Abstract
Locomotion is an integral component of most animal behaviors and many human diseases and disorders are associated with locomotor deficits, but little is known about the genetic basis of natural variation in locomotor behavior. Locomotion is a complex trait, with variation attributable to the joint segregation of multiple interacting quantitative trait loci (QTL), with effects that are sensitive to the environment. We assessed variation in a component of locomotor behavior (locomotor reactivity) in a population of 98 recombinant inbred lines of Drosophila melanogaster and mapped four QTL affecting locomotor reactivity by linkage to polymorphic roo transposable element insertion sites. We used complementation tests of deficiencies to fine map these QTL to 12 chromosomal regions and complementation tests of mutations to identify 13 positional candidate genes affecting locomotor reactivity, including Dopa decarboxylase (Ddc), which catalyzes the final step in the synthesis of serotonin and dopamine. Linkage disequilibrium mapping in a population of 164 second chromosome substitution lines derived from a single natural population showed that polymorphisms at Ddc were associated with naturally occurring genetic variation in locomotor behavior. These data implicate variation in the synthesis of bioamines as a factor contributing to natural variation in locomotor reactivity.
LOCOMOTION is an integral component of most animal behaviors: movement is required for localization of food and mates, escape from predators, defense of territory, and response to stress. Locomotor behavior can also be regarded as a major component of fitness, given its central role in survival and reproduction (Gilchrist et al. 1997). Many human neurological diseases (e.g., Parkinson's disease and Huntington's disease) are associated with locomotor deficits, and hyperactivity and hypoactivity are, respectively, associated with activity disorders and depression (American Psychiatric Association 1994). Locomotion is a complex behavior, and variation in nature is likely attributable to the joint segregation of multiple interacting quantitative trait loci (QTL), with effects that are sensitive to the environment. Thus, understanding the genetic architecture of locomotor behavior is important from the dual perspectives of evolutionary biology and human health. However, our current knowledge falls short of the level of detail with which we ultimately seek to describe variation in this trait: Which genes contribute to natural variation in locomotion? What are their homozygous, heterozygous, epistatic, and pleiotropic effects? How do these effects vary in a natural range of environments? And what are the molecular polymorphisms responsible for natural allelic variation? Although to date no complex trait in any organism has been dissected at this level of detail, the greatest opportunity for success will come from genetic analysis of model organisms with excellent genetic and genomic resources, such as Drosophila melanogaster. Further, basic biological processes, including development of the nervous system, are evolutionarily conserved between flies and mammals (Adams et al. 2000). Thus, the same genes affecting Drosophila locomotion may well be relevant in humans.
Indeed, Drosophila provides a model for several human neurodegenerative diseases with marked locomotor impairments. Parkinson's disease is the most common neurodegenerative movement disorder and is associated with progressive degeneration of nigrostriatal dopaminergic neurons and accumulation of Lewy bodies, neuronal cytoplasmic inclusions containing ubiquitinated proteins (Olanow and Tatton 1999). Consistent with a causal role of dopamine in the pathogenesis of Parkinson's disease, treatment with the precursor L-dopa ameliorates the symptoms in early stages of the disease. Dopamine has also been implicated in locomotion of mice (Zhou and Palmiter 1995) and Drosophila.
In Drosophila, tyrosine is converted to L-dopa by tyrosine hydroxylase (encoded by pale), which is then decarboxylated by Dopa decarboxylase (encoded by Dopa decarboxlyase, Ddc) to yield dopamine (Monastirioti 1999). Mutations with reduced levels of dopamine, as well as pharmacological interventions altering dopamine levels, are consistently associated with alterations in locomotor activity, although the direction of the effect is variable. Analysis of lines artificially selected for increased and decreased locomotor activity in an open field environment (Connolly 1966, 1967) indicated an inverse relationship between level of activity and dopamine levels (Tunnicliff et al. 1969). Pharmacologically increasing the level of dopamine in the highly active selected line was associated with a reduction in locomotor activity, but not in the control line, indicating an genotype-by-environment interaction (Connolly et al. 1971). tyrosinase 1 (tyr1) mutants have reduced levels of dopamine and increased levels of spontaneous locomotor activity (Burnell and Daly 1982; Meehan and Wilson 1987). Conversely, mutations in pale are associated with reduced locomotion, and the effect is rescued by administering L-dopa (Pendleton et al. 2002).
The neurotransmitters serotonin (5-hydroxytryptamine, 5-HT), octopamine (the invertebrate homolog of noradrenaline), and γ-aminobutyric acid (GABA) also affect Drosophila locomotion. eagle (eg) mutants have reduced numbers of 5-HT cells in the ventral nerve cord and are inactive (Lundell and Hirsch 1998). inactive (iav) mutants have reduced octopamine levels and are hypoactive (O'Dell and Burnet 1988). Pharmacological treatment of flies with GABA transport inhibitors decreases locomotor activity in a dose-dependent fashion, while cotreatment with a GABA antagonist restores locomotor activity, consistent with the inverse relationship between GABA levels and locomotion observed in vertebrates (Leal and Neckameyer 2002).
The mushroom bodies and components of the central complex (which consists of a network of four neuropilar regions: the protocerebral bridge, fan-shaped body, ellipsoid body, and the paired noduli) control Drosophila locomotor behavior. mushroom body miniature (mbm) mutations lack Kenyon cells and exhibit increased locomotor activity, as do animals in which the mushroom bodies have been chemically or genetically ablated (Martin et al. 1998). Mutations in seven genes with neuroanatomical alterations in the structure of the central complex walk more slowly than wild-type flies. They are less active or quickly lose activity or fail to start walking or flying under circumstances in which wild-type flies would readily do so (reviewed by Strauss 2002). These genes include no bridge (nob, Strauss et al. 1992; Martin et al. 1999), ocelliless (oc, Martin et al. 1999), central complex (cex, Martin et al. 1999), highwire (hiw, Wan et al. 2000), eyeless (ey, Callaerts et al. 2001), single minded (sim, Pielage et al. 2002), C31 (Strauss 2002), and C141 (Strauss 2002). These observations are consistent with the hypothesis that the central complex is required for maintenance of walking activity, while the mushroom bodies act as inhibitors of locomotion (Martin et al. 1999; Strauss 2002).
Locomotor activity is not a single trait, but can (and has) been examined according to the time course of behavior (from minutes to days), differentiating among speed, amount, and directionality of activity and taking external stimuli such as the presence of food or stress of handling into account. Flies tend to react vigorously to a handling disturbance; the activity immediately following such a disturbance (Meehan and Wilson 1987), the time to settle to the endogenous level of activity, the level of expression of locomotor activity at steady state (Connolly 1967), and the amount, speed, and direction of activity at any of these time points (Burnet et al. 1988; Gotz 1989; Strauss et al. 1992; Strauss and Heisenberg 1993) are all measures of activity. However, the various aspects of locomotor behavior are not perfectly correlated, implicating different genetic mechanisms underlying particular measures of activity. For example, the lines selected for activity in an open field environment (Connolly 1966) differed in baseline activity but not in the change in activity over time following introduction to a novel environment (Connolly 1967). Amount and speed of activity are under independent genetic control (Burnet et al. 1988). Further, some mutations affect activity only in a context-dependent manner. Mutations at the period (per) locus affect the circadian rhythm, but not the total amount of activity (Konopka and Benzer 1971). Variants of the foraging (for) gene affect the distance larvae and adults travel away from a food source, but these differences are not expressed in the absence of food (Sokolowski 1980; deBelle and Sokolowski 1987; Pereira and Sokolowski 1993).
There is abundant naturally segregating variation for locomotor activity in Drosophila (Connolly 1966, 1967; van Dijken and Scharloo 1979; Burnet et al. 1988), yet we know virtually nothing of the genes and pathways responsible for this variation. There is a trade-off between quantifying all the aspects of locomotor behavior outlined above and assessing behavior from a large number of strains. Quantitative genetic analysis requires the latter; therefore, we developed a rapid and high-throughput assay to quantify the level of activity immediately following a disturbance (sensu Meehan and Wilson 1987), which we call locomotor reactivity. This is a general behavioral response that includes many constituent and more specific behaviors. Thus, genetic variation in locomotor reactivity could arise from variation in genes affecting the many biological processes affecting the development of the structures required for the execution, perception, and response to the stimulus. Our approach is to use quantitative genetic analysis of natural variants as an unbiased screen for genes and pathways affecting variation in locomotor behavior. Having identified these genes, one can subsequently examine the allelic effects in detail by determining which particular aspects of locomotor behavior are affected. We recognize that this approach is at variance with (and anathema to) that of behavioral geneticists whose goal is to identify genes specifying the neural circuits that provide the potential for specific behaviors, where it is necessary to define and measure highly specific components of behavior (Baker et al. 2001).
We assessed variation in locomotor reactivity in a population of recombinant inbred (RI) lines derived from the wild-type strain Oregon-R (Ore-R) and a strain selected for low male-mating propensity, 2b (Kaidanov 1990), and mapped four QTL affecting this trait by linkage to roo transposable element markers (Nuzhdin et al. 1997). We refined the QTL map positions using quantitative complementation tests of deficiencies, followed by complementation tests of mutations of positional candidate genes (Long et al. 1996; Mackay and Fry 1996; Pasyukova et al. 2000; Fanara et al. 2002; De Luca et al. 2003; Harbison et al. 2004; Moehring and Mackay 2004) to identify 13 candidate genes contributing to the observed variation between the Ore and 2b parental strains. One of the implicated candidate genes is Ddc, which catalyzes both the decarboxylation of L-dopa to dopamine and 5-hydroxytryptophan to serotonin in Drosophila (Monastirioti 1999; Steward 2000). We used linkage disequilibrium mapping (LD) on a sample of 164 alleles from a single population to show that molecular polymorphisms in Ddc are associated with natural variation in locomotor behavior.
MATERIALS AND METHODS
Genome scan for QTL affecting locomotor behavior:
Drosophila stocks:
We mapped QTL affecting variation in locomotor behavior between two isogenic strains: 2b, a derivative of a line originally selected for low male-mating propensity (Kaidanov 1990), and Ore-R, a standard wild-type and unrelated stock (Lindsley and Zimm 1992). We used a mapping population of 98 RI lines previously derived from these strains (Nuzhdin et al. 1997). The genotype of each RI line was determined using 80 roo transposable element markers with polymorphic insertion sites that differ between the parental lines and the fourth chromosome visible mutation shaven (also known as sparkling), giving 81 informative markers with an average spacing of ∼3 cM (Nuzhdin et al. 1997). The roo transposable element markers are located at cytological positions 1B, 3E, 4F, 5D, 6E, 7D, 7E, 9A, 10D, 11C, 11D, 12E, 14C, 15A, 16D, 17C, 19A, 21E, 22F, 27B, 29F, 30AB, 30D, 33E, 34EF, 35B, 38A, 38E, 43A, 43E, 46A, 46C, 48D, 49D, 50B, 50D, 50F, 57C, 57F, 60E, 61A, 63A, 65A, 65D, 67D, 68B, 68C, 69D, 70C, 71E, 72A, 73D, 76A, 76B, 77A, 77E, 78D, 82D, 85A, 85F, 87B, 87E, 87F, 88E, 89B, 91A, 91D, 92A, 93A, 93B, 94D, 96A, 96F, 97D, 97E, 98A, 99A, 99B, 99E, and 100A. We maintained all stocks in vials containing 10 ml cornmeal–agar–molasses medium at 25° under 12-hr light–dark cycles.
Assay to quantify locomotor behavior:
We placed single 3- to 7-day-old adult flies, collected under CO2 exposure, into vials containing 5 ml of standard cornmeal–agar–molasses media and left them overnight to acclimate to their new environment. To quantify locomotor reactivity, we subjected each fly to a mechanical disturbance by tapping the vial twice against a table and recorded the amount of time the fly was active in the 30 sec immediately following the disturbance. This was accomplished by starting a timer and a stopwatch at the beginning of the assay. The timer indicated the duration of the assay. The stopwatch was started and stopped when the fly was active or immobile, respectively, for the duration of the assay. The measure of locomotor reactivity was thus a score ranging from 0 to 30, representing the time in seconds that the fly moved within the assay period, as recorded by the stopwatch. All measurements were taken in a behavioral chamber at the same time of day (8 am–12 pm, 2–6 hr after lights on) under constant temperature (25°) and humidity (75%). Activity scores were obtained for 20 males and 20 females for each genotype tested in a randomized design.
Quantitative genetic analysis:
We evaluated variation in locomotor reactivity among the RI lines using two-way analysis of variance (ANOVA) according to the mixed model Y = μ + L + S + L × S + E, where μ is the overall mean, L and S are the main effects of line (random) and sex (fixed), L × S is the genotype-by-sex interaction term, and E is the within-genotype environmental variance. Broad-sense heritability (H2) for locomotor reactivity in this population of RI lines was computed as 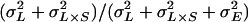 , where
, where 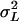 is the among-line variance component,
is the among-line variance component, 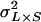 is the variance attributable to the line × sex interaction, and
is the variance attributable to the line × sex interaction, and 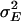 is the within-line environmental variance component. The genetic correlation (rGS) across sexes was calculated as
is the within-line environmental variance component. The genetic correlation (rGS) across sexes was calculated as 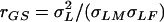 , where
, where 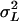 is the among-line variance component from the analysis pooled across both sexes, and σLM and σLF are the square roots of the among-line variance components from the separate analyses of each sex. These ANOVAs and those described below were computed using SAS software (SAS Institute 1988) using the GLM and VARCOMP procedures.
is the among-line variance component from the analysis pooled across both sexes, and σLM and σLF are the square roots of the among-line variance components from the separate analyses of each sex. These ANOVAs and those described below were computed using SAS software (SAS Institute 1988) using the GLM and VARCOMP procedures.
QTL mapping:
We mapped QTL affecting locomotor reactivity by linkage to roo polymorphic markers, using composite interval mapping (CIM) (Zeng 1994) of RI line means as implemented by QTL Cartographer software (Basten et al. 1994, 1999). QTL intervals were detected testing the hypothesis that there is no QTL for activity within the interval between two markers, using multiple regression to control for effects of other QTL. The likelihood-ratio (LR) test statistic is −2 ln(L0/L1), where (L0/L1) is the ratio of the null hypothesis (there is no QTL within the test interval) to the alternate hypothesis (there is a QTL within the test interval). Marker cofactors were chosen for the trait using forward selection–backward elimination stepwise regression. LR test statistics were computed every 0.1 cM using a conditioning window of 10 cM, such that only markers 10 cM away from the markers flanking the test interval were included in the model. To control for multiple testing, we randomly permuted the trait and marker data 1000 times. We computed the maximum LR across all intervals for each permuted data set to generate an empirical distribution of LR statistics under the null hypothesis, where there is no association between the genotypic and phenotypic data. LR statistics computed from the original data that exceeded the 950th LR statistic determined for the permuted data were considered to be significant at an experiment-wise 5% type I significance level (Churchill and Doerge 1994; Doerge and Churchill 1996).
Tests for epistasis:
All possible pairwise epistatic interactions between markers were evaluated using ANOVAs on RI line means according to the model Y = μ + Mi + Mj + S + Mi × S + Mj × S + Mi × Mj + Mi × Mj × S + E, where Mi and Mj are the two focal markers evaluated for epistasis, S is the main effect of sex, and E is the error variance. We evaluated the significance of the Mi × Mj and Mi × Mj × S interactions, using the conservative Bonferroni correction for multiple tests and a Q = 0.05 false-positive discovery rate criterion (Storey and Tibshirani 2003).
High-resolution QTL mapping:
Drosophila stocks:
Deficiency and mutant stocks used in complementation tests were obtained from the Bloomington Drosophila Stock Center and are listed in supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/.
Deficiency complementation mapping:
We used deficiency complementation mapping (Pasyukova et al. 2000) to map the four QTL affecting locomotor reactivity with higher resolution. We chose 61 deficiency stocks (Df/Bal, where Bal indicates a dominantly marked Balancer chromosome), uncovering QTL regions affecting locomotor reactivity. We extended the deficiency complementation analysis to the markers immediately adjacent to those defining the 2-LOD support intervals, because QTL boundaries as determined by CIM can fluctuate slightly, depending on the model parameters (Pasyukova et al. 2000). We crossed males of each of the deficiency stocks to both Ore-R and 2b virgin females and measured locomotor activity, as described above, for 20 males and 20 females from each of the four F1 genotypes of each cross (Df/Ore, Df/2b, Bal/Ore, and Bal/2b). The design was completely randomized for each QTL region. Only females (n = 20/F1 genotype) were tested for the X chromosome deficiencies. All deficiencies exhibiting quantitative failure to complement Ore-R and 2b QTL (see below) were then retested in exactly the same manner, bringing the total sample size to 320 individuals tested for each significant deficiency (40 individuals/sex × two sexes × four F1 genotypes).
Statistical analysis:
The test for quantitative failure to complement compares the difference in mean activity levels of the Df/Ore and Df/2b flies to the difference between the activity of Bal/Ore and Bal/2b individuals. We infer quantitative complementation if the difference in mean activity between Df/Ore and Df/2b genotypes is equal to the difference in mean activity between the Bal/Ore and Bal/2b genotypes. We infer quantitative failure to complement when the difference in activity of the Df genotypes (Df/Ore–Df/2b) is greater than the difference in activity of the Bal genotypes (Bal/Ore–Bal/2b). The statistical analysis of the complementation data is a three-way, fixed-effects ANOVA model: Y = μ + S + L + G + S × L + S × G + L × G + S × L × G + E, where μ is the overall mean, S is the main effect of sex, L is the main effect of the parental line (Ore-R or 2b), G is the main effect of the genotype (Df or Bal), and E is the error variance. Significance of the L × G (and/or S × L × G) interaction is indicative of quantitative failure to complement (and/or sex-specific failure to complement), provided that subsequent tests reveal that (Df/Ore–Df/2b) > (Bal/Ore–Bal/2b). We repeated the complementation tests for deficiencies exhibiting significant failure to complement in the initial experiment. These ANOVAs included a term for the random effect of replicates nested within S × L × G. The QTL locations were initially defined by the breakpoints of the deficiencies exhibiting significant failure to complement, pooled over both replicates. These locations were further delimited by the right and left breakpoints of overlapping nonsignificant (complementing) deficiencies to the left and right, respectively, of the significant deficiency (Pasyukova et al. 2000).
Complementation tests to candidate genes:
Quantitative complementation tests of 27 mutations at 26 candidate genes in the QTL regions defined by deficiency mapping were used to identify putative candidate genes within each QTL. All mutant stocks were heterozygous over balancers, with the exception of nobKS49. The experimental design, sample sizes, and analyses were the same as described above for the deficiency complementation tests, expect for nobKS49, where the control cross utilized the Berlin strain in which the nob mutation was induced. All candidate genes that exhibited significant failure to complement on the initial test were retested, as described above for the deficiency complementation tests.
Natural variation in locomotor reactivity:
Drosophila stocks:
One of the positional candidate genes corresponding to a QTL affecting locomotor reactivity was Ddc (2-53.9, 37C1). Previously, we established isofemale lines from gravid females collected from the Raleigh, North Carolina, farmer's market in 1999. We extracted 164 isogenic second chromosomes from these lines and substituted them into the genetic background of the highly inbred Samarkand (Sam) strain by standard techniques using balancer chromosomes (Lyman et al. 1999; DeLuca et al. 2003). In addition, we substituted the chromosome containing a γ-ray-induced, recessive lethal null Ddc allele (Ddc27) into the Sam genetic background.
Complementation tests:
We used complementation tests to assess whether naturally occurring Ddc alleles potentially contributed to quantitative variation in locomotor reactivity. We crossed each of the 164 second chromosome substitution lines (C2i, i = 1–164) to both Sam; Ddc27 and Sam; Ddc+ stocks (where Ddc+ denotes the Sam Ddc allele) and collected Sam; Ddc27/C2i and Sam; Ddc+/C2i F1 individuals, respectively, from each cross. We measured locomotor reactivity, as described above, for 20 males and 20 females of each F1 genotype. The experimental design was nearly balanced, with a total sample size of 13,068 rather than 13,120 individuals. The experimental design was completely randomized. We analyzed these data using the three-way factorial, mixed-model ANOVA: Y = μ + L + S + C + L × S + L × C + S × C + L × S × C + E, where μ is the overall mean, L is the random main effect of the C2 line, S is the fixed main effect of sex, C is the fixed main effect of the cross (Ddc27or Ddc+), L × S, L × C, S × C, and L × S × C are the interaction terms, and E is the error variance. Significance of the L × C interaction indicates that the difference in the phenotype between second chromosome alleles varies between the mutant and control backgrounds and that wild-derived alleles fail to complement the Ddc mutant allele for locomotor reactivity. Similarly, significance of the L × S × C interaction indicates sex-specific failure of the wild-derived alleles to complement the Ddc mutant allele for locomotor reactivity. In either case, Ddc is a potential candidate gene affecting natural variation in locomotor reactivity.
The total genetic variance of locomotor activity (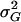 ) in this population was estimated as
) in this population was estimated as 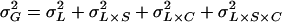 , where
, where 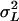 is the among-line variance component,
is the among-line variance component, 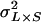 is the variance attributable to the line–sex interaction,
is the variance attributable to the line–sex interaction, 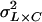 is the line–cross interaction variance, and
is the line–cross interaction variance, and 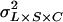 is the three-way line–sex–cross interaction variance. The total phenotypic variance (
is the three-way line–sex–cross interaction variance. The total phenotypic variance (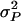 ), was estimated as
), was estimated as 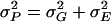 , where
, where 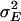 is the environmental variance component.
is the environmental variance component.
Genotype–phenotype associations:
Previously, we genotyped the 164 wild-derived Ddc alleles from the Raleigh population for 36 molecular markers [31 single nucleotide polymorphisms (SNPs) and 5 insertion/deletion polymorphisms] (DeLuca et al. 2003). Two of the marker pairs were in perfect LD, giving 34 different association tests (DeLuca et al. 2003). We assessed whether molecular polymorphisms at Ddc were associated with variation in locomotor reactivity by three-way factorial ANOVA of line means, according to the fixed effects model Y = μ + S + C + M + S × C + S × M + C × M + S × C × M + E, where μ is the overall mean, S is the main effect of sex, C is the main effect of the cross, M is the main effect of the marker (or haplotype), S × C, S × M, C × M, and S × C × M are the interaction terms, and E is the error variance. Significance of the M term indicates that the difference in phenotype between marker alleles varies, suggesting an allelic association in locomotor reactivity with a particular marker. In addition to the single-marker analysis, we also performed a three-way ANOVA to test the effect of global (across all 36 polymophisms) haplotype variation in Ddc on locomotor reactivity using the following model: Y = μ + S + C + H + S × C + S × H + C × H + S × C × H + E, where the main effects of sex and cross are as described above, and H represents the fixed effect of haplotype class (1–103). Significance of the H term indicates differences in phenotype between the global Ddc haplotype classes, suggesting an effect of molecular variation in Ddc on variation of locomotor reactivity.
We used permutation tests to determine empirical distributions under the null hypotheses of no association between Ddc genotypes and locomotor reactivity. To assess whether any of the individual markers were significantly associated with variation in locomotor reactivity, we permuted the trait phenotypes among the 36 markers 1000 times and performed single-marker association analyses on the permuted data. We recorded the lowest P-value for the effect of marker from each permutation run. The 50th lowest P-value corresponds to α = 0.05 under the null hypothesis; any marker with a P-value less than this is individually significant (De Luca et al. 2003). To assess whether we observed more nominally significant marker–phenotype associations than expected by chance, we recorded the number of significant associations at P < 0.05, P < 0.025, and P < 0.01 for each permuted data set and determined whether the observed number of significant markers at each P-value (0.05, 0.025, and 0.01) exceeded the expected number determined by permutation. Finally, to assess whether the observed association of locomotor reactivity with Ddc population haplotypes was less than expected by chance, we permuted the trait phenotypes among the global haplotypes 1000 times and recorded the lowest P-value for the effect of haplotype for each permuted data set.
RESULTS
Genome scan for QTL affecting locomotor behavior:
Quantitative genetic variation in locomotor behavior:
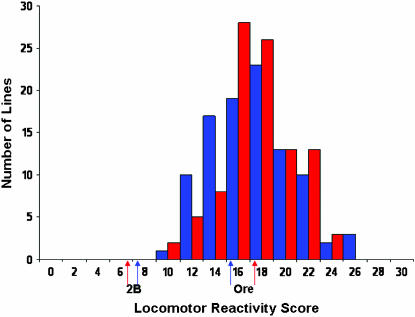
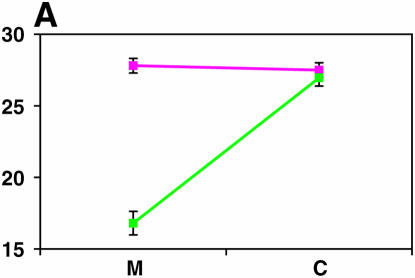
We observed a highly significant difference in locomotor reactivity behavior between the Ore-R and 2b isogenic lines (F1,96 = 156.68, P < 0.0001; Figure 1). The mean (±SE) level of locomotor reactivity of Ore-R was 16.6 ± 0.79 sec (15.3 ± 0.75 sec for males and 18.0 ± 0.83 sec for females), more than twice the mean level of reactivity of the 2b strain, 6.8 ± 0.77 sec (7.9 ± 0.70 sec for males and 6.0 ± 0.84 sec for females). Not surprisingly, there was highly significant genetic variation in locomotor reactivity among the RI lines derived from Ore-R and 2b (F97,3724 = 9.50, P < 0.0001; Figure 1). In addition, the line–sex interaction term was significant (F97,3724 = 1.53, P = 0.0007), indicating variation in sex dimorphism of reactivity in this population of RI lines. The estimate of broad-sense heritability of locomotor reactivity in the RI lines was H2 = 0.26. The correlation in locomotor reactivity between the sexes (rGS = 0.82 ± 0.058) was high and positive, but significantly different from unity. Thus, the effects of genes affecting this trait are not identical in males and females.
Figure 1.—
Variation in locomotor reactivity for males (blue) and females (red). Mean reactivity scores are depicted for the parental strains, Ore-R and 2b, and for the mapping population of 98 RI lines.
QTL mapping:
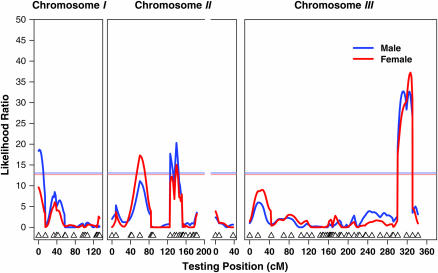
Given the significant genetic variation among the RI lines in locomotor reactivity, we used QTL mapping to identify genomic regions responsible for the observed differences in activity between the parental lines. We used CIM to map four QTL affecting reactivity levels based on permutation-derived significance thresholds at cytological locations 1B–3E, 27B–29E, 30D–38A, and 99A–100A (Figure 2 and Table 1). Consistent with the inference of sex-specific effects on locomotor reactivity from the quantitative genetic analyses, the 1B–3E QTL was formally significant only in males; the 27B–29E QTL was significant only in females; and the remaining two QTL had similar effects in both sexes. The physical regions encompassed by the QTL range from 2600 to 9200 kb and include between 574 and 1553 positional candidate genes (Table 1). It is possible that additional QTL affect variation in locomotor reactivity in this mapping population, but their effects are too small to be detected, given the sample size used in this experiment.
Figure 2.—
QTL for locomotor reactivity. Triangles on the x-axis depict the locations of roo transposable element markers in centimorgans. The y-axis is the LR test statistic. The horizontal lines denote the LR statistics corresponding to permutation-derived experiment-wise 5% significance thresholds, shown separately for males and females.
TABLE 1.
Summary of QTL mapping results
| QTL | Marker | 2-LOD interval | Male LRa | Male a | Maleba/σP | Female LRa | Female a | Femaleba/σP | kbc | No. of locid |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1B | 1B–3E | 18.69* | −1.70 | −0.284 | 9.62 | −0.83 | −0.138 | 3200 | 712 |
| 2 | 27B | 27B–29E | 10.42 | 1.001 | 0.167 | 17.30* | 1.13 | 0.188 | 3200 | 368 |
| 3 | 35BC | 30D–38A | 19.38* | −1.54 | −0.257 | 15.04* | −1.13 | −0.189 | 9200 | 1553 |
| 4 | 99A | 98A–100A | 32.70* | 2.16 | 0.361 | 37.23* | 1.66 | 0.277 | 2600 | 574 |
Peak LR (* denotes significant LR).
Additive effect (a) of QTL (Ore-R − 2b) in phenotypic standard deviation units (σP).
Size of QTL region given by the 2-LOD support intervals in kilobase pairs.
Number of genes within 2-LOD support intervals of each QTL based on Drysdale et al. (2005).
Epistasis:
We tested for pairwise epistatic interactions between all possible marker combinations. We observed four blocks of significant epistatic interactions between markers 21E and 65D–67D; 50D–50F and 65A; 50D–50F and 91A–92A; and 61A–65A and 72A–88E. Six pairs of markers in these blocks were significant using a conservative Bonferroni correction for multiple tests, and the rest were significant using a false discovery rate (Storey and Tibshirani 2003) criterion of Q ≤ 0.05, corresponding to a P-value of ≤ 0.0011. None of the blocks of markers contributing to epistatic interactions had significant additive QTL effects in the CIM analysis.
High-resolution QTL mapping:
Deficiency complementation mapping:
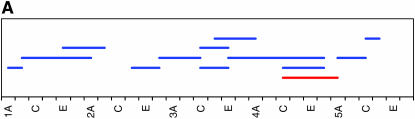
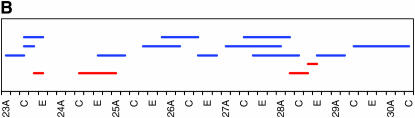
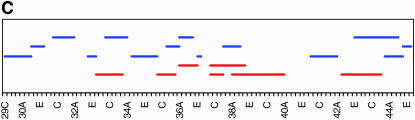
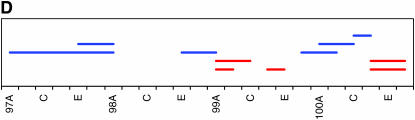
As noted above, our initial genome scan localized QTL affecting locomotor reactivity to relatively broad genomic regions. Therefore, we implemented deficiency complementation mapping using 61 deficiency stocks spanning the QTL intervals (supplemental Table 1 at http://www.genetics.org/supplemental/) to map the QTL with higher resolution (Table 2, Figure 3). We extended the deficiency complementation analysis to the markers immediately adjacent to those defining the 2-LOD support intervals, because QTL boundaries as determined by CIM can fluctuate slightly, depending on the model parameters (Pasyukova et al. 2000). The QTL from 1B–3E was refined to a region just adjacent to original QTL, from 4F11–5A2. The peak on the second chromosome spanning 27B to 29E fractionated into three smaller QTL, including 23E1–23E6, 24C3–24E4, and 28C1–28E7. The QTL peak spanning 30D to 38A reduced into six smaller regions, including 32F1–33B3, 35B10–35D1, 36D1–36E2, 37B2–37F1, 38B1–40B1, and 42B3–42E1. The QTL with a large effect on the third chromosome spanning 98A to 100A split into two regions, 99A6–99C1 and 100D2–100F5. Overall, the four main QTL that spanned over 18.2 Mb fractionated into 12 much smaller regions that harbor between 29 and 407 positional candidate genes (Table 2). This range is skewed because of the 407-gene region from 38B1 to 40B1, which spans the centromere and for which additional deficiencies were not available at the time of testing. Including this region, the average number of positional candidate genes identified using deficiency complementation mapping is 101.8 loci/QTL. This reduced number greatly facilitates the task of choosing candidate genes for further study. While each refined QTL region could represent the effect of a single gene, it is also possible that there are closely linked genes within the QTL regions that contribute to variation in locomotor reactivity.
TABLE 2.
High-resolution QTL mapping from complementation tests of deficiencies
| Chromosome | Deficiency name | P-value L × G | QTL | No. of locib |
|---|---|---|---|---|
| X | Df(1)JC70/FM7c | 0.0004 | 4F11–5A2 | 43 |
| 2 | Df(2L)S2590/CyO | 0.0039 | 23D2–23E3 | 53 |
| 2 | Df(2L)ed-dp/SM1 | 0.0024 | 24C3–24E4 | 73 |
| 2 | Df(2L) Trf-C6R31/CyO | 0.0078 | 28C1–28E7 | 106 |
| Df(2L)XE-2750/CyO | 0.0004 | |||
| 2 | Df(2L)Prl/CyO | 0.0001 | 32F1–33B3 | 58 |
| 2 | Df(2L)TE35BC-24/CyO | 0.0001a | 35B10–35D1 | 37 |
| 2 | Df(2L)H20/CyO | 0.0064a | 36D1–36E2 | 29 |
| 2 | Df(2L)prA16/CyO | 0.0001a | 37B2–37F1 | 175 |
| DF(2l)TW158/CyO | 0.0022 | |||
| 2 | Df(2L)TW161/CyO | 0.0001a | 38B1–40B1 | 407 |
| 2 | Df(2R)ST1/CyO | 0.0008 | 42B3–42E1 | 87 |
| 3 | Df(3R)Dr-rv1/TM3 | 0.0001 | 99A6–99C1 | 87 |
| Df(3R)01215/TM3 | 0.0083 | |||
| 3 | Df(3R)faf-BP/TM6B | 0.0048 | 100D2–100F5 | 66 |
| Df(3R)04661/TM3 | 0.0003a |
Significant L × G × S interaction in addition to significant L × G term.
Number of genes in QTL intervals based on Drysdale et al. (2005).
Figure 3.—
High-resolution QTL mapping using complementation tests of deficiencies. Each horizontal bar shows the cytological breakpoints of the tested deficiencies (supplemental Table 1 at http://www.genetics.org/supplemental/). Red bars indicate deficiencies exhibiting quantitative failure to complement QTL affecting locomotor behavior, and blue bars indicate quantitative complementation. (A–D) The four genomic regions to which QTL affecting locomotor reactivity were mapped. The scale denotes sections and subsections of polytene chromosomes. (A) 1B–5F. (B) 23A–30C. (C) 29C–44F. (D) 97A–100F.
Mutant complementation mapping:
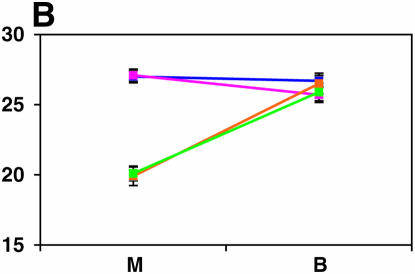
We used complementation tests to 27 mutations at 26 positional candidate genes within QTL defined by deficiency complementation mapping to identify mutations that fail to complement the Ore-R and 2b QTL alleles for locomotor reactivity (supplemental Table 2 at http://www.genetics.org/supplemental/). We preferentially chose candidate genes for complementation tests that were known to function in the nervous system, were required for normal development (including leg development genes), were involved in response to stress, and/or were previously implicated in behavior. Mutations in the following 13 genes failed to complement Ore-R and 2b QTL alleles for locomotor reactivity: nob, turtle (tutl), TBP-related factor (Trf), crooked legs (crol), shuttlecraft (stc), Catecholamines up (Catsup), Lim3, Ddc, derailed (drl), tyr1, Aconitase (Acon), teashirt (tsh), and Drop (Dr) (Figure 4, supplemental Table 3 at http://www.genetics.org/supplemental/). Among the 13 mutations implicated in this study only nob, tyr1, and Dr have been previously found to affect locomotor behavior. The other 10 loci are novel candidate genes affecting locomotor reactivity. We did not test candidate genes in the 23E1–23E6 QTL, as no genes in this region fulfilled our criteria (all are predicted genes of unknown function). Further, none of the mutations at positional candidate genes corresponding to the QTL at 36D1–36E2 and 42B3–42E1 exhibited failure to complement.
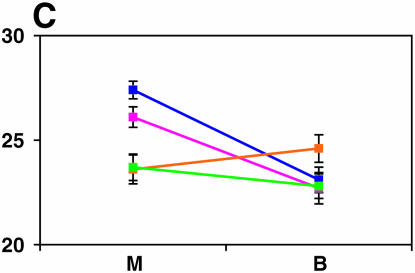
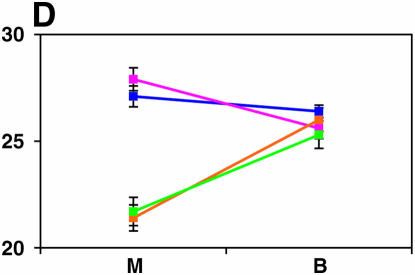
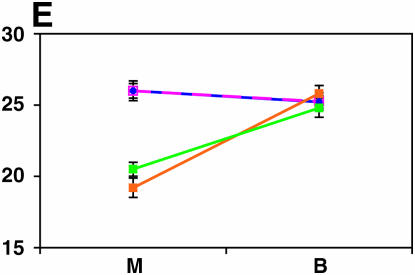
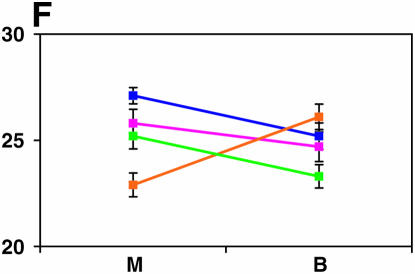
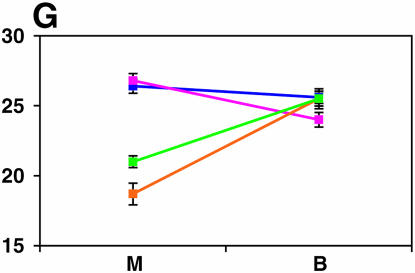
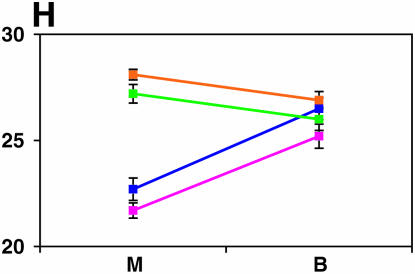
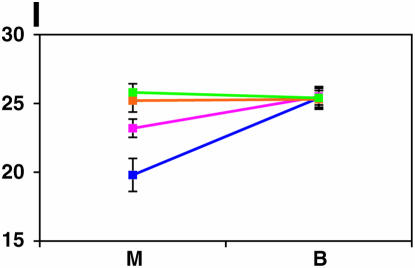
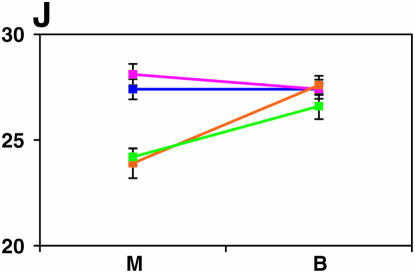
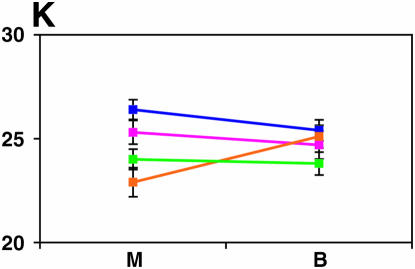
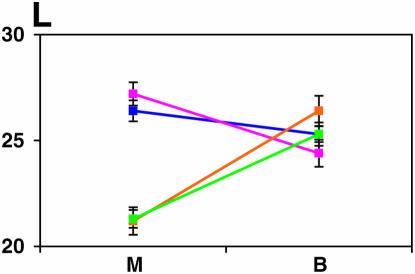
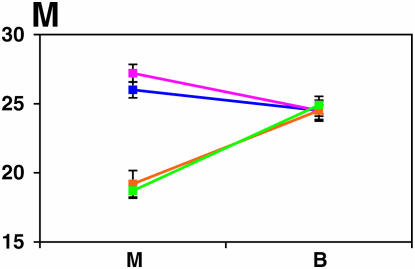
Figure 4.—
Mutant complementation tests. Means and standard errors of locomotor reactivity (in seconds, y-axis) are given for the genes that exhibited failure to complement Ore-R and 2b alleles, separately for crosses to mutants (M) and balancers (B) (x-axis). Males (blue) from the crosses to Ore-R. Females (pink) from the crosses to Ore-R. Males (gold) from the crosses to 2b. Females (green) from the crosses to 2b. (A) nobKS49. (B) tutl01085. (C) Trf1. (D) crol04418. (E) stc05441. (F) Catsup1. (G) Lim31. (H) Ddc27. (I) drl2. (J) tyr11. (K) Acon07054. (L) tsh04319. (M) Dr1.
Linkage disequilibrium mapping:
Complementation tests:
Ddc was one of the candidate genes affecting variation in locomotor reactivity between Ore-R and 2b. We conducted complementation tests to determine whether Ddc could potentially be associated with naturally occurring variation in locomotor reactivity. We crossed a Ddc null allele (Ddc27) and a wild-type Ddc allele, both of which had been substituted into the inbred Sam genetic background, to a panel of 164 wild-derived isogenic second chromosomes extracted from the Raleigh population, which had also been substituted into the Sam background. The analysis of these data is given in Table 3. There is considerable naturally segregating genetic variation in locomotor reactivity attributable to genes on the second chromosome, as shown by the highly significant effect of the second chromosome substitution line. In addition, the significant main effect of cross indicates that the mean locomotor reactivity of lines differs according to whether they are heterozygous for the null or for the wild type Ddc allele: in fact, mean locomotor reactivity is reduced in the Ddc27 background. The major terms of interest from this analysis are those involving the interaction between line and cross (L × C and L × S × C), both of which are highly significant. These are the complementation terms, which indicate that the difference in means between the Ddc27 and Ddc+ alleles varies among the chromosome substitution lines and that the sex dimorphism of the complementation effects also varies among the wild-derived chromosomes. That is, there is genetic interaction between the Ddc27 and Ddc+ chromosomes and the QTL affecting locomotor reactivity on the wild-derived chromosomes.
TABLE 3.
ANOVA of complementation tests of isogenic second chromosomes to Ddc27 and Ddc+ alleles
| Source | d.f. | MSa | F | P | σ2 b |
|---|---|---|---|---|---|
| Line (L) | 163 | 183.007 | 3.95 | <0.0001 | 1.7150 |
| Sex (S) | 1 | 4.292 | 0.27 | 0.6072 | Fixed |
| Cross (C) | 1 | 821.598 | 18.82 | <0.0001 | Fixed |
| L × S | 163 | 16.175 | 1.20 | 0.1267 | 0.0627 |
| L × C | 163 | 43.661 | 3.23 | <0.0001 | 0.7544 |
| S × C | 1 | 37.528 | 2.78 | 0.0976 | Fixed |
| L × S × C | 163 | 13.520 | 1.57 | <0.0001 | 0.2500 |
| Error | 12,412 | 8.612 | 8.6142 |
From type III sums of squares.
Variance component.
These data do not prove that Ddc is associated with natural variation in locomotor reactivity, but rather that Ddc cannot be excluded as a potential candidate gene, as would have been the case if the interactions between line and cross were not significant. This is because the Ddc27 and Ddc+ alleles are not on co-isogenic chromosomes. Therefore, there are several possible and not mutually exclusive interpretations of the observed quantitative failure to complement: (1) allelic interactions between naturally occurring Ddc alleles and the mutant and wild-type Ddc alleles; (2) epistatic interactions between other QTL on the wild second chromosomes affecting locomotor reactivity and the mutant and wild-type Ddc alleles; and (3) interactions between any of the QTL affecting locomotor reactivity that differ between the chromosomes harboring the Ddc27 and Ddc+ alleles and the QTL affecting variation in locomotor reactivity on the wild second chromosomes. If the former interpretation is true, then we expect that molecular polymorphism at Ddc will be associated with naturally occurring variation in locomotor reactivity.
Genotype–phenotype associations:
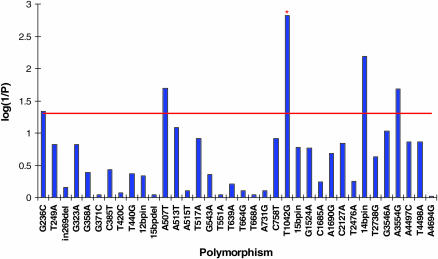
We tested this hypothesis directly by evaluating associations between polymorphic markers at Ddc in this population of 164 second chromosome lines and variation in locomotor reactivity. Five markers exhibited an association with locomotor reactivity at the nominal 5% significance level: G236C (P = 0.0458) and A507T (P = 0.0201) in the promoter region, T1042G in intron 1 (P = 0.0015), In2622Del in intron 3 (P = 0.0064), and A3554G, a synonymous SNP in exon 4 (P = 0.0209) (Figure 5). The T1042G polymorphism was formally significant at an experiment-wise P = 0.05 both using a conservative Bonferroni-corrected significance threshold (P = 0.0015, based on 34 different association tests) and using a significance threshold derived by permutation. We also used permutation to test whether there were more significant marker-trait associations than expected by chance. The permutation-derived probabilities of observing five marker-trait associations at P < 0.05, four marker-trait associations at P < 0.025, and two marker-trait associations at P < 0.01 were, respectively, P = 0.071, P = 0.039, and P = 0.040. Thus, we observed more marker-trait associations at P < 0.025 than expected by chance, also implicating Ddc as a candidate gene affecting natural variation in locomotor reactivity. The four markers significantly associated with variation in locomotor behavior at P < 0.025 are in strong LD with each other. Estimates of LD were significant at P < 0.0001 (Fisher's exact test) between A507T and T1042G, A507T and In2622Del, T1042G and A3554G, and In2664Del and A3554G (DeLuca et al. 2003). Indeed, LD extends throughout the 5.5-kb region including the Ddc transcription unit (DeLuca et al. 2003). There are 103 population haplotypes at Ddc formed by considering all 36 polymorphic sites in the sample of 164 alleles, and these haplotypes are strongly associated with variation in locomotor reactivity (P = 9.7 × 10−14). This association is highly significant (P < 0.001) on the basis of a permutation test.
Figure 5.—
Association of polymorphic markers at Ddc with variation in locomotor reactivity among 164 second chromosome substitution lines. The 36 polymorphic markers at Ddc are given on the x-axis, and P-values, transformed to log(1/P), from the ANOVA tests of association of the markers with locomotor reactivity are given on the y-axis. The red horizontal line denotes the nominal 5% significance level. Marker T1042G, denoted by *, is formally significant on the basis of a permutation test for individual makers, as well as the Bonferroni-corrected experiment-wise 5% significance level.
DISCUSSION
Genetic architecture of locomotor behavior:
We mapped four additively acting QTL affecting variation in locomotor reactivity between Ore-R and 2b. However, the genetic architecture of locomotor behavior in these strains is much more complex than suggested by the initial QTL mapping, since the four QTL detected by recombination mapping fractionated into a minimum of 12 QTL on the basis of quantitative complementation tests of deficiencies (Pasyukova et al. 2000), and there was evidence for epistasis between gene regions without main QTL effects. Further, several of the QTL defined by deficiency mapping harbored multiple candidate genes as indicated by complementation tests to mutants. It should be noted that there were a few small gaps in the deficiency coverage and that only a small fraction of positional candidate genes were tested for complementation to mutations, either because mutations were unavailable or because they did not have high priority according to our criteria for inclusion in this study. Similar complexity has been observed in previous analyses of variation in longevity (Pasyukova et al. 2000; Mackay et al. 2005), starvation resistance (Harbison et al. 2004), and mating behavior (Moehring and Mackay 2004) in these lines. In addition, the number of QTL detected in any line–cross analysis is always a lower bound to the number of QTL affecting naturally occurring variation in the trait (Mackay 2001). More QTL, with smaller effects, contributing to variation in locomotor behavior between Ore-R and 2b could be detected by increasing the number of recombinants as well as the number of individuals tested per recombinant genotype. Also, these strains represent only a subset of naturally occurring variation, and expanding the study to other strains is likely to uncover additional QTL that do not segregate in these strains. When one adds to these considerations the fact that locomotor reactivity is only one of many measures of locomotor behavior, the inescapable conclusion is that a large number of loci must affect natural variation in this important suite of traits. This, in turn, implies substantial pleiotropy between the genetic architectures of multiple complex traits, as previously inferred from the analysis of effects of P-element-induced mutations affecting olfactory behavior (Anholt et al. 1996), sensory bristle number (Norga et al. 2003), and resistance to starvation stress (Harbison et al. 2004).
Quantitative complementation tests:
The major challenge to understanding the genetic basis of complex traits in most species has been the difficulty in mapping individual genes corresponding to QTL by recombination alone (Mackay 2001). In Drosophila, we can circumvent this difficulty by utilizing complementation tests of deficiencies to rapidly map QTL to small genomic regions and complementation tests of mutations to identify positional candidate genes corresponding to QTL (Long et al. 1996; Mackay and Fry 1996; Pasyukova et al. 2000; Fanara et al. 2002; DeLuca et al. 2003; Harbison et al. 2004; Moehring and Mackay 2004). These approaches are not without caveats. Until recently, deficiency mapping has been limited because there are gaps in deficiency coverage, and the breakpoints have been ascertained only cytologically. More importantly, deficiencies have been induced in a heterogeneous collection of genetic backgrounds and are often marked with recessive and dominant mutations, and the Balancer chromosome is from an unrelated genetic background with additional mutations. The heterogeneous genetic backgrounds pose a problem for comparing effects across overlapping deficiencies, since variable effects could be due to multiple linked QTL or differences in genetic background. Recently, DrosDel and Exelixis deficiencies have been generated in co-isogenic backgrounds without additional mutations, have molecularly defined breakpoints (Parks et al. 2004), and will prove a major boon for future deficiency complementation mapping efforts. Similarly, the drawback for using quantitative complementation tests of mutations at positional candidate genes has been the limited number of mutations at positional candidate genes embraced by a QTL (ideally, one would like to perform complementation tests to all positional candidate genes), because those that exist are not typically derived in an isogenic background. The Exelixis collection of co-isogenic P and piggyBac insertions (Thibault et al. 2004) and the ongoing effort to mutate all Drosophila genes (Spradling et al. 1999; Bellen et al. 2004) will greatly enhance future efforts to identify all positional candidate genes that fail to complement QTL alleles.
Candidate genes affecting variation in locomotor behavior between Ore-R and 2b:
We have identified 13 candidate genes affecting variation in locomotor reactivity between Ore-R and 2b using complementation tests of mutations at positional candidate genes (supplemental Table 3 at http://www.genetics.org/supplemental/). Of these, three have been previously implicated to affect adult locomotion (nob, tyr1, and Dr). nob mutants are inactive and have neuroanatomical defects in the protocerebral bridge and fan-shaped body of the central complex, the brain region required for maintenance of locomotor activity (Strauss et al. 1992; Martin et al. 1999; Hitier et al. 2000; Strauss 2002). tyr1 affects catecholamine biosynthesis. tyr1 mutations have reduced levels of phenol oxidase (Pentz et al. 1990) and dopamine (Burnell and Daly 1982) and are hyperactive (Burnell and Daly 1982; Meehan and Wilson 1987). Mutations at Dr are “uncoordinated.” Dr is a highly pleiotropic homeobox gene that encodes a RNA polymerase II transcription factor involved in brain and muscle development and in establishing the dorsal identity of neuroblasts (Isshiki et al. 1997; Jurata et al. 2000).
Six candidate genes have mutant phenotypes consistent with an involvement in regulating locomotion, although effects on locomotor behavior have not been quantified previously (tutl, drl, Catsup, Lim3, Ddc, and Acon). tutl plays an essential role in establishing neural circuits for coordinated motor output: tutl mutations exhibit an abnormal response to tactile stimulation, an inability to regain an upright position from an inverted position, and an inability to fly (Bodily et al. 2001). drl encodes a protein tyrosine kinase, which acts in embryos to guide axons across the midline in the anterior commissure and represses crossing the midline in the posterior commissure. Derailed may have a similar function in commissure choice in adults, where it is required for appropriate projections between mushroom bodies (Bonkowsky et al. 1999). drl mutants show structural brain defects in the adult central complex and mushroom bodies and have defects in olfactory learning and memory (Bolwig et al. 1995).
Variation in levels of dopamine (Tunnicliff et al. 1969; Connolly et al. 1971; Burnell and Daly 1982; Meehan and Wilson 1987; Pendleton et al. 2002) and other biogenic amine neurotransmitters (O'Dell and Burnet 1988; Lundell and Hirsch 1998) has been associated with variation in Drosophila locomotor activity. Four of the candidate genes affecting variation in locomotor reactivity between Ore-R and 2b affect dopamine and biogenic amine synthesis. Catsup, Ddc, and Lim3 are members of the Ddc cluster (Stathakis et al. 1995). Mutations in many of the genes in this cluster show defects in cuticle formation, sclerotinization (hardening) or melanization of the cuticle, and formation of melanotic pseudotumors, all of which are hallmarks of abnormal catecholamine metabolism (Wright 1996). Catsup encodes a metal ion transporter and is a negative regulator of tyrosine hydroxylase, which catalyzes the rate-limiting step of dopamine biosynthesis (Stathakis et al. 1999). Mutations in Catsup have significantly elevated tyrosine hydroxylase activity, resulting in unusually high levels of dopamine. Ddc is required in catecholamine and serotonin biosynthesis (Monastirioti 1999). Lim3 is a homeobox gene that encodes an RNA polymerase II transcription factor required for motor neuron development (Thor et al. 1999; Hobert and Westphal 2000). Acon encodes a product involved in amino acid biosynthesis and the tricarboxylic acid cycle and is putatively required for biogenic amine synthesis (Drysdale et al. 2005).
The remaining four genes are novel candidate genes affecting locomotor behavior (Trf, crol, stc, and tsh). These genes all encode RNA polymerase II transcription factors that have been implicated in nervous system development. Trf is expressed in the embryonic central nervous system and in adult male germ cells (Davidson 2003). Trf mutants are male sterile and exhibit an ether-induced leg-shaking behavioral phenotype (Crowley et al. 1993). crol is required for proper leg development and ecdysone-regulated gene expression during metamorphosis (D'Avino and Thummel 1998). Mutants of crol specifically affect the number of ecdysone-induced genes, including genes controlling motor axon guidance and synaptogenesis (Kraut et al. 2001). stc is the only known homolog of human transcription factor NF-X1 (Stroumbakis et al. 1996). stc is expressed in the embryonic central nervous system, where it is required for motor neuron development. tsh is a homeobox gene required for trunk segment identity and interacts with the Wnt-signaling pathway to modulate the response of Wnt signaling (Gallet et al. 1998; Sharpe et al. 2001).
These results show that “wild-type” allelic variation at genes previously identified through mutational analysis contributes to quantitative genetic variation, and conversely, quantitative genetic analysis is an effective method for functional genome annotation, identifying novel loci affecting behavior (Sokolowski 2001). Mutations at many essential genes are homozygous lethal; quantitative complementation tests are particularly useful in determining effects of these genes on complex traits.
All of the candidate genes affecting differences in locomotor reactivity between Ore-R and 2b have pleiotropic effects during development. Thus, these genes may act throughout development to lay the blueprint of the nervous system that is responsible for manifesting adult behavior. Further, consistent with our inference of substantial pleiotropy among the genetic architectures of multiple complex traits, many of the candidate genes affecting variation in locomotor reactivity in these lines have also been implicated as candidate genes affecting other complex traits. Ddc (DeLuca et al. 2003), stc (Pasyukova et al. 2004), Catsup (Mackay et al. 2005), and Lim3 (Mackay et al. 2005) are all candidate genes affecting variation in longevity between Ore-R and 2b, and crol is associated with variation in starvation stress resistance between these strains (Harbison et al. 2004).
There is some overlap of genes found in this QTL screen and those that have been implicated from mutational screens, but there are many genes with mutational effects on locomotor behavior (e.g., per, ey, oc, and hiw) not detected in the QTL screen for variation in locomotor reactivity between Ore and 2b. This highlights the relationship between mutational and quantitative genetic approaches, where the former approach identifies genes involved in the manifestation of the normal behavior and the latter identifies genes affecting natural variation in the behavior (Mackay 2001). These approaches are complementary because (1) genes required to produce the behavior may be so critical they do not vary in nature due to the selective constraint against changes that would greatly decrease overall reproductive fitness; (2) natural variants may specifically affect behavior, but the behavioral phenotypes may not be detectable in a mutagenesis study because null mutations are lethal or have a pronounced effect on a different phenotype; and (3) quantitative methods can detect genes only with naturally occurring allelic variation in the founder population.
Mutagenesis studies have identified single genes affecting locomotion, providing insight into the genetic framework in which the locomotor circuit is built. The definition of quantitative trait genes affecting locomotion will further characterize the components responsible for variation in nature, enhancing the picture of how genes and environment interact to produce complex behavior. It should be emphasized that, to understand behavior, one may have to go beyond understanding the function of individual components (genes and cells) and venture into the territory of neural function and emergent properties (Hobert 2003). More specifically, we ultimately need to determine how the temporal and spatial expression of neural networks produces movements characterizing adult behavior (Sokolowski 2001).
Linkage disequilibrium mapping:
Identifying candidate genes contributing to variation in behavior between two strains conveys no information about the contribution of the candidate genes to naturally occurring variation in the trait. LD mapping is necessary to assess the contribution of variation at candidate genes to natural variation in locomotor behavior. We observed more significant associations of polymorphic markers at Ddc with variation in locomotor reactivity than expected by chance and also observed that population haplotypes derived from all genotyped markers were significantly associated with variation in locomotor behavior. Although LD in Drosophila generally decays rapidly with physical distance in normal regions of recombination (Long et al. 1998; Robin et al. 2002), there was considerable LD throughout the 5.5-kb region, including the Ddc transcription unit (DeLuca et al. 2003). Since genotypes of all polymorphic sites were not determined, and LD did not decay markedly within the surveyed region, all we can deduce is that one or more polymorphisms at Ddc, or polymorphisms in LD with polymorphisms at Ddc, affect naturally occurring variation in locomotor behavior. Future efforts to identify causal polymorphisms must obtain complete sequences of all Ddc alleles, extending sufficiently 5′ and 3′ of the coding region for LD to decay across the sequenced region. In addition, larger samples will be required to detect rare and informative recombinants. Nevertheless, these data implicate variation in the synthesis of bioamines as a factor contributing to natural variation in locomotor reactivity.
A previous study identified three molecular markers in Ddc that were associated with variation in life span among the same set of chromosomes (DeLuca et al. 2003). None of the markers associated with variation in locomotor behavior were associated with variation in life span. Catsup is another positional candidate gene affecting dopamine synthesis, locomotor reactivity, and longevity. Similarly, different molecular polymorphisms in Catsup have been shown to be independently associated with natural variation in longevity and locomotor behavior (Carbone et al. 2006). Thus, analysis of natural alleles of highly pleiotropic genes will prove invaluable in parsing the mechanisms underpinning the effects on constituent traits (Sokolowski 2001).
Acknowledgments
We thank Susan Harbison for help with statistical analysis, Christy Dilda for constructing the Ddc27 chromosome substitution line, and Robert Anholt for comments on the manuscript. This work was supported by predoctoral training fellowships from the National Institutes of Health (NIH) (T32 GM08443) and North Carolina State University to K.W.J., NIH grant F32 GM066603 to T.J.M., and NIH grant R01 GM45146 to T.F.C.M. This is a publication of the W. M. Keck Center for Behavioral Biology.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association, 1994. Diagnostic and Statistical Manual of Mental Disorders, Ed. 4. American Psychiatric Press, Arlington, VA.
- Anholt, R. R. H., R. F. Lyman and T. F. C. Mackay, 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics 143: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. S., B. J. Taylor and J. C. Hall, 2001. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell 105: 13–24. [DOI] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z. B. Zeng, 1994. Zmap—a QTL cartographer, pp. 65–66 in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, edited by C. Smith, J. S. Gavora, B. Benkel, J. Chesnias and W. Fairfull. Organizing Committee, 5th World Congress on Applied Genetics Applied to Livestock Production, Guelph, Ontario, Canada.
- Basten, C. J., B. S. Weir and Z. B. Zeng, 1999. QTL Cartographer, Version 1.13. Department of Statistics, North Carolina State University, Raleigh, NC.
- Bellen, H. J., R. W. Levis, G. Liao, Y. J. W. Carlson, G. Tsang et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily, K. D., C. M. Morrison, R. B. Renden and K. Broadie, 2001. A novel member of the Ig superfamily, turtle, is a CNS-specific protein required for coordinated motor control. J. Neurosci. 21: 3113–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwig, G. M., M. Del Vecchio, G. Hannon and T. Tully, 1995. Molecular cloning of linotte in Drosophila: a novel gene that functions in adults during associative learning. Neuron 15: 829–842. [DOI] [PubMed] [Google Scholar]
- Bonkowsky, J. L., S. Yoshikawa, D. D. O'Keefe, A. L. Scully and J. B. Thomas, 1999. Axon routing across the midline controlled by Drosophila Derailed receptor. Nature 402: 540–543. [DOI] [PubMed] [Google Scholar]
- Burnell, A. M., and B. A. Daly, 1982. Spontaneous locomotor activity and dopamine levels in tyr-1, pp. 361–370 in Advances in Genetics, Development and Evolution of Drosophila, edited by S. Lakovaara. Plenum Press, New York.
- Burnet, B., L. Burnet, K. Connolly and N. Williamson, 1988. A genetic analysis of locomotor activity in Drosophila melanogaster. Heredity 61: 111–119. [Google Scholar]
- Callaerts, P., S. Leng, J. Clements, C. Benassayag, C. Cribbs et al., 2001. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J. Neurobiol. 46: 73–88. [DOI] [PubMed] [Google Scholar]
- Carbone, M. A., K. W. Jordan, R. F. Lyman, S. T. Harbison, J. Leips et al., 2006. Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosophila. Curr. Biol. 16: 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 13: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, K., 1966. Locomotor activity in Drosophila. II. Selection for active and inactive strains. Anim. Behav. 14: 444–449. [DOI] [PubMed] [Google Scholar]
- Connolly, K., 1967. Locomotor activity in Drosophila. III. A distinction between activity and reactivity. Anim. Behav. 15: 149–152. [DOI] [PubMed] [Google Scholar]
- Connolly, K., G. Tunnicliff and J. T. Rick, 1971. The effects of γ-hydroxybutyric acid on spontaneous locomotor activity and dopamine level in a selected strain of Drosophila melanogaster. Comp. Biochem. Physiol. 40B: 321–326. [DOI] [PubMed] [Google Scholar]
- Crowley, T. E., T. Hoey, J. K. Liu, Y. N. Jan, L. Y. Jan et al., 1993. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature 361: 557–561. [DOI] [PubMed] [Google Scholar]
- Davidson, I., 2003. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 28: 391–398. [DOI] [PubMed] [Google Scholar]
- D'Avino, P. P., and C. S. Thummel, 1998. crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125: 1733–1745. [DOI] [PubMed] [Google Scholar]
- deBelle, J. S., and M. B. Sokolowski, 1987. Heredity of rover/sitter: alternative foraging strategies of Drosophila melanogaster larvae. Heredity 59: 73–83. [Google Scholar]
- DeLuca, M., N. V. Roshina, G. L. Geiger-Thornsberry, R. F. Lyman, E. G. Pasyukova et al., 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby and FlyBase Consortium, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395 (http://flybase.org). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanara, J. J., K. O. Robinson, S. M. Rollmann, R. R. Anholt and T. F. C. Mackay, 2002. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162: 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet, A., A. Erkner, B. Charroux, L. Fassano and S. Kerridge, 1998. Trunk specific modulation of Wingless signaling in Drosophila by Teashirt binding to Armadillo. Curr. Biol. 8: 893–902. [DOI] [PubMed] [Google Scholar]
- Gilchrist, G. W., R. B. Huey and L. Partridge, 1997. Thermal sensitivity of Drosophila melanogaster: evolutionary responses of adults and eggs to laboratory natural selection at different temperatures. Physiol. Zool. 70: 403–414. [DOI] [PubMed] [Google Scholar]
- Gotz, K. G., 1989. Visual guidance in Drosophila, pp. 391–407 in Development and Neurobiology of Drosophila, edited by O. Siddiqui, P. Babu, L. M. Hall and J. C. Hall. Plenum Press, New York.
- Harbison, S. T., A. H. Yamamoto, J. J. Fanara, K. K. Norga and T. F. C. Mackay, 2004. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166: 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitier, R., A. F. Simon, F. Savarit and T. Preat, 2000. no-bridge and linotte act jointly at the interhemispheric junction to build up the adult central brain of Drosophila melanogaster. Mech. Dev. 99: 93–100. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2003. Introduction: behavioral genetics—the third century. J. Neurobiol. 54: 1–3. [DOI] [PubMed] [Google Scholar]
- Hobert, O., and H. Westphal, 2000. Functions of LIM-homeobox genes. Trends Genet. 16: 75–83. [DOI] [PubMed] [Google Scholar]
- Isshiki, T., M. Takeichi and A. Nose, 1997. The role of msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development 124: 3099–3109. [DOI] [PubMed] [Google Scholar]
- Jurata, L. W., J. B. Thomas and S. L. Pfaff, 2000. Transcriptional mechanisms in development of motor control. Curr. Opin. Neurobiol. 10: 72–79. [DOI] [PubMed] [Google Scholar]
- Kaidanov, L. Z., 1990. The rules of genetic alteration of Drosophila melanogaster inbred lines determined by selection. Arch. Biol. Nauka 42: 131–148. [Google Scholar]
- Konopka, R. J., and S. Benzer, 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain of function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. [DOI] [PubMed] [Google Scholar]
- Leal, S. M., and W. S. Neckameyer, 2002. Pharmacological evidence for GABAergic regulation of specific behaviors in Drosophila melanogaster. J. Neurobiol. 50: 245–261. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Deigo.
- Long, A. D., S. L. Mullanay, T. F. C. Mackay and C. H. Langley, 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics 144: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., R. F. Lyman, C. H. Langley and T. F. C. Mackay, 1998. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell, M. J., and J. Hirsch, 1998. Eagle is required for the specification of serotonin neurons and other neuroblast 7–3 progeny in the Drosophila CNS. Development 125: 463–472. [DOI] [PubMed] [Google Scholar]
- Lyman, R. F., C. Q. Lai and T. F. C. Mackay, 1999. Linkage disequilibrium mapping of molecular polymorphisms at the scabrous locus associated with naturally occurring variation in bristle number in Drosophila melanogaster. Genet. Res. 74: 303–311. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., and J. D. Fry, 1996. Polygenic mutation in Drosophila melanogaster: genetic interactions between selection lines and candidate trait loci. Genetics 144: 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F. C., N. V. Roshina, J. W. Leips and E. G. Pasyukova, 2005. Complex genetic architecture of Drosophila longevity, pp. 181–216 in Handbook of the Biology of Aging, Ed. 6, edited by E. J. Masaro and S. N. Austad. Elsevier, New York.
- Martin, J. R., R. Ernst and M. Heisenberg, 1998. Mushroom bodies suppress locomotor activity in Drosophila melanogaster. Learn. Mem. 5: 179–191. [PMC free article] [PubMed] [Google Scholar]
- Martin, J. R., T. Rabbe and M. Heisenberg, 1999. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J. Comp. Physiol. A 185: 277–288. [DOI] [PubMed] [Google Scholar]
- Meehan, M. J., and R. Wilson, 1987. Locomotor activity in the Tyr-1 mutant of Drosophila melanogaster. Behav. Genet. 17: 503–512. [DOI] [PubMed] [Google Scholar]
- Moehring, A. J., and T. F. C. Mackay, 2004. The quantitative genetic basis of male mating behavior in Drosophila melanogaster. Genetics 167: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti, M., 1999. Biogenic amine systems in the fruit fly. Microsc. Res. Tech. 45: 106–121. [DOI] [PubMed] [Google Scholar]
- Norga, K. K., M. C. Gurganus, C. L. Dilda, A. Yamamoto, R. F. Lyman et al., 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13: 1388–1397. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, C. L. Dilda, Z. B. Zeng and T. F. C. Mackay, 1997. Sex specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dell, K., and B. Burnet, 1988. The effect of locomotor activity and reactivity of the hypoactive and inactive mutations in Drosophila melanogaster. Heredity 61: 199–207. [Google Scholar]
- Olanow, C. W., and W. G. Tatton, 1999. Etiology and pathogenesis of Parkinson's disease. Annu. Rev. Neurosci. 22: 123–144. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Pasyukova, E. G., C. Vieira and T. F. C. Mackay, 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156: 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova, E. G., N. V. Roshina and T. F. C. Mackay, 2004. Shuttle craft: a candidate quantitative trait gene for Drosophila lifespan. Aging Cell 3: 297–307. [DOI] [PubMed] [Google Scholar]
- Pendleton, R. G., A. Rasheed, T. Sardina and T. Tully, 2002. Effects of tyrosine hydroxylase mutants on locomotor activity in Drosophila: a study in functional genomics. Behav. Genet. 32: 89–94. [DOI] [PubMed] [Google Scholar]
- Pentz, E. S., B. C. Black and T. R. Wright, 1990. Mutations affecting phenol oxidase activity in Drosophila: quicksilver and tyrosinase-1. Biochem. Genet. 28: 151–171. [DOI] [PubMed] [Google Scholar]
- Pereira, H. S., and M. B. Sokolowski, 1993. Mutations in the larval foraging gene affect adult locomotory behavior after feeding in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 90: 5044–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage, J., G. Steffes, D. C. Lau, B. A. Parente, S. T. Crews et al., 2002. Novel behavioral and developmental defects associated with Drosophila single-minded. Dev. Biol. 249: 283–299. [DOI] [PubMed] [Google Scholar]
- Robin, C., R. F. Lyman, A. D. Long, C. H. Langley and T. F. C. Mackay, 2002. hairy: a quantitative trait locus for Drosophila bristle number. Genetics 162: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute, 1988. SAS/SYSTAT User's Guide, 4/e. SAS Institute, Cary, NC.
- Sharpe, C., N. Lawrence and A. M. Arias, 2001. Wnt signaling: a theme with nuclear variations. BioEssays 23: 311–318. [DOI] [PubMed] [Google Scholar]
- Sokolowski, M. B., 1980. Foraging strategies of Drosophila melanogaster: a chromosomal analysis. Behav. Genet. 10: 291–302. [DOI] [PubMed] [Google Scholar]
- Sokolowski, M. B., 2001. Drosophila: genetics meets behaviour. Nat. Rev. Genet. 2: 879–890. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D. G., E. S. Pentz, M. E. Freeman, J. Kullman, G. R. Hankins et al., 1995. The genetic and molecular organization of the Dopa decarboxlyase gene cluster of Drosophila melanogaster. Genetics 141: 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D. G., D. Y. Burton, W. E. McIvor, S. Krishnakumar, T. R. F. Wright et al., 1999. The Catecholamines Up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tyrosine hydroxylase activity. Genetics 153: 361–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward, O., 2000. Functional Neuroscience. Springer Publishers, New York.
- Storey, J., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, R., 2002. The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 12: 633–638. [DOI] [PubMed] [Google Scholar]
- Strauss, R., and M. Heisenberg, 1993. A higher control center of locomotor behavior in the Drosophila brain. J. Neurosci. 13: 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, R., U. Hanesch, M. Kinkelin, R. Wolf and M. Heisenberg, 1992. No-bridge of Drosophila melanogaster: portrait of a structural brain mutant of the central complex. J. Neurogenet. 8: 125–155. [DOI] [PubMed] [Google Scholar]
- Stroumbakis, N. D., Z. Li and P. P. Tolias, 1996. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic central nervous system. Mol. Cell. Biol. 16: 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Thor, S., S. G. E. Andersson, A. Tomlinson and J. B. Thomas, 1999. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature 397: 76–80. [DOI] [PubMed] [Google Scholar]
- Tunnicliff, G., J. T. Rick and K. Connelly, 1969. Locomotor activity in Drosophila. V. A comparative biochemical study of selectively bred populations. Comp. Biochem. Physiol. 29: 1239–1245. [DOI] [PubMed] [Google Scholar]
- van Dijken, F. R., and W. Scharloo, 1979. Divergent selection on locomotor activity in Drosophila melanogaster. I. Selection response. Behav. Genet. 9: 543–553. [DOI] [PubMed] [Google Scholar]
- Wan, H. I., A. DiAntonio, R. D. Fetter, K. Bergstrom and R. Strauss, 2000. Highwire regulates synaptic growth in Drosophila. Neuron 26: 313–329. [DOI] [PubMed] [Google Scholar]
- Wright, T. R. F., 1996. Phenotypic analysis of the Dopa decarboxlyase gene cluster in Drosophila melanogaster. J. Heredity 87: 175–190. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. Y., and R. D. Palmiter, 1995. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell 83: 1197–1209. [DOI] [PubMed] [Google Scholar]