suppressor (Kohli et al. 1979). In the presence of thiamine, the nmt1 promoter is repressed (Maundrell 1993) and this level of Chk1 expression does not alter cell cycle progression. Upon withdrawal of thiamine, the derepression of the nmt1 promoter after four to five cell cycles leads to a G2 delay; cells are viable but enter mitosis at an increased cell length as a result of a longer G2 period. A small percentage of the cells die under these conditions, leading to pink colonies on agar plates containing the vital dye phloxin B, compared to wild-type colonies, which are white under these conditions (Figure 1A). This strain was then mutagenized with nitrosoguanidine for several independent screens (A–E) totaling ∼300,000 colonies, and mutants were selected that were either resistant or supersensitive to nmt1-driven Chk1 in the absence of thiamine, although they remained identical to parental cells in the presence of thiamine (see materials and methods). Twelve mutants that were resistant to Chk1 overexpression were isolated. Eleven of these were alleles of rad24, which encodes a 14-3-3 isoform that mediates Chk1 signaling via interaction with Chk1 and its substrates (Chen et al. 1999; O'Connell et al. 2000), and will be described in another article.
Figure 1.—
Isolation of mutants supersensitive to Chk1 overexpression. (A) The indicated strains were streaked onto minimal media containing phloxin B together with thiamine (nmt1 promoter repressed) or without thiamine (nmt1 promoter derepressed), and the plates were incubated at 30° for 4 days. Note that the residual growth of mutants on plates lacking thiamine is from a heavy inoculum from a plate containing thiamine and also that the nmt1 promoter takes about five cell cycles to fully derepress. These inviable cells are stained with phloxin B and cannot be propagated on a fresh plate. (B) Cells were grown to midlogarithmic phase in the presence of thiamine, washed three times in thiamine-free medium, and then grown in the absence of thiamine for 18 hr at 30°. Cells were fixed with formaldehyde and stained with DAPI. Cytoplasmic staining is from mitochondrial genomes. Bar, 10 μm. (C) The same cultures as in B were harvested and snap frozen. Extracts were prepared and analyzed by Western blotting using anti-Chk1 polyclonal antibodies, which can detect only overexpressed Chk1. The filter was stripped and reprobed with antitubulin antibodies.
Of the supersensitive class, three strong mutants (B3, C47, and D8) were isolated from three independent screens (B, C, and D) that were inviable on plates lacking thiamine, although they phenocopied the parental strain in the presence of thiamine (Figure 1A). Genetic analysis showed that these mutations were in the same gene, shown below to be cdr1. These cells were examined by microscopy and shown to arrest as highly elongated cells in the absence of thiamine (Figure 1B), which was confirmed to be a G2 cell cycle arrest by flow cytometry of DNA (not shown). By Western blotting of extracts with affinity purified anti-Chk1 antibodies we confirmed that the Chk1 supersensitive mutants did not arise via upregulation of Chk1 expression, which was unchanged compared to controls (Figure 1C).
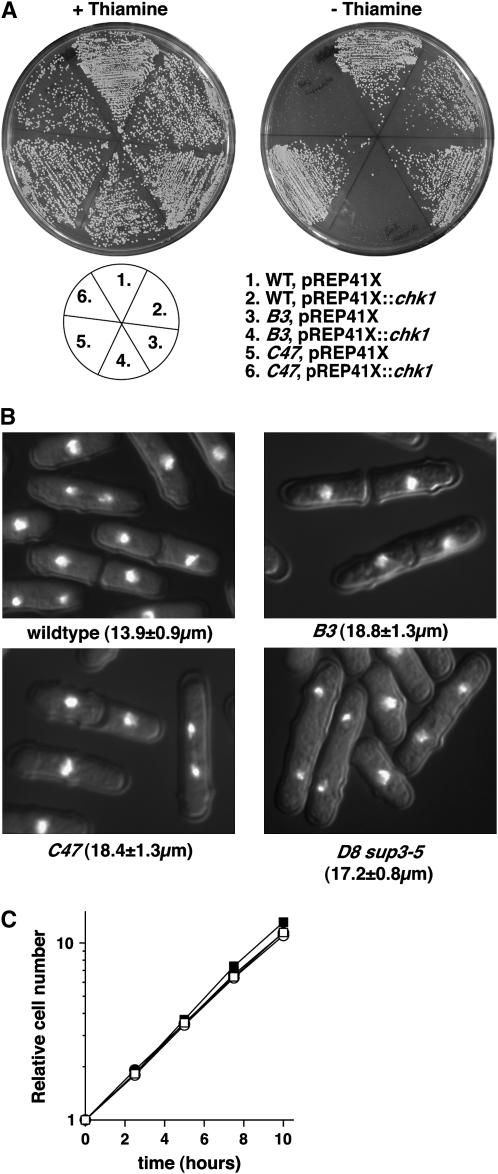
We next crossed these strains to wild-type cells to isolate the single mutations away from the integrated sup3-5 nmt1∷chk1, which was easily achieved for B3 and C47. To confirm that the single mutants retained the hypersensitivity to Chk1, they were transformed with a plasmid in which Chk1 is expressed from an attenuated nmt1 promoter (pREP41X∷chk1) that, as an episome, directs ∼10-fold lower levels of Chk1 expression compared to the single integrated wild-type nmt1 promoter (Forsburg 1993). In wild-type cells, this level of Chk1 expression results in a modest G2 delay, although the level was lethal to the B3 and C47 strains (Figure 2A). For D8, tetrad analysis showed that this mutation was lethal in the absence of the sup3-5 nmt1∷chk1 construct; germinated spores arrested as highly elongated, uninucleated cells (not shown). As this strain was isolated on the basis of hypersensitivity to Chk1, we hypothesized that this lethality may be due to the absence of the sup3-5 marker used to select the nmt1∷chk1 integration. This was indeed the case, and D8 could be propagated in cells containing sup3-5 but not nmt1∷chk1. We transformed this strain with pREP41X∷chk1, and this too was lethal in the absence of thiamine (not shown).
Figure 2.—
Normal cell cycle progression of the supersensitive mutants in the absence of Chk1 overexpression. (A) B3 and C47 were separated from sup3-5 nmt1∷chk1 by tetrad dissection and transformed with pREP41X∷chk1 (medium strength nmt1) together with vector and wild-type controls. Sensitivity to Chk1 overexpression in the presence and absence of thiamine was assayed as described in Figure 1. (B) Midlogarithmic cultures of the indicated strains were fixed with formaldehyde and stained with DAPI, and the mean length at division (±standard deviation, n = 100) was measured with an eyepiece micrometer. (C) Wild type (□), B3 (•), C47 (▪), and D8 sup3-5 (○) were grown to midlogarithmic phase, and cell cycle progression was assayed by triplicate cell number determination using a Coulter counter for 10 hr.
An important component of our screen was that, for mutants to be retained for further analysis, they needed to be normal in the absence of Chk1 overexpression. To confirm that this was the case, we assayed cell cycle progression in the single mutants. Although each mutant divided at a cell length of ∼18 μm compared to wild-type cells, which divide at 14 μm (Figure 2B), by monitoring cell numbers in exponentially growing cultures we found that each mutant had a normal cell doubling time (Figure 2C). Therefore, the slightly elongated nature of these cells is a result of a larger birth size rather than an extended growth period without division. We therefore conclude that these strains are specifically supersensitive to Chk1 signaling.
We next assessed whether these mutations were dominant or recessive. We were surprised to observe that each heterozygous diploid retained the supersensitivity to Chk1 expression, albeit to a slightly lesser extent than the haploids on plates (supplemental Figure 1 at http://www.genetics.org/supplemental/). Microscopy and FACS confirmed that these heterozygotes retained G2 arrest upon Chk1 overexpression, whereas wild-type diploid cells overexpressing Chk1 underwent a modest delay (supplemental Figure 1B at http://www.genetics.org/supplemental/ and data not shown). Thus, each allele is dominant for supersensitivity to Chk1, which could result either from a gain-of-function or from dominant interference with a wild-type allele.
The supersensitive mutants are dominant-negative alleles of cdr1:
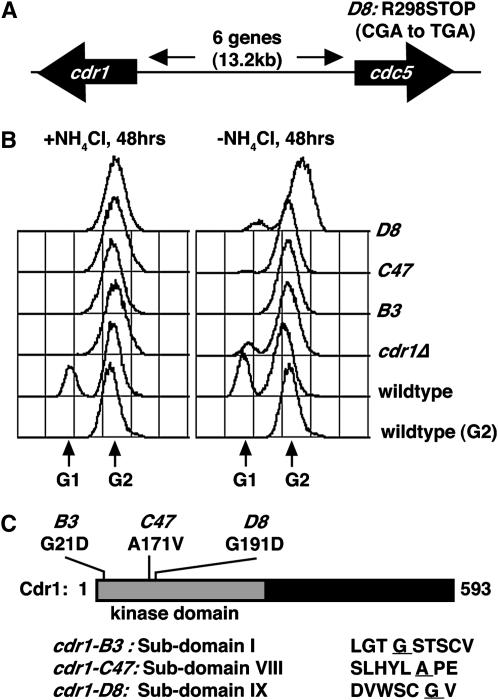
As these alleles are dominant for supersensitivity to Chk1, we could not identify the gene by conventional complementation screens. However, the lethal cell cycle arrest of D8 that was suppressed by sup3-5 was recessive in diploids (data not shown). sup3-5 is a tRNA suppressor that inserts serine residues opposite UGA termination codons (Kohli et al. 1979), and thus we knew the nature of the lethal mutation in D8. We devised a plasmid shuffle screen to replace an episomal sup3-5 allele with genomic clones (see materials and methods). From this screen we obtained several clones of cdc5, an essential gene required for pre-mRNA splicing (Ohi et al. 1994). Sequencing of the cdc5 gene in the D8 strain indeed showed a CGA-to-TGA mutation (Figure 3A), truncating Cdc5 after amino acid 298 of 757. We were somewhat surprised by this finding, however, as no link between Cdc5 and Chk1 signaling is apparent. Moreover, we failed to find mutations in cdc5 in the B3 or C47 strains, even though no wild-type recombinants were recovered from crosses between different Chk1 supersensitive alleles (>20,000 spores tested). Linkage analysis could not separate the recessive lethality of D8 from its dominant supersensitivity to Chk1. We concluded, therefore, that the supersensitivity to Chk1 must arise from a mutation very tightly linked to cdc5, and not from the cdc5 mutation. That is, the D8 strain fortuitously contained two linked mutations: one in cdc5 and one in another gene that was the cause of the supersensitivity to Chk1.
Figure 3.—
The Chk1 supersensitive mutants are alleles of cdr1. (A) A plasmid shuffling screen (see materials and methods) was used to identify a termination codon in cdc5 (CGA to TGA) in D8 cells, but cdc5 mutations were not present in B3 or C47. The cdr1 locus is located 13.2 kb to the left of cdc5 on chromosome I. (B) The mutants exhibit a changed division response (Cdr) phenotype. Cultures were started at 2 × 106 cells/ml and grown for 48 hr to saturation in the presence or absence of NH4Cl, fixed with 70% ethanol, and processed for DNA content determination by FACS. Note the absence of a G1 peak in cdr1 mutants and the cdr1Δ control. (C) Mutations in the Cdr1 catalytic domain (subdomains I, VIII, and IX; substituted amino acid is underlined) were present in our cdr1 mutants.
While sequencing nearby candidate genes, we noted that saturated cultures of the supersensitive mutants were not the characteristically small cells seen in saturated wild-type cultures. Under conditions of limited nutrition, which results from either culture saturation or can be induced by nitrogen withdrawal, S. pombe cells advance the timing of mitotic entry, and as G2 is the major growth period, these cells become smaller and eventually undergo G1 cell cycle arrest (Young and Fantes 1987). FACS analysis confirmed an absence of G1 cell cycle arrest in the mutant strains (Figure 3B), which is reminiscent of cells lacking either of two related protein kinases, Cdr1 and Cdr2, which derive their name from this changed division response (Young and Fantes 1987). Inspection of the genes within the vicinity of cdc5 showed that cdr1 was separated from cdc5 by six genes >13.2 kb (Figure 3A). Normally this distance would be separable by recombination and cdr1 has not been implicated in checkpoint signaling, but we nevertheless sequenced cdr1 in each of the mutants. Indeed, each allele contained single amino acid substitutions in largely invariant residues of the first, eighth, and ninth kinase subdomains (Figure 3C). Thus, the mutations are alleles of cdr1 and were therefore named cdr1-B3, -C47, and -D8. Given the highly conserved nature of the catalytic subdomains within serine/threonine protein kinases (Hanks et al. 1988) and the dominance of the supersensitive to the Chk1 phenotype and the Cdr phenotype in the saturated cultures, these are likely to be dominant-negative cdr1 alleles.
The cdr1 mutants were then crossed to a number of checkpoint and cell cycle mutants to establish whether a normal pattern of signaling was present in these cells and to attempt to decipher where in the checkpoint pathway Chk1 signaling is upregulated. Although Cdr1 is known to regulate Wee1 (Coleman et al. 1993; Parker et al. 1993; Wu and Russell 1993), we needed to address whether this was the sole cause of the supersensitive phenotype. Cell cycle delay/arrest resulting from Chk1 overexpression is independent of upstream checkpoint rad genes (Walworth et al. 1993). Similarly, the cdr1 mutants did not require upstream components such as Rad3, the 9-1-1 complex, or the BRCT-domain protein Crb2 to exhibit supersensitivity to Chk1 overexpression (not shown). However, cells lacking the Cdc2 Y15 kinase Wee1 or the interacting 14-3-3 protein Rad24 are largely insensitive to Chk1 overexpression (O'Connell et al. 2000). Double mutants were therefore constructed between each cdr1 allele and wee1Δ or rad24Δ, and sensitivity to Chk1 overexpression from the integrated nmt1∷chk1 allele was assayed. Both mutations suppressed the supersensitivity of the dominant cdr1 mutants, which were capable of forming colonies on medium lacking thiamine (supplemental Figure 2 at http://www.genetics.org/supplemental/) and were no longer elongated (not shown). These data confirm that the cdr1 mutants exert their phenotype at the level of Wee1.
The dominant alleles of cdr1 bypass G2 DNA damage checkpoint defects:
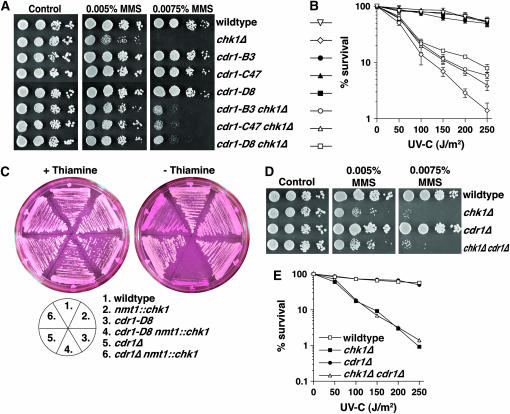
We next constructed double mutants between each cdr1 allele and rad3Δ, rad17Δ, crb2Δ, and chk1Δ (without nmt1∷chk1). Figure 4 shows that the cdr1 mutations indeed partially suppressed the hypersensitivity of chk1Δ cells to the alkylating agent MMS and to UV-C irradiation. Similar partial suppression was also observed with rad3Δ, rad17Δ, and crb2Δ double mutants (not shown). The cdr1 mutants themselves showed wild-type sensitivities to a range of DNA-damaging agents. This was expected, given their sensitized response to Chk1 overexpression, which, if anything, should further enforce the checkpoint response to DNA damage.
Figure 4.—
Dominant-negative cdr1 mutations partially suppress the MMS and UV-C hypersensitivity of chk1Δ. (A) Ten-fold serial dilutions of the indicated strains were spotted onto YES agar with a range of concentrations of MMS, and colonies were allowed to form over 4 days at 30°. (B) UV-C survival assays of the same series of strains shown in A. (C) Plate assays of sensitivity to Chk1 overexpression were carried out as described in Figure 1. Note that cdr1Δ shows an intermediate sensitivity to Chk1 compared to wild-type and cdr1-D8 controls. (D) MMS sensitivity and (E) UV-C survival was assayed as in A and B. Note that cdr1Δ does not partially suppress chk1Δ.
While Wee1 is implicated in checkpoint signaling in several systems, Cdr1 has been implicated in cell cycle control only in response to limited nutrition, which does not occur in any of the experiments using Chk1 overexpression or checkpoint activation in midlogarithmic cultures. We therefore wanted to assess whether the dominant-negative cdr1 alleles exerted their effects only on Cdr1, or whether other related kinases may also be dominantly interfered with. We favored the latter hypothesis, as we did not obtain recessive alleles of cdr1 (or cdr2) in our screen. Although cdr1Δ cells are somewhat more sensitive to Chk1 overexpression than wild-type cells, they do form colonies on media lacking thiamine, although they are stained with phloxin B, indicating the presence of some dead cells (Figure 4C). Further, cdr1Δ was not capable of suppressing the MMS or UV-C hypersensitivity of chk1Δ cells (Figure 4, D and E). Therefore, these alleles dominantly interfere with molecules other than Cdr1 alone.
Biochemical analysis of Cdr1 mutants:
Cdr1 exists in cells as multiple phospho-isoforms generated, at least in part, by autophosphorylation (Wu et al. 1996). We did not observe changes in the multiple Cdr1 species upon Chk1 activation by irradiation or overexpression (data not shown), showing that at least autophosphorylation was unaffected. There have been several reports regarding phosphorylation of Wee1 by Cdr1 using recombinant proteins produced in insect cells (Coleman et al. 1993; Parker et al. 1993; Wu and Russell 1993), but not using Cdr1 isolated from S. pombe extracts. We attempted to reproduce these experiments using immunoprecipitated endogenous or overexpressed Cdr1 and bacterially expressed Wee1 as substrate, but were unable to demonstrate significant kinase activity under any condition. Thus, while it is possible that Cdr1 activity is modulated by checkpoint signaling, this does not involve changes in Cdr1 protein levels or autophosphorylation. Although autophosphorylation assays are less quantitative than when excess substrate is provided, these observations lend further support to the notion that these mutations dominantly interfere with Cdr1-mediated Wee1 regulation.
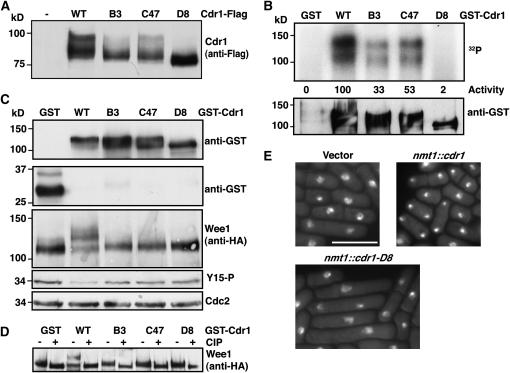
Each Cdr1 mutant protein showed altered migration on SDS–PAGE, the most extreme being cdr1-D8 (Figure 5A), which is similar to that described for the kinase-dead cdr1-K41A mutation (Wu et al. 1996). It is notable that cdr1-D8 gave the strongest suppression of chk1Δ and the other checkpoint mutants (Figure 4) and consistently had the strongest sensitivity to Chk1 upon replica plating. In vitro autophosphorylation assays using GST-Cdr1 fusion proteins purified from S. pombe extracts showed a significant reduction of activity in cdr1-B3 and -C47 and almost complete loss of activity in cdr1-D8 (Figure 5B).
Figure 5.—
Regulation of Wee1 by Cdr1. (A) Each cdr1 allele was tagged with His6Flag3 at the 3′-end of its endogenous locus. Proteins were immunoprecipitated with anti-Flag M2 beads and detected with M2 antibodies. (B) GST-tagged wild-type and mutant Cdr1 were expressed from the nmt1 promoter and purified on glutathione sepharose, and autophosphorylation was assayed in vitro from 50% of the recovered material. “Activity” is the arbitrary units determined by a Bio-Rad FX phosphorimager of the 32P signal, and levels of GST-Cdr1 in the assays were determined by Western blotting. (C) GST and GST-Cdr1 fusion proteins were expressed from the nmt1 promoter for 15 hr at 30° in cells expressing Wee1-His6HA3 from the endogenous locus. Anti-GST Western blots show GST and GST-Cdr1 levels, and Wee1 is detected with anti-HA antibodies. Identical effects on Wee1 were obtained with untagged Cdr1 constructs. Total Cdc2 and Y15 Cdc2 were detected from the same extracts. (D) Cell extracts were prepared as in C, and Wee1-His6HA3 was recovered on Ni-NTA agarose. Samples were split and treated with calf intestinal phosphatase (CIP) or mock treated. (E) Cdr1 and Cdr1-D8 were expressed from the nmt1 promoter in wild-type cells for 18 hr at 30°, fixed, and stained with DAPI. Bar, 10 μm. Overexpressed Cdr1-B3 and -C47 behave identically to Cdr1-D8 (data not shown).
Cdr1 is known to negatively regulate Wee1 via phosphorylation of residues in the C-terminal catalytic domain of Wee1 (Coleman et al. 1993; Parker et al. 1993). We next monitored phosphorylation of endogenous Wee1 in cells overproducing wild-type and mutant Cdr1 proteins from the nmt1 promoter. Wee1 migrates as a prominent band with a slower migrating smear that is due to phosphorylation (O'Connell et al. 1997). Wild-type Cdr1 overexpression resulted in a significant reduction in Wee1 mobility that was confirmed to be hyperphosphorylation using calf-intestinal phosphatase (Figure 5, C and D). Overexpression of Cdr1-B3 or -C47 did not affect Wee1 phosphorylation, whereas overexpression of Cdr1-D8 consistently reduced basal phosphorylation of Wee1 (Figure 5C). Moreover, wild-type Cdr1 overexpression results in a “wee” phenotype (Wu et al. 1996) and a reduction in Cdc2 Y15 phosphorylation (Figure 5C). Conversely, the overexpression of the dominant alleles resulted in a Wee1-dependent cell cycle delay (Figure 5E and data not shown). Together these data show that the cdr1 alleles isolated in our screen show poor kinase activity toward Wee1 in vivo and exert dominant-negative effects acting through Wee1 regulation.
Cdr1 overexpression results in DNA damage checkpoint defects:
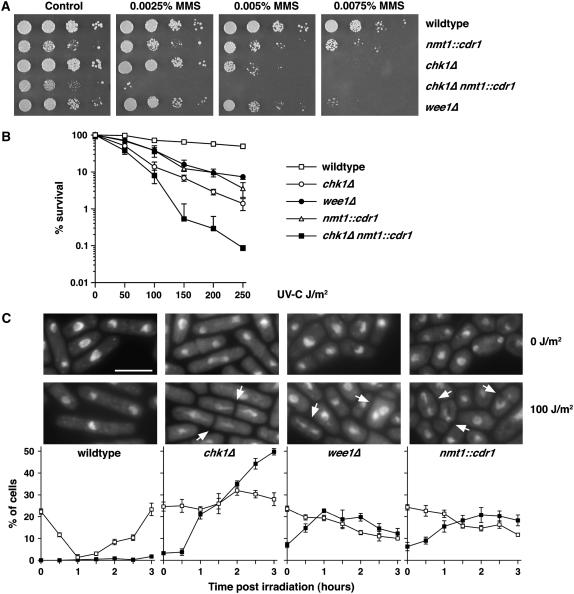
As the above data show that Cdr1 functions downstream of Chk1, most likely through regulation of Wee1, we asked whether upregulation of Cdr1 could overcome a G2 DNA damage checkpoint in otherwise wild-type cells. Expression of Cdr1 from the nmt1 promoter causes a “wee” phenotype due to shortening of G2 by Wee1 downregulation (Wu et al. 1996). We found that this level of overexpression also caused a hypersensitivity to both MMS and UV-C (Figure 6, A and B). The degree of sensitivity was similar to that of wee1Δ cells. Like wee1Δ (Walworth et al. 1993), Cdr1 overexpression also exacerbated the sensitivity of chk1Δ cells. While wee1Δ cells do show some cell cycle delay in response to DNA damage (Barbet and Carr 1993), the kinetics of cell cycle arrest are somewhat delayed (Raleigh and O'Connell 2000), especially when considering the shortened G2 period of these cells. Both wee1Δ and nmt1∷cdr1 cells showed a significant percentage of “cut” cells following irradiation (Figure 6C), which, in S. pombe, is indicative of progression through mitosis prior to completion of DNA repair. Overexpression of wild-type or mutant Cdr1 in wee1Δ cells did not affect the response to DNA damage compared to vector controls (data not shown), confirming that the effects of Cdr1 overexpression are via negative regulation of Wee1. Moreover, Cdr1 overexpression suppressed the G2 delay caused by Chk1 overexpression (not shown). Together, these data show that upregulation of Cdr1 by overexpression can cause checkpoint defects through downregulation of Wee1.
Figure 6.—
Cdr1 overexpression results in a partially defective checkpoint response. (A) MMS sensitivity and (B) UV-C survival assays were performed as in Figure 4. Note that nmt1∷cdr1 cells exhibit a sensitivity similar to that of wee1Δ cells and that Cdr1 overexpression, while somewhat toxic, substantially enhances the MMS and UV-C hypersensitivity of chk1Δ cells. (C) The indicated strains were mock irradiated (0 J/m2) or UV-C irradiated (100 J/m2) and grown at 30°. At 30-min intervals for 3 hr, samples were taken and fixed with formaldehyde. Fixed samples were stained with DAPI, and the percentage of cells that were binucleate (□) or exhibited an aberrant mitosis (cut phenotype, ▪) were counted in triplicate (±standard deviation, n = 100). A drop in binucleates indicates a cell cycle delay, and aberrant mitoses (arrowed) are indicative of checkpoint failure. Bar, 10 μm.
DISCUSSION
Recent years have seen rapid advances in our understanding of cell cycle checkpoints. Although a detailed description of the highly conserved events leading to Chk1 activation has been elucidated, a description of mechanisms by which the checkpoint arrest is terminated remains somewhat rudimentary. Here we have identified a hitherto unknown function for Cdr1, and most likely other Cdr1-related kinases, in the negative regulation of the G2 DNA damage checkpoint.
Cdr1 was first identified as nim1, a truncated clone of cdr1 that was a high-copy suppressor of the temperature-sensitive G2 arrest of the cdc25-22 mutant (Russell and Nurse 1987; Feilotter et al. 1991). The original mutants in cdr1, and a related kinase encoded by cdr2, were identified in a screen for mutants that failed to arrest as small G1 cells in response to nitrogen deprivation (Young and Fantes 1987). Subsequent experiments using recombinant proteins showed that Cdr1 phosphorylates the catalytic domain of Wee1 and suppresses its kinase activity against Y15 of Cdc2 (Coleman et al. 1993; Parker et al. 1993; Wu and Russell 1993). No direct physical interaction has been reported for the endogenous Wee1 and Cdr1 proteins, which may be technically challenging, as these proteins are labile and of low abundance. Despite this, the above in vitro biochemical data support the genetic experiments in S. pombe that indicated that cdr1 acts in opposition to wee1 to advance mitotic entry under conditions of nitrogen starvation, and overexpression studies showed that no other cellular targets exist (Lundgren et al. 1991).
In addition to this well-defined role in nutritional signaling, the evidence that Cdr1 negatively regulates checkpoint signaling is as follows: First, the dominant-negative cdr1 alleles sensitize cells to Chk1 overexpression. Second, these mutations suppress the DNA damage hypersensitivity of chk1Δ and mutations in upstream components required for Chk1 activation. We failed to obtain evidence for checkpoint regulation of Cdr1, which also is not regulated through the cell cycle (Russell and Nurse 1987; Wu et al. 1996). However, as Chk1 activity is induced by checkpoint regulation (den Elzen and O'Connell 2004), the ratio of Cdr1:Chk1 activity toward Wee1 changes under these conditions. As with cdr1Δ (Russell and Nurse 1987; Feilotter et al. 1991), each dominant allele was synthetically lethal with cdc25-22 (data not shown), which is also consistent with a lack of restraint on Wee1 activity. Finally, overexpression of Cdr1 renders cells hypersensitive to DNA damage and exacerbates the sensitivity of chk1Δ cells. In this regard, Cdr1 overexpression mimics the effects of wee1Δ in the context of checkpoint signaling. Although wee1Δ cells retain the ability to delay G2 progression in response to DNA damage, they are considerably DNA damage hypersensitive and show an induction of mitotic defects following DNA damage. Thus, as wee1Δ cells are defective in successfully completing mitosis following DNA damage, we consider this to be a physiologically defective response despite the residual G2 delay, which is in keeping with data showing that Wee1 homologs are involved in checkpoint signaling in Drosophila, Xenopus, and humans (Price et al. 2000; Lee et al. 2001; Li et al. 2002; Stumpff et al. 2004; Stanford and Ruderman 2005). Although Cdr1 overexpression has been shown to cause a “wee” phenotype (Russell and Nurse 1987; Wu et al. 1996), the only function for Cdr1 determined previously from the cdr1Δ cells had been an inability to arrest in G1 upon nitrogen starvation (Young and Fantes 1987; Feilotter et al. 1991). All the experiments presented here, except those presented in Figure 3B, have been performed with midlogarithmic cultures with adequate nutrients. Here we show that cdr1Δ does not suppress the sensitivity of chk1Δ to DNA damage and is not as sensitive to Chk1 overexpression as the dominant-negative cdr1 alleles. These data explain why our screen did not identify recessive cdr1 alleles and imply that the phenotypes of these alleles involve interference with additional molecules. Cdr2 is a Cdr1-related kinase that also regulates Wee1 under conditions of nitrogen starvation, although it has a Wee1-independent role in the regulation of cytokinesis (Breeding et al. 1998; Kanoh and Russell 1998). Budding yeast homologs of Cdr1/2 are similarly involved in cytokinesis (McMillan et al. 1999; Shulewitz et al. 1999), but have not been shown to regulate DNA damage responses, although the DNA damage checkpoint in this organism does not signal via the Wee1 homolog Swe1. We assayed the suppression of MMS and UV-C sensitivity of cdr2Δ chk1Δ and cdr1Δ cdr2Δ chk1Δ, and in no case did we observe the suppression obtained with the dominant-negative alleles (data not shown). There are several other S. pombe protein kinases with significant similarity to Cdr1 (P = 10−47–10−63 by BLAST): Kin1, a protein kinase involved in cell polarity (Levin and Bishop 1990), and Ppk25, Ssp2, and Ppk9, protein kinases of unknown function (Bimbo et al. 2005). As the dominant-negative proteins do not accumulate, it is not clear how they exert their dominant effect. This conceivably could be through sequestration of Wee1, although we were not able to show a stable interaction between Wee1 and either wild-type or mutant Cdr1 (data not shown). Alternatively, the mutants could interfere with Cdr1/2- and/or Chk1-mediated Wee1 phosphorylation through transient interaction with Wee1 or upstream regulators, such as Rad24. However, as the dominant alleles suppress chk1Δ, a direct effect on Chk1 is not likely. While we did see changes in the mobility of Wee1 in these mutants with and without overexpression (Figure 5C and data not shown), with the multiple kinases impinging on Wee1, a detailed map of phosphorylation will be required to confirm this hypothesis. Nevertheless, our observations confirm the importance of Wee1 regulation in the conveyance of Chk1-dependent checkpoint signals through to Cdc2 to allow time for DNA repair prior to mitotic commitment.
At this stage, the identity of the human homolog of Cdr1 is not clear, although there are several related proteins. However, the high level of conservation of the regulation of the G2/M transition and the G2 DNA damage checkpoint between S. pombe and humans suggests that a functional homolog should exist, and it will be important to establish whether it, too, antagonizes Chk1 signaling. Several studies of human cells have shown that inhibition of Chk1 can greatly sensitize tumor cells to genotoxic stresses, particularly in cells lacking p53 function. Thus, there has been great interest in identifying Chk1 inhibitors to be used as a targeted therapy, particularly in chemoresistant tumors. Should Cdr kinases antagonize Chk1 in human cells, then agonists of Cdr kinases may represent an alternative mechanism to achieve this end.
Acknowledgments
The authors gratefully acknowledge J. Raleigh who was involved in the early aspects of this work. We thank S. Forsburg, P. Young, and H. Shinagawa for reagents and C. Tapia-Alveal, E. Outwin, and B. Laurent for critical reading of the manuscript. This work was supported by National Institutes of Health/National Cancer Institute grant CA100076 and a fellowship from the Spanish Secretaría de Estado de Educación y Universidades and assisted by the Fondo Social Europeo (T.C.) and a scholarship of the Leukemia and Lymphoma Society (M.O.).
References
- Barbet, N. C., and A. M. Carr, 1993. Fission yeast wee1 protein kinase is not required for DNA damage-dependent mitotic arrest. Nature 364: 824–827. [DOI] [PubMed] [Google Scholar]
- Barbet, N., W. J. Muriel and A. M. Carr, 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114: 59–66. [DOI] [PubMed] [Google Scholar]
- Barr, F. A., H. H. Sillje and E. A. Nigg, 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5: 429–440. [DOI] [PubMed] [Google Scholar]
- Bimbo, A., Y. Jia, S. L. Poh, R. K. Karuturi, N. den Elzen et al., 2005. Systematic deletion analysis of fission yeast protein kinases. Eukaryot. Cell 4: 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeding, C. S., J. Hudson, M. K. Balasubramanian, S. M. Hemmingsen, P. G. Young et al., 1998. The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell 9: 3399–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., T. H. Liu and N. C. Walworth, 1999. Association of Chk1 with 14–3-3 proteins is stimulated by DNA damage. Genes Dev. 13: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, T. R., Z. Tang and W. G. Dunphy, 1993. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell 72: 919–929. [DOI] [PubMed] [Google Scholar]
- den Elzen, N. R., and M. J. O'Connell, 2004. Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J. 23: 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy, W. G., 1994. The decision to enter mitosis. Trends Cell Biol. 4: 202–207. [DOI] [PubMed] [Google Scholar]
- Feilotter, H., P. Nurse and P. G. Young, 1991. Genetic and molecular analysis of cdr1/nim1 in Schizosaccharomyces pombe. Genetics 127: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, A. M., L. S. White, P. J. Donovan and H. Piwnica-Worms, 2005. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol. Cell. Biol. 25: 2853–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S. L., 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21: 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari, B., A. Blasina, M. N. Boddy, C. H. McGowan and P. Russell, 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V., K. Furuya and A. M. Carr, 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Rep. 4: 1227–1239. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., A. M. Quinn and T. Hunter, 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52. [DOI] [PubMed] [Google Scholar]
- Kanoh, J., and P. Russell, 1998. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell 9: 3321–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely, J., S. B. Haase, P. Russell and J. Leatherwood, 2000. Functions of fission yeast orp2 in DNA replication and checkpoint control. Genetics 154: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, J., T. Kwong, F. Altruda, D. Soll and G. Wahl, 1979. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J. Biol. Chem. 254: 1546–1551. [PubMed] [Google Scholar]
- Latif, C., N. R. Elzen and M. J. O'Connell, 2004. DNA damage checkpoint maintenance through sustained Chk1 activity. J. Cell Sci. 117: 3489–3498. [DOI] [PubMed] [Google Scholar]
- Lee, J., A. Kumagai and W. G. Dunphy, 2001. Positive regulation of Wee1 by Chk1 and 14–3-3 proteins. Mol. Biol. Cell 12: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., and J. M. Bishop, 1990. A putative protein kinase gene (kin1+) is important for growth polarity in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87: 8272–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Y. Wang, Y. Sun and T. S. Lawrence, 2002. Wild-type TP53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat. Res. 157: 322–330. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., B. Furnari, O. Mondesert and P. Russell, 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14–3-3 protein. Nature 397: 172–175. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., J. Kanoh and P. Russell, 2001. Nuclear exclusion of Cdc25 is not required for the DNA damage checkpoint in fission yeast. Curr. Biol. 11: 50–54. [DOI] [PubMed] [Google Scholar]
- Lu, X., T. A. Nguyen and L. A. Donehower, 2005. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle 4: 1060–1064. [PubMed] [Google Scholar]
- Lundgren, K., N. Walworth, R. Booher, M. Dembski, M. Kirschner et al., 1991. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Maundrell, K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- McMillan, J. N., M. S. Longtine, R. A. Sia, C. L. Theesfeld, E. S. Bardes et al., 1999. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19: 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Morikawa, H., T. Morishita, S. Kawane, H. Iwasaki, A. M. Carr et al., 2004. Rad62 protein functionally and physically associates with the smc5/smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 24: 9401–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P., 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- O'Connell, M. J., J. M. Raleigh, H. M. Verkade and P. Nurse, 1997. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 16: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, M. J., N. C. Walworth and A. M. Carr, 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10: 296–303. [DOI] [PubMed] [Google Scholar]
- Ohi, R., D. McCollum, B. Hirani, G. J. Den Haese, X. Zhang et al., 1994. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 13: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, L. L., S. A. Walter, P. G. Young and H. Piwnica-Worms, 1993. Phosphorylation and inactivation of the mitotic inhibitor wee1 by the nim1/cdr1 kinase. Nature 363: 736–738. [DOI] [PubMed] [Google Scholar]
- Price, D., S. Rabinovitch, P. H. O'Farrell and S. D. Campbell, 2000. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics 155: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh, J. M., and M. J. O'Connell, 2000. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J. Cell Sci. 113: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse, 1987. The mitotic inducer nim1+ functins in a regulatory network of protein kinase homologs controlling the initiation of mitosis. Cell 49: 569–576. [DOI] [PubMed] [Google Scholar]
- Shulewitz, M. J., C. J. Inouye and J. Thorner, 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, C. S., R. G. Syljuasen, J. Falck, T. Schroeder, L. Ronnstrand et al., 2003. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3: 247–258. [DOI] [PubMed] [Google Scholar]
- Stanford, J. S., and J. V. Ruderman, 2005. Changes in regulatory phosphorylation of Cdc25C Ser287 and Wee1 Ser549 during normal cell cycle progression and checkpoint arrests. Mol. Biol. Cell 16: 5749–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff, J., T. Duncan, E. Homola, S. D. Campbell and T. T. Su, 2004. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr. Biol. 14: 2143–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth, N., S. Davey and D. Beach, 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363: 368–371. [DOI] [PubMed] [Google Scholar]
- Wu, L., and P. Russell, 1993. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature 363: 738–741. [DOI] [PubMed] [Google Scholar]
- Wu, L., K. Shiozaki, R. Aligue and P. Russell, 1996. Spatial organization of the Nim1-Wee1-Cdc2 mitotic control network in Schizosaccharomyces pombe. Mol. Biol. Cell 7: 1749–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z., Z. Chen, A. H. Gunasekera, T. J. Sowin, S. H. Rosenberg et al., 2003. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 278: 21767–21773. [DOI] [PubMed] [Google Scholar]
- Young, P., and P. Fantes, 1987. Schizosaccharomyces pombe mutants affected in their division response to starvation. J. Cell Sci. 88: 295–304. [DOI] [PubMed] [Google Scholar]