Abstract
In Saccharomyces cerevisiae, silencers flanking the HML and HMR loci initiate the establishment of transcriptional silencing. We demonstrate that the activity of a silencer pertaining to its potency and directionality is dependent on its genomic position. The context of the HML-E silencer is more permissive to silencer function than that of HML-I or HMR-E, despite that HML-E and HML-I are only 3.3 kb apart. The apparent strength and directionality of a silencer in a particular location is affected by other silencing elements (silencers and protosilencers) present in its context. We show that at the HML locus, at least four silencing elements engage in multiple functional interactions that contribute to the activities of the silencers. Notably, these dispersed silencing elements can synergize to silence genes located not only inside, but also outside the HML sequence that harbors them. Moreover, the relative positions and orientations of these elements are important for silencing, indicating that they belong to an intricate silencing network.
IN the yeast Saccharomyces cerevisiae, the HML and HMR loci and regions near the telomeres are transcriptionally silenced via the formation of a repressive chromatin structure that is akin to metazoan heterochromatin (Moazed 2001a; Grewal and Moazed 2003). Silencing at the HM loci is initiated by small specialized flanking sequences called the HML-E and -I and HMR-E and -I silencers that are composed of various combinations of two or three of the binding sites for Abf1p, Rap1p, and origin recognition complex for DNA replication (ORC) (Figure 1A) (Rusche et al. 2003). The silencer-binding proteins initiate silencing by recruiting the Sir silencing complex consisting of Sir2p–Sir4p through physical interactions. Sir2p is an NAD-dependent histone deacetylase that is believed to be responsible for the characteristic histone hypoacetylation associated with silent chromatin (Moazed 2001b; Rusche et al. 2003). Once recruited to the silencer, the Sir complex is thought to deacetylate nearby nucleosomes. As the Sir complex self-interacts and preferentially binds hypoacetylated histones (Carmen et al. 2001; Liou et al. 2005), additional Sir complexes are recruited to the newly deacetylated nucleosomes. The nucleosome-bound Sir complex then starts another round of nucleosome deacetylation and Sir complex binding. In this manner, the Sir complex is proposed to promote autonomous stepwise propagation along an array of nucleosomes, leading to the formation of transcriptionally silent chromatin across the HM loci (Moazed 2001a; Grewal and Moazed 2003; Rusche et al. 2003). The telomeric repeats contain multiple binding sites for Rap1p that can recruit Sir proteins, and propagation of the Sir proteins inward along the chromosome leads to the formation of silent chromatin in regions near the telomeres (Rusche et al. 2003).
Figure 1.—
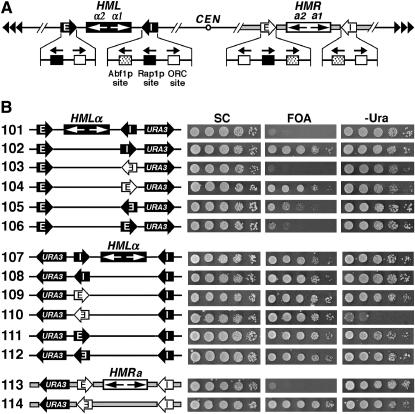
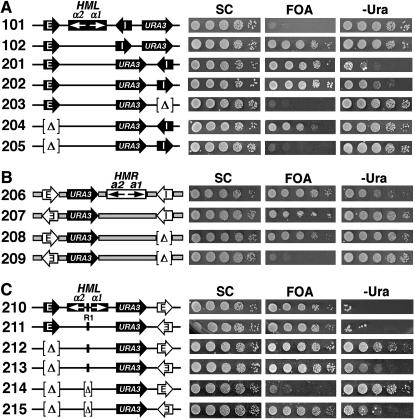
The apparent directionality of a silencer observed in a specific experiment depends on the genomic location of the silencer as well as the sensitivity of the silencing assay. (A) Schematics of the HML and HMR loci on chromosome III in S. cerevisiae. The HML-E and HML-I silencers are shown as solid arrows with white letters, and the HMR-E and HMR-I silencers are shown as open arrows with black letters. The direction of each silencer sequence is drawn as pointing toward the HMLα or HMRa genes, but is not necessarily the functional direction. The binding sites for Abf1p, Rap1p, and ORC in the silencers as well as their 5′ → 3′ directions are indicated. In HML-E, 16 bp centromere-proximal to the ORC site is a putative Sum1p-binding site whose deletion has been shown to reduce the activity of a weakened HML-E silencer (Irlbacher et al. 2005). The α1 and α2 genes at the HML locus and the a1 and a2 genes at HMR are also indicated. CEN, centromere. Tandem arrowheads, telomeric repeats. The regions spanning the HMR locus and flanking sequences are indicated by thick shaded lines. (B) Effects of the genomic contexts of HML-I and HML-E on the directionality of silencers. Left, the modified HML or HMR loci in strains 101–114 (see materials and methods for their construction). The silencers, HM genes, and the URA3 reporter gene are indicated. Cells of each strain were grown to late log phase and serial 10-fold dilutions were spotted on test plates and allowed to grow for 3 days. SC, synthetic complete medium. 5-FOA, SC supplemented with 1 mg/ml 5-fluoroorotic acid (5-FOA). −Ura, SC depleted of uracil. Growth phenotypes of each strain are shown on the right.
Although yeast silencers function through a common mechanism, they exhibit different efficiencies in silencing. At the HMR locus, HMR-E can silence the HMRa genes on its own, whereas HMR-I plays no, or at most an auxiliary, role (Brand et al. 1985; Rivier et al. 1999). At the HML locus, on the other hand, either HML-E or HML-I alone is sufficient to silence the HMLα genes (Mahoney and Broach 1989). When ectopically inserted near the MAT locus, HMR-E was shown to be stronger than HML-E, which itself was stronger than HML-I in silencing activity (Shei and Broach 1995). A possible explanation for the hierarchy of the silencers concerning their potencies of silencing is that each silencer consists of a unique sequence. Although silencers are all composed of combinations of binding sites for Abf1p, Rap1p, and ORC, the actual sites for a specific factor in different silencers are variants of a consensus sequence and therefore may have distinct affinities for the corresponding factor. In fact, it has been shown that Rap1p binds HMR-E more tightly than HML-I (Boscheron et al. 1996). Moreover, the organization of the factor-binding sites regarding the spacing of the sites is unique in each silencer. These may collectively contribute to the difference in the potency of silencing among silencers.
The HML and HMR loci are located relatively close (∼11 and 23 kb) to the left and right telomeres of chromosome III, respectively. Yeast telomeres tend to cluster at the nuclear periphery, thereby forming subnuclear compartments/foci that are rich in Sir proteins. (Taddei and Gasser 2004). Consequently, the HM silencers are believed to be in the vicinity of Sir-rich foci due to their proximity to telomeres. In accord with this, moving HM loci to locations far away from the telomeres, or placing them in plasmids, generally results in a reduction or elimination of silencing (Feldman et al. 1984; Thompson et al. 1994; Shei and Broach 1995; Maillet et al. 1996). Moreover, there is evidence for a direct interaction between HML and the left telomere of chromosome III, which requires the HML silencers (Lebrun et al. 2003).
Two silencers separated by a distance of up to 4 kb can cooperate to bring about silencing in the region they bracket that is stronger than that promoted by either silencer alone (Feldman et al. 1984; Boscheron et al. 1996; Rivier et al. 1999; Sekinger and Gross 1999). Moreover, a single binding site for any of the silencer-binding proteins can enhance the activity of a distant silencer without acting as a silencer on its own and is therefore referred to as a protosilencer (Boscheron et al. 1996). In known cases of silencer–protosilencer interactions, silencing of genes located in the region flanked by the silencer and protosilencer was found to be increased (Boscheron et al. 1996; Cheng and Gartenberg 2000; Lebrun et al. 2001). Silencers and protosilencers are collectively called silencing elements. How two silencing elements collaborate to induce stronger silencing has not been resolved.
The current model for the establishment of silencing implies that initiation of silencing does not discriminate against either direction. This is consistent with the finding that the HMR-E silencer silences the a1 gene at HMR in an orientation-independent manner (Brand et al. 1985). However, this notion is challenged by our discovery that the HML-I silencer acts only in one direction despite its close similarity to HMR-E in structure (Figure 1A) (Bi et al. 1999). Moreover, HMR-E ectopically inserted near the MAT locus preferentially functions in one direction (Shei and Broach 1995). What then determines the directionality of a silencer? It is possible that inherent structural characteristics of a silencer determine its directionality in silencing. Every silencer is clearly asymmetric with an order of ORC–Rap1p–Abf1p sites for both HML-I and HMR-E and Rap1p–ORC sites for HML-E (Figure 1A). We have recently shown that such a structural asymmetry leads to an asymmetric organization of nucleosomes around the silencer (Zou et al. 2006). Consistent with the notion that a continuous array of nucleosomes is required for the spreading of silent chromatin (Rusche et al. 2003; Bi et al. 2004), we found that the different patterns of nucleosome distribution on the two sides of a silencer coincide with unequal potentials for silencing (Zou et al. 2006). These results suggest that the two sides of a silencer are inherently associated with unequal potentials for transcriptional silencing.
In this work, we explored why silencers appeared to exhibit distinct directionalities and potencies in silencing as reported in different studies. To avoid potential complications that might result from comparing data obtained using different assays, we examined the activities of silencers using the same URA3 reporter gene in the same context of HML-E, HML-I, or the HMR-E silencer. Our results revealed that the function of a silencer regarding both its directionality and potency is dependent on its genomic context, as a consequence of functional interactions between the silencer and other silencing elements present in its surroundings. We found that the context of the HML-E silencer was significantly more permissive to the function of a silencer than that of HML-I and HMR-E. We also showed that at the HML locus, at least four dispersed silencing elements could synergize to silence genes located not only inside, but also outside the HML sequence. Moreover, the relative positions and orientations of these elements were important for silencing, indicating that they formed an intricate silencing network.
MATERIALS AND METHODS
Plasmids and strains:
Plasmid pUC26 was made by inserting the BamHI–HML–BamHI fragment (coordinates 9666–16,263 of chromosome III) into pUC19. Plasmid pRS416-HMR was made by inserting the HindIII–HMR–HindIII sequence (289,227–294,210) into pRS416. A 1.1-kb URA3 sequence (Bi et al. 1999) was inserted at the EcoRV site (15,411) of pUC26 to make p101. The HML-I sequence (14,561–14,838) in pUC26 was replaced by a HindIII site to make pYC61. URA3 was inserted at the EcoRV site of pYC61 to make p303. HML-I in the opposite direction was inserted at HindIII of p303, resulting in p102. The HMR-E sequence (291,276–291,539) in either orientation was inserted at the HindIII site of p303, resulting in p103 and p104, respectively. The HML-E sequence (11,172–11,295) in either orientation was inserted at the HindIII site of p303, resulting in p105 and p106, respectively. URA3 was inserted at the Bsu36I site (10,979) of pUC26, resulting in p111. The AflII–HML-E–EcoRI fragment of p111 was inverted to make p112 and replaced by HML-I or HMR-E in either direction to make 107–110, respectively. Plasmid p113 was made by inserting URA3 at the SpeI site (291,110) of pRS416-HMR. The HMR-E sequence in p113 was replaced by a BamHI site, resulting in p114m. An inverted HMR-E was inserted at the BamHI site of p114m, making p114. The PstI–HML–URA3–PstI fragment in p101 or p102 was inverted, making p115 or p116, respectively.
Plasmid p201 was derived from pUC26 by inserting URA3 at its PvuII site (14,441). The HML-I sequence of p210 was replaced by a HindIII site, resulting in p203. An inverted HML-I was inserted at the HindIII site of p203, making p202. The HML-E sequence in p201 and -202 was replaced by an AflII site, making p204 and -205, respectively. The HMR-E sequence in either direction was inserted at the HindIII site of p203, making p210 and p211, respectively. The HML-I silencer in p204 was replaced by HMR-E in either direction, making p212 and 213, respectively. The UASα sequence (12,985–13,279) of p212 and p213 was replaced by a HindIII site, making p214 and p215, respectively.
The HML-E sequence in p101–p104 was replaced by an AflII site, making p301, p302, p304, and p305, respectively. The HML-I sequence in p101 was replaced by a HindIII site, making p303. The HML-I sequence in p109 or p110 was replaced by a HindIII site, making p306 and p307, respectively. UASα was deleted from p102, p104, and p305, making p401, p402, and p403, respectively. The Rap1p-binding site R2 (14,538–14,550) was deleted from p101–p104, making p405, p406, p408, and p409, respectively. R2 was deleted from p403, making p404. HML-I plus R2 was inserted at the HindIII site of p303, making p407. The AflII–HML–HindIII sequence in p101 and p102 was inverted, making p410 and 411, respectively. The AflII–HML–HindIII sequence in p101–p104 was replaced by the HMR sequence (291,539–293,577), making p501–p504, respectively. Plasmid pMB21 is an integration plasmid containing SIR3-TRP1 and SUP4-o (Bi et al. 1999).
All strains used in this study were derived from DMY2 (MATa ura3-52 leu2-3,112 ade2-1 lys1-1 his5-2 can1-100 sir3∷LEU2) (Mahoney and Broach 1989). Strains 101s–112s, 115s, 116s, 201s–205s, 210s–215s, 301s–307s, 401s–411s, and 501s–504s were constructed by transforming strain DMY2 to Ura+ with BamHI-digested plasmids p101–p112, p115, p116, p201–p205, p210–215, p301–p307, p401–p411, and p501–p504, respectively. Strains 113s, 114s, and 206s–209s were constructed by transforming DMY2 to Ura+ with HindIII-digested plasmids p113, p114, and p206–p209, respectively. These strains were rendered SIR3+ by integrating pMB21 at TRP1 in the genome, resulting in strains 101–116, 201–215, 301–307, 401–411, and 501–504, respectively, as illustrated in Figures 1–7. Strains 101r–104r and 303r–305r were derived from 101–104 and 303–305, respectively, by replacing the coding sequence of PPR1 with kanMX. The relevant genotypes of all strains were confirmed by Southern blotting.
Figure 2.—
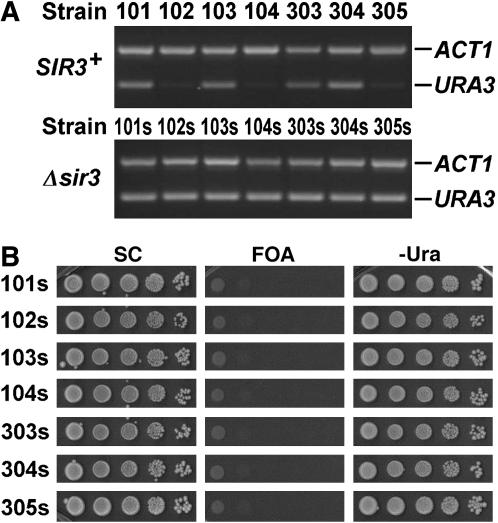
Sir-dependent URA3 silencing in representative strains. (A) The abundance of URA3 mRNA in strains 101–104 and 303–305 and their sir3− derivatives 101s–103s and 303s–305s grown to log phase in SC medium was measured by RT–PCR. The abundance of ACT1 mRNA in each strain was simultaneously measured as an internal control. (B) Shown are growth phenotypes of the sir3− strains 101s–103s and 303s–305s on SC, 5-FOA, and −Ura.
Figure 3.—
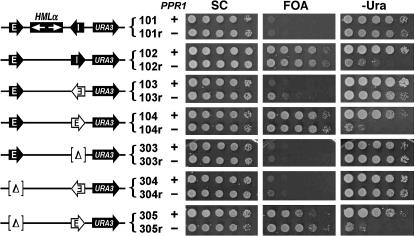
Examination of silencer function by measuring basal URA3 silencing in the absence of PPR1. The growth phenotypes of strains 101–104 and 303–305 and their ppr1− derivatives 101r–104r and 303r–305r on SC, 5-FOA, and −Ura are shown.
Figure 4.—
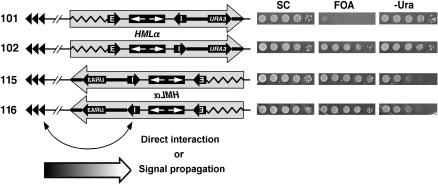
Effect of inverting HML and flanking sequences on the activity of HML-I. A sequence (large shaded arrow) encompassing HML and ∼1 kb of left (zigzag line) and right (thick line) flanking sequences, as well as URA3 inserted 600 bp from HML-I, in strains 101 and 102 was inverted to make 115 and 116. Growth phenotypes of strains 101, 102, 115, and 116 on SC, 5-FOA, and −Ura media are shown on the right. Possible means of interaction between the left telomere of chromosome III (TEL III-L) and silencers at HML are shown at the bottom. Curved arrow, direct interaction. Large arrow at the bottom, propagation of a signal along the chromosome.
Figure 5.—
Silencing elements in the genomic context of a silencer regulate its potency and directionality. (A) HML-E affects the apparent directionality of HML-I. Left, strains used. Right, growth phenotypes of these strains on SC, 5-FOA, and −Ura media. [Δ] denotes the deletion of a sequence. (B) HMR-I affects the apparent directionality of HMR-E. Left, strains used. Right, growth phenotypes. (C) Effect of UASα on the directionality of HMR-E in the context of HML-I. The Rap1p-binding site (designated R1) in UASα (upstream activating sequence of the α1 and α2 genes) is indicated by a bar in strains 210–213. Note that R1 is highlighted only in certain relevant strains in this report. UASα was deleted in strains 214 and 215. Growth phenotypes of strains 210–215 are shown on the right.
Figure 6.—
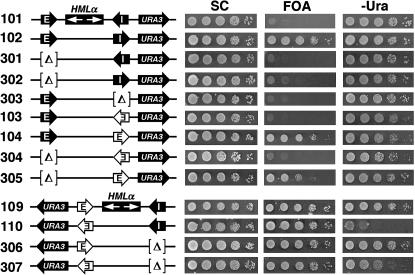
Two silencers separated by a sizable distance can synergize to promote silencing outside the region they bracket. Schematics of the modified HML loci in strains 301–307 as well as 101–104, 109, and 110 are shown on the left. Symbols used are as described in Figures 1 and 2. Growth phenotypes of these strains are shown on the right.
Figure 7.—
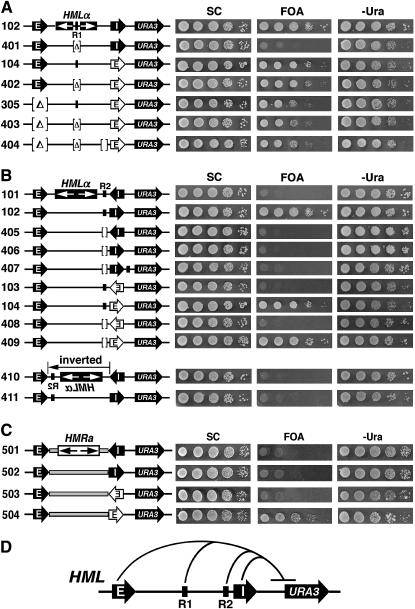
Multiple dispersed silencing elements can cooperate to promote silencing outside the region harboring them. (A) The protosilencer R1 within the HML sequence is involved in silencing outside of HML. Left, strains used. Right, growth phenotypes. (B) The Rap1p-binding site near HML-I in HML (designated R2) is involved in silencing outside of HML. Left, strains used. Strains 410 and 411 are identical with 101 and 102, respectively, except that the HML sequence bracketed by the HML-E and -I silencers is inverted. Note that R2 is highlighted only in certain relevant strains in this report. Right, growth phenotypes. (C) Effects of replacing the HML sequence with the HMR sequence on silencing outside of HML. The HMR sequence excluding the HMR silencers (coordinates 291,539–293,532 of chromosome III) indicated by shaded rectangles was used to replace the HML sequence excluding the HML silencers in strains 101–104, making strains 501–504. (D) Four dispersed silencing elements at HML synergize to promote silencing outside HML. The HML-E and inverted HML-I silencers as well as the R1 and R2 protosilencers were all required for the silencing of URA3 located to the right of HML. This is a summary of results concerning strains 102, 302, 303, 401, and 406 described in Figures 1 and 5–7.
RT–PCR:
Cells were grown at 30° to log phase in SC medium. Total RNA was isolated from ∼5 × 107 cells and 0.4 μg of RNA was used for multiplex RT–PCR with a SuperScript III one-step RT–PCR system with Platinum Taq DNA Polymerase (Invitrogen, San Diego). The proper concentration of RNA template for RT–PCR had been determined to be in the linear range by serial dilutions. Oligo(dT)(20) was used for the cDNA synthesis step and additional dNTPs were added to accommodate multiple PCR products. URA3 and ACT1 ORF primers were used to generate PCR products of 300–550 bp that were fractionated on 2.0% agarose gels.
RESULTS
The apparent directionality of a silencer measured in an experiment is dependent on the sensitivity of the assay used:
The URA3 gene is a widely used reporter for studying transcriptional silencing (van Leeuwen and Gottschling 2002). Ura3p is involved in pyrimidine biosynthesis and can convert 5-fluoroorotic acid (5-FOA) to a toxic metabolite (Boeke et al. 1987). In the presence of uracil, URA3 is expressed at a low basal level, whereas uracil depletion activates the Ppr1p transactivator, thereby increasing the expression of URA3 to a higher activated level (Losson and Lacroute 1981). Therefore, while cell growth on 5-FOA-containing medium (which also contains uracil) indicates the repression of the basal level transcription of URA3, lack of growth on medium depleted of uracil (−Ura) reflects the silencing of activated expression of URA3 (van Leeuwen and Gottschling 2002). In the absence of Ppr1p, URA3 cannot be expressed at the higher activated level even when uracil is depleted from the medium. Therefore, for ppr1− cells, growth on 5-FOA and lack of growth on −Ura media would both indicate the silencing of basal expression of URA3.
By measuring the repression of basal expression of URA3 inserted near HML-I we showed previously that HML-I initiated silencing in only one direction (Figure 1B, compare growth phenotypes of strains 101 and 102 on 5-FOA) (Bi et al. 1999). HMR-E ectopically placed in the position of HML-I also functioned unidirectionally (Figure 1B, growth phenotypes of strains 103 and 104 on 5-FOA) (Zou et al. 2006). We showed here that HML-E inserted in the position of HML-I only weakly silenced basal URA3 expression in one direction (Figure 1B, 105 and 106 on 5-FOA).
To complement the assay of URA3 silencing by monitoring cell growth on 5-FOA, we also directly measured the level of URA3 mRNA, using RT–PCR in a representative set of strains used in this work. URA3 mRNA was abundant in strains 101, 103, 303, and 304, but was barely detectable in strains 102, 104, and 305 (Figure 2A). These results are consistent with the growth phenotypes of strains 101–104 and 303–305 on 5-FOA (Figure 1B, 101–104 on 5-FOA; Figure 6, 303–305 on 5-FOA). Note that URA3 silencing measured in this work was strictly Sir dependent as deletion of SIR3 completely abolished URA3 silencing (Figure 2A, compare URA3 RNA in 102, 104, and 305 to that in their sir3− derivatives 102s, 104s, and 305s, respectively; Figure 2B, lack of growth of the sir3− strains on 5-FOA; and data not shown).
We also examined the directionality of silencing of silencers, using activated expression of URA3 as the reporter. Interestingly, none of the silencers in either direction (in the context of HML-I) was able to silence the activated expression of URA3 as indicated by the robust growth of all strains 101–106 on −Ura (Figure 1B). Since the activated expression of URA3 is mediated by the trans-activator Ppr1p, it should not occur if PPR1 is deleted. Therefore, silencing of URA3 under noninducing conditions could be better examined in the absence of PPR1. We deleted PPR1 from the aforementioned set of seven representative strains (Figure 2A), resulting in strains 101r–104r and 303r–305r (Figure 3, left). URA3 silencing in these strains was measured by monitoring their growth phenotypes on 5-FOA and −Ura and comparing them to that in their PPR1+ parents (Figure 3). As shown in Figure 3, unlike strains 102 and 104, their ppr1− derivatives 102r and 104r were not able to grow on −Ura, which confirmed that Ppr1p-independent basal expression, but not Ppr1p-dependent activated expression of URA3, could be silenced by HML-I or HMR-E orientated toward the URA3 gene.
Results from the above experiments suggest that the outcome of a silencing experiment depends on the sensitivity of the reporter gene to the silencing machinery. Throughout this report, both the basal and the activated expressions of URA3 were used as silencing reporters to examine the potency and directionality of silencers.
The apparent potency and directionality of a silencer are dependent on its genomic context:
To investigate whether the directionality of a silencer is influenced by its genomic context, we compared the behaviors of a silencer in the contexts of the HML-I, HML-E, and HMR-E silencers (Figure 1B). The basal expression of URA3 inserted to the left (telomere-proximal) of HML was strongly silenced by HML-E in either orientation (Figure 1B, strains 111 and 112 on 5-FOA). This was in contrast to the weak and unidirectional silencing by HML-E transplaced in the position of HML-I (Figure 1B, 105 and 106 on 5-FOA). HML-I and HMR-E in the context of HML-E also efficiently silenced URA3 in an orientation-independent manner (Figure 1B, 107–110 on 5-FOA). Therefore, the context of HML-E, opposite to that of HML-I, allows/facilitates silencers to function robustly and bidirectionally. Notably, whereas HML-E or HML-I in either direction was not able to silence the activated expression of URA3 (Figure 1B, growth of 107, 108, 111, and 112 on −Ura), HMR-E was able to do so in one orientation (toward URA3) but not the other (Figure 1B, compare 109 and 110 on −Ura). Therefore, the directional nature of HMR-E silencing was revealed only when activated expression of URA3 was used as the reporter. These results suggest that, compared to the context of HML-I, the context of HML-E increases that efficiency of silencing on both sides of a silencer so that basal URA3 expression could be silenced independent of the orientation of the silencer. However, the strengths of silencing on the two sides of the silencer were likely still unequal. This was manifested by the fact that HMR-E silenced activated URA3 expression in only one direction. We also examined the function of HMR-E in its native position at HMR using URA3 as the reporter. URA3 inserted to the left of HMR-E was not silenced (Figure 1B, lack of growth of strain 113 on 5-FOA). However, URA3 was silenced when HMR-E was inverted to face URA3 (Figure 1B, 114 on 5-FOA). On the other hand, HMR-E in either direction was not able to silence activated expression of URA3 (Figure 1B, 113 and 114 on −Ura).
In summary, the above results suggest that each silencer is inherently unidirectional (presumably due to its asymmetric structural features), but may promote bidirectional silencing if placed in a context that can increase the efficiency of silencing on the “disfavored” side of the silencer to a level comparable to that on the other side. They also show that the apparent directionality of a silencer measured in a particular experiment depends on the sensitivity/resolution of the assay employed.
High activity of a silencer in the context of HML-E is correlated with its relative proximity to the telomere:
Results presented in Figure 1B demonstrated that the context of HML-E is clearly more permissive to silencer function than those of HML-I and HMR-E (e.g., compare 103 and 113 to 109 on 5-FOA). It was possible that the flanking sequences of HML-E acted to enhance silencing by HML-E or any other silencer ectopically inserted there, whereas the flanking sequences of HML-I or HMR-E failed to do so. In an attempt to test this hypothesis, we inverted the sequence encompassing the entire HML locus plus ∼1 kb flanking sequence on each side (Figure 4, zigzag and thick lines), together with the URA3 gene inserted to the right of HML-I in strains 101 and 102 (Figure 4, large shaded arrow), resulting in strains 115 and 116 (Figure 4). Interestingly, although this inversion did not change the local context of HML-I, or the position and orientation of URA3 relative to HML-I (Figure 4, left, compare 115 and 116 to 101 and 102, respectively), it greatly improved URA3 silencing by HML-I in either orientation (Figure 4, compare 115 to 101 on 5-FOA, as well as 115 and 116 to 101 and 102, respectively, on −Ura). Because the telomere-proximal flanking sequence of the resident HML-E (Figure 4, zigzag line) was not in the vicinity of HML-I in strains 115 or 116, it was probably not involved in the enhancement of the activity of HML-I.
Inversion of HML and its flanking sequences (indicated by the large shaded arrows in Figure 4) shortened the distance between HML-I and the left telomere of chromosome III (TEL III-L) by ∼3.3 kb. It is possible that TEL III-L enhances silencer function at HML by directly interacting with the silencer (Figure 4, bottom), which is in line with the evidence for physical contact between HML and TEL III-L (Lebrun et al. 2003). However, if such a looping model were correct, then it would be hard to imagine why TEL III-L enhanced the function of the endogenous HML-E but not HML-I, despite that the two silencers were only 3.3 kb apart. On the other hand, it is also possible that a signal that could positively regulate silencing (e.g., the Sir complex) propagates from TEL III-L toward HML (Figure 4, bottom, large arrow). Because this putative signal may gradually lose strength along its path, it may have a stronger effect on HML-E (or any silencer ectopically placed there) than on HML-I.
The directionality of a silencer can be altered by other silencing elements in its context:
The fact that silencers exhibited distinct directionalities in different chromosomal locations prompted us to investigate what in the context of a silencer regulated its function. We focused on possible elements at the HML locus that can influence the silencing activities of the resident or ectopic silencers. It has been shown previously that the HML-E and -I silencers could cooperate to promote stronger silencing in the HML sequence they bracketed, and the Rap1p-binding site within UASα (the shared regulatory region of the α1 and α2 genes) could serve as a protosilencer to enhance the silencing by HML-E (Feldman et al. 1984; Boscheron et al. 1996; Cheng and Gartenberg 2000). We show below that silencer–silencer and silencer–protosilencer interactions can affect the apparent directionality of a silencer.
As shown in Figure 5A, the basal expression of URA3 inserted between the α1 gene and HML-I was strongly silenced independent of the orientation of HML-I (strains 201 and 202 on 5-FOA). The activated expression of URA3 was strongly silenced when HML-I was orientated toward URA3, but was only moderately silenced when HML-I was in the opposite direction (Figure 5A, 201 and 202 on −Ura). Therefore, URA3 experienced stronger silencing when located within HML than when located to the right of HML-I, but the directionality of HML-I was not changed (Figure 5A, compare 201 and 202 to 101 and 102, respectively, on −Ura). To test whether URA3 silencing in strains 201 and 202 resulted from combined actions of HML-E and -I, we examined the effect of deleting either silencer on silencing. Deletion of HML-I abolished URA3 silencing, demonstrating that HML-E alone was not sufficient to silence URA3 (Figure 5A, 203). In the absence of HML-E, HML-I in its natural direction (toward α1) silenced basal URA3 expression, but failed to do so in the opposite direction (Figure 5A, strains 204 and 205). HML-I in either direction was not able to silence activated expression of URA3 in the absence of HML-E (Figure 3A, 204 and 205 on −Ura). These results indicate that the inability of HML-I to silence URA3 on its ORC side (Figure 5A, 205) could be overcome with the assistance of HML-E (Figure 3A, 202). As HML-E alone was not able to silence URA3 (Figure 5A, 203), silencing of URA3 in strain 202 reflected a synergistic interaction between HML-E and HML-I. These results confirmed that HML-I was a unidirectional silencer (Figure 5A, compare 204 and 205 on 5-FOA) and also demonstrated that HML-E could alter the apparent directionality of HML-I (when the basal expression of URA3 was used as the silencing reporter) (Figure 5A, compare 201 and 202 to 204 and 205 on 5-FOA, respectively).
We next examined whether the apparent directionality of HMR-E was affected by HMR-I at the HMR locus. The basal expression of URA3 inserted between HMR-E and the a2 gene was strongly silenced (Figure 5B, robust growth of 206 on 5-FOA). This was independent of the orientation of HMR-E (Figure 5B, compare 207 and 206 on 5-FOA). However, when HMR-I was deleted, HMR-E silenced URA3 only in one direction (Figure 5B, compare 208 and 209 on 5-FOA). Therefore, HMR-I could alter the apparent directionality of HMR-E (when the basal expression of URA3 was used as the silencing reporter). On the other hand, HMR-E significantly silenced the activated expression of URA3 in one direction only when HMR-I was present (Figure 5B, compare 206 and 207 to 208 and 209 on −Ura, respectively).
We also examined if the apparent directionality of HMR-E ectopically inserted at HML was affected by the resident silencing elements. Strains 210 and 211 were derived from 201 by replacing HML-I with HMR-E in opposite directions (Figure 5C, left). Robust silencing of both the basal and the activated expression of URA3 was observed in these strains, which was independent of the orientation of HMR-E (Figure 5C, compare 210 and 211 on 5-FOA and −Ura, respectively). Deletion of HML-E had no effect on the silencing of basal URA3 expression regardless of the direction of HMR-E (Figure 5C, 212 and 213 on 5-FOA). However, it greatly reduced the silencing of activated expression of URA3 when HMR-E was orientated away from it (Figure 5C, compare 212 and 210 on −Ura). As a consequence, HMR-E appeared to act unidirectionally (Figure 5C, compare 212 and 213 on −Ura). Further deletion of UASα containing a Rap1p binding site (designated R1) reduced the efficiency of silencing by HMR-E orientated away from URA3 but not that of HMR-E in the opposite orientation (Figure 5C, compare 214 and 215 to 212 and 213, respectively). Therefore, both the apparent potency and the directionality of HMR-E were affected by the HML-E silencer and the protosilencer R1 at HML. We conclude that the apparent potency and directionality of a silencer are regulated by other silencing elements in its context.
Dispersed silencing elements can act in synergy to promote silencing outside the region harboring them:
Results from the above experiments revealed collaborative interactions among silencers and protosilencers at HML and HMR. These and previous examples of such interactions concerned the silencing of a reporter gene located between the two participating silencing elements (Figure 5; Feldman et al. 1984; Boscheron et al. 1996; Rivier et al. 1999; Cheng and Gartenberg 2000; Lebrun et al. 2001). Here we describe clear cases of two silencers separated by the 3.3-kb HML sequence functioning in synergy to silence a gene located outside HML. As shown in Figure 4, in strain 102 bearing HML-E and the inverted HML-I, URA3 inserted to the right of HML was silenced. However, neither HML-I nor HML-E alone was able to silence URA3 (Figure 6, 302 and 303 on 5-FOA). Therefore, URA3 silencing in strain 102 requires the synergistic interaction of HML-E and inverted HML-I. On the other hand, HML-E and HML-I in its native orientation failed to work together to silence URA3 (Figure 6, strain 101). Robust URA3 silencing was retained when HML-I in strain 102 was replaced by HMR-E orientated toward URA3 (Figure 6, strain 104 on 5-FOA). Deletion of HML-E decreased URA3 silencing (Figure 6, compare 305 to 104 on 5-FOA), indicating that HML-E enhanced the silencing activity of HMR-E in strain 104. These results demonstrate that a silencer in the position of HML-I can collaborate with the HML-E silencer at a distance of 3.3 kb to silence a gene located to the right of the HML sequence. The HMR-E silencer placed in the context of HML-E did not require the assistance of HML-I to fully silence the basal expression of URA3 located to the left of HML (Figure 6, 5-FOA, compare 306 and 307 to 109 and 110, respectively). However, HML-I helped HMR-E promote stronger silencing of the activated expression of URA3 (Figure 6, compare 307 to 110 on −Ura). In summary, these results clearly demonstrated that two silencers separated by the HML sequence can cooperate to silence genes outside of HML more efficiently.
As the Rap1p site R1 in the middle of HML could cooperate with an ectopic HMR-E to silence URA3 within HML (Figure 5C, compare 214 to 212 on 5-FOA), we wondered whether R1 could also collaborate with silencers to silence URA3 outside of HML. To address this question, we deleted UASα containing R1 from strain 102, resulting in strain 401 (Figure 7A). The failure of strain 401 to grow on 5-FOA (Figure 7A) indicated that R1 was also required for URA3 silencing in strain 102. On the other hand, R1 did not seem to be required for URA3 silencing mediated by HMR-E (Figure 7A, compare strains 402 to 104, as well as 403 to 305), which is consistent with the notion that HMR-E is stronger than HML-I.
The fact that R1 was required for the silencing of URA3 outside of HML in strain 102 raised the question of whether the other known Rap1p site within HML was also required. This site (referred to as R2) resides near HML-I (Figure 7B) and only weakly binds Rap1p in vitro (Boscheron et al. 1996). R2 is apparently not required for the silencing of HMLα by the resident HML-I (Feldman et al. 1984). However, we showed that deletion of R2 abolished the silencing of URA3 inserted to the right of the inverted HML-I silencer (Figure 7B, compare strains 102 and 406 on 5-FOA). Therefore, R2, like R1, is also required for the silencing of URA3 outside of HML. On the other hand, R2 was not required for the unidirectional silencing activity of the HMR-E silencer (Figure 7B, compare 408 and 409 to 103 and 104, respectively). In fact, even when all of HML-E, R1, and R2 were simultaneously deleted, HMR-E in place of HML-I was still able to significantly silence URA3 (Figure 7A, 404 on 5-FOA).
To verify the importance of the HML sequence bearing R1 and R2 in aiding the silencing of genes outside HML, we replaced the entire 3.3-kb HML sequence bracketed by HML-E and HML-I with the 2.0-kb HMR sequence that bears no known protosilencers in strains 101–104, resulting in 501–504, respectively (Figure 7C, left). URA3 silencing by the inverted HML-I silencer was abolished (Figure 7C, 502), which was consistent with the fact that R1 and R2 in HML were both necessary for URA3 silencing in strain 102 (Figure 7, A and B, compare 401 and 406 to 102). On the other hand, HMR-E in place of HML-I still significantly silenced URA3 in a unidirectional manner (Figure 7C, strains 503 and 504), which is in line with the fact that HMR-E activity was relatively independent of R1 and R2 (Figure 7, A and B, compare 402 and 409 to 104).
Taken together, the above data demonstrate that dispersed silencing elements can synergize to silence a gene located outside of the region harboring them. Remarkably, in strain 102, at least four silencing elements (HML-E, HML-I, R1, and R2) are required for the silencing of URA3 inserted outside HML (Figures 6 and 7, compare strains 302, 303, 401, and 406 to 102).
The relative positions of the silencing elements are important for their functional interactions:
As at least four silencing elements participated in URA3 silencing in strain 102, it was possible that these elements all contribute to the putative buildup of the “strength” of silencing (perhaps in the form of the abundance of Sir proteins) over a threshold for silencing to occur. If this were the case, then the relative positions of the auxiliary protosilencers R1 and R2 might not be important for silencing. We tested this by precisely inverting the HML sequence in strain 102, making strain 411 (Figure 7B). In strain 411, sequences within HML that were previously close to HML-I were now distant from it, and vice versa. It was clear that inversion of HML abolished URA3 silencing (Figure 7B, compare 411 to 102 on 5-FOA). One possibility was that R2 had to be physically close to HML-I for productive collaborations between the silencing elements. However, relocating R2 to a position near the right side of HML-I also abolished URA3 silencing (Figure 7B, compare strain 407 to 102). Therefore, the mere proximity of R2 to HML-I was not sufficient for its role in promoting URA3 silencing. The above results demonstrated that the relative position of R2 is important for its role in facilitating silencing in strain 102 and suggested that R2 was part of an intricate silencing network consisting of HML-E, HML-I, R1, and R2 (Figure 7D).
DISCUSSION
The principal mechanism for how yeast silencers initiate the formation of transcriptionally silent chromatin has been elucidated (Moazed 2001a; Grewal and Moazed 2003). However, the issue regarding whether and how silencers function in an orientation-dependent or -independent manner has not been completely resolved (Brand et al. 1985; Shei and Broach 1995; Bi et al. 1999). We have recently obtained evidence indicating that a silencer promotes asymmetric positioning of nucleosomes around it, leading to unequal silencing potentials on the two sides (Zou et al. 2006). In other words, silencers are by nature unidirectional. It is therefore puzzling that the same silencer (e.g., HMR-E) could act in an orientation-independent fashion as measured in one experiment, but appear unidirectional in another (Shei and Broach 1995; Brand et al. 1985). Results presented in this work provide two explanations for this conundrum.
The first concerns the distinct sensitivities of silencing assays used to measure silencer activity in different studies. The efficiency of silencing of a particular reporter gene by a silencer depends on the potency of the silencer as well as the strength of the promoter of the reporter gene that is inversely correlated with its sensitivity to the silencing machinery (van Leeuwen and Gottschling 2002). Using both the basal and the activated expression of URA3 as reporters, we found that a silencer may (i) silence both the basal and the activated expressions, (ii) silence the basal but not the activated expression, or (iii) silence neither the basal nor the activated expression. As a consequence, the directionality of a silencer examined in a specific experiment in this work depended on whether the basal or the activated expression of URA3 was used as the silencing reporter. For example, HMR-E inserted in the position of HML-E silenced the basal expression of URA3 on the left of HML in an orientation-independent manner, but silenced activated URA3 expression in only one direction (Figure 1B). Moreover, although HMR-E was able to silence the a1 gene at HMR independent of its orientation (Brand et al. 1985), we found it silenced basal URA3 expression in one direction, but failed to silence activated URA3 expression in either direction (Figure 1B). In addition, despite the directional behavior of HML-I detected by assaying URA3 silencing, the endogenous CHA1 gene located on the disfavored side of HML-I is still subject to Sir-dependent repression in native strains (Moreira and Holmberg 1998). In summary, the directional nature of the function of a silencer may be revealed only by using a silencing assay with sufficient resolution, and, regarding the silencing of native genes, the apparent directionality of a native silencer in its native context is not absolute.
The other explanation was based on our finding that the function of a silencer is influenced by its local genomic context. The directional nature of a silencer may be revealed in a “neutral” context, but may also be “masked” in a context that can increase silencing on the inherently disfavored side of the silencer to a level comparable to that of the preferred side.
We have demonstrated in this report that the activity of a silencer concerning both its potency and directionality can be affected by other silencing elements (silencers, protosilencers, and telomeres) that are present in its context. For instance, the HML-E silencer was able to increase silencing on the disfavored ORC side of HMR-E (in the context of HML-I) to a level comparable to that on its Abf1p side (Figure 5C). In other words, HML-E served to transform HMR-E into a bidirectional silencer. This is one of many examples observed in this work regarding two silencing elements cooperating to promote stronger silencing in the region bracketed by them. Moreover, we also showed that two silencers separated by the 3.3-kb HML sequence could synergize to silence a gene located outside of the region bordered by them (Figure 6). In addition, we found evidence that being close to the left telomere of chromosome III enhances the efficiency of silencing by a silencer (Figure 4).
The mechanism(s) underlying functional interactions among silencing elements have not been resolved, but several models have been proposed (Boscheron et al. 1996; Bi et al. 1999; Fourel et al. 2002). The fact that two silencers separated by up to several kilobases are able to cooperate to silence a reporter located between them can be explained by assuming that convergent spreading of Sir proteins emanating from the silencers is additive or synergistic so that silent chromatin established between the silencers is stronger than that formed by either silencer alone. In support of this model, it was shown that silent chromatin formed by two bracketing HMR-E silencers had a higher density of positioned nucleosomes and thus a more compact chromatin structure than that formed by a single HMR-E (Reimer and Buchman 1997). However, this interpretation does not apply to silencer–protosilencer cooperation since a protosilencer is not able to autonomously initiate silencing. On the other hand, because a protosilencer is actually a binding site for Abf1p, Rap1p, or ORC that can position nucleosomes (Yu and Morse 1999; Lipford and Bell 2001; Bi et al. 2004; Yarragudi et al. 2004; Zou et al. 2006), it is conceivable that a protosilencer helps position nucleosomes in the region between it and the silencer in a configuration that is more favorable for the spread of Sir proteins from the silencer (Boscheron et al. 1996). Along this line, we think that the presence of a protosilencer on the disfavored side of a silencer may alter the putatively inhibitory pattern of nucleosome positioning and allow silencing to occur more efficiently on this side, thereby masking the inherent directionality of the silencer.
The two models discussed above cannot readily explain how a silencer cooperates with another silencer or protosilencer to promote stronger silencing in areas outside the region containing the silencing elements. Ample evidence suggests that yeast telomeres cluster at the nuclear periphery, creating discrete subnuclear compartments that are rich in Sir proteins (Maillet et al. 1996; Taddei and Gasser 2004). Recent evidence indicates that HML silencers physically associate with the left telomere of chromosome III and therefore with a Sir-rich compartment (Lebrun et al. 2003). Therefore, functional cooperation between two silencers could be the consequence of a mutual enhancement of their tethering to a Sir-rich compartment. We have presented a striking example of four silencing elements (silencers HML-E and HML-I and protosilencers R1 and R2) working together to silence a distal gene (summarized in Figure 7D). It is possible that all silencing elements act, at least in part, by contributing to the tethering of the locus to a Sir-rich subnuclear compartment.
Acknowledgments
We thank Lance Johnson and Vyna Nauyen for assistance. This work was supported by National Institutes of Health grant R01 GM62484 to X.B.
References
- Bi, X., M. Braunstein, G. J. Shei and J. R. Broach, 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96: 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, X., Q. Yu, J. J. Sandmeier and Y. Zou, 2004. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24: 2118–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. R. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175. [DOI] [PubMed] [Google Scholar]
- Boscheron, C., L. Maillet, S. Marcand, M. Tsai-Pflugfelder, S. M. Gasser et al., 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15: 2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz and K. Nasmyth, 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41: 41–48. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., L. Milne and M. Grunstein, 2001. Acetylation of the yeast histone H4 terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277: 4778. [DOI] [PubMed] [Google Scholar]
- Cheng, T. H., and M. R. Gartenberg, 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14: 452–463. [PMC free article] [PubMed] [Google Scholar]
- Feldman, J. B., J. B. Hicks and J. R. Broach, 1984. Identification of sites required for repression of a silent mating type locus in yeast. J. Mol. Biol. 178: 815–834. [DOI] [PubMed] [Google Scholar]
- Fourel, G., E. Lebrun and E. Gilson, 2002. Protosilencers as building blocks for heterochromatin. BioEssays 24: 828–835. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and D. Moazed, 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- Irlbacher, H., J. Franke, T. Manke, M. Vingron and A. E. Ehrenhofer-Murray, 2005. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 19: 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, E., E. Revardel, C. Boscheron, R. Li, E. Gilson et al., 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, E., G. Fourel, P. A. Defossez and E. Gilson, 2003. A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol. Cell. Biol. 23: 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz and D. Moazed, 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527. [DOI] [PubMed] [Google Scholar]
- Lipford, J. R., and S. P. Bell, 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7: 21–30. [DOI] [PubMed] [Google Scholar]
- Losson, R., and F. Lacroute, 1981. Cloning of a eukaryotic regulatory gene. Mol. Gen. Genet. 184: 394–399. [DOI] [PubMed] [Google Scholar]
- Mahoney, D. J., and J. R. Broach, 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9: 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet, L., C. Boscheron, M. Gotta, S. Marcand, E. Gilson et al., 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10: 1796–1811. [DOI] [PubMed] [Google Scholar]
- Moazed, D., 2001. a Common themes in mechanisms of gene silencing. Mol. Cell 8: 489–498. [DOI] [PubMed] [Google Scholar]
- Moazed, D., 2001. b Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13: 232–238. [DOI] [PubMed] [Google Scholar]
- Moreira, J. M., and S. Holmberg, 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 17: 6028–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer, S. K., and A. R. Buchman, 1997. Yeast silencers create domains of nuclease-resistant chromatin in a SIR4-dependent manner. Chromosoma 106: 136–148. [DOI] [PubMed] [Google Scholar]
- Rivier, D. H., J. L. Ekena and J. Rine, 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Sekinger, E. A., and D. S. Gross, 1999. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 18: 7041–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shei, G. J., and J. R. Broach, 1995. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol. Cell. Biol. 15: 3496–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., and S. M. Gasser, 2004. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim. Biophys. Acta 1677: 120–128. [DOI] [PubMed] [Google Scholar]
- Thompson, J. S., L. M. Johnson and M. Grunstein, 1994. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol. Cell. Biol. 14: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, F., and D. E. Gottschling, 2002. Assays for gene silencing in yeast. Methods Enzymol. 350: 165–186. [DOI] [PubMed] [Google Scholar]
- Yarragudi, A., T. Miyake, R. Li and R. H. Morse, 2004. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 9152–9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., and R. H. Morse, 1999. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y., Q. Yu and X. Bi, 2006. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]