Abstract
Subfunctionalization is the process by which a pair of duplicated genes, or paralogs, experiences a reduction of individual expression patterns or function while still reproducing the complete expression pattern and function of the ancestral gene. Two germin-like protein (GLP)-encoding genes, GerB and GerF, are paralogs that belong to a small gene family in barley (Hordeum vulgare). Both genes share high nucleotide sequence similarity in coding and noncoding regions and encode identical apoplastic proteins. The use of RNA gel blots, coupled with single-stranded conformation polymorphism (SSCP) analysis of RT–PCR products, elucidated the developmental and tissue-specific expression patterns of each gene. Individual expression patterns provided evidence of both overlapping redundancy and early subfunctionalization. GerB is predominantly expressed in developing shoots, while GerF is predominantly expressed in seedling roots, developing spikes, and pericarp/testa. GerF promoter deletion studies located a region (−356/−97) responsible for high promoter activity and showed the ability of GerB and GerF upstream regions to drive gfp expression in coleoptiles, epicarps, and lemma/palea of developing spikes. The observed expression patterns are consistent with proposed roles in plant development and defense mechanisms for this gene family. These roles may explain why redundancy has been selectively maintained in this duplicate gene pair.
THE evolution of plant genomes has been shaped by the occurrence of multiple gene duplication events (Paterson 2004; Blanc and Wolfe 2004a). Under the classical model of duplicated gene evolution, these events represented an opportunity for one of the copies, or paralogs, to diverge and potentially acquire novel functions (Ohno 1970). A modern view, aimed at explaining the high retention rates observed for paralogs in eukaryotic genomes, introduced the concept of gene preservation by complementary degenerative mutations, or subfunctionalization (Force et al. 1999). Accumulation of mutations in paralogs may result in: (1) evolution of one copy to a nonfunctional pseudogene (pseudogenization), (2) divergence of one copy to acquire a new biological function (neofunctionalization), (3) reduction of expression patterns in both copies while still maintaining the complete expression pattern of the ancestral gene (subfunctionalization), and (4) functional retention of both paralogs to increase the level of gene product (Force et al. 1999; Gu et al. 2003; Osborn et al. 2003).
Genes have multiple regulatory elements that govern their spatial and temporal patterns of expression. Mutations in these elements can affect independent subfunctions, making both gene duplicates complementary and essential (Force et al. 1999). Thus, accumulation of complementary degenerative mutations may explain why certain genes are retained as duplicates (Force et al. 1999; Lynch and Force 2000). Complete loss-of-function mutations in different regulatory elements will result in qualitative subfunctionalization of paralogs (Force et al. 1999). This is best exemplified when an ancestral gene expressed in two tissues duplicates and diverges into two paralogs, each expressed in only one of those tissues (spatial divergence). Reduction-of-expression mutations, on the other hand, will result in quantitative subfunctionalization of both paralogs so that both are required to supply sufficient protein product (Force et al. 1999).
Recent studies revealed that gene loss has been the most likely fate of duplicated genes in Arabidopsis thaliana (Maere et al. 2005). In spite of this, high retention rates of certain groups of duplicated genes have been observed. These include genes involved in development, transcription, signal transduction, secondary metabolism, and response to biotic stresses (Maere et al. 2005). Rates were lower, however, for gene duplicates involved in transcription and signal transduction when originated by small duplication events, probably due to dosage effects (Birchler et al. 2001, 2005). Thus, retention of duplicated genes not only correlates with their function but also depends on whether they originated from whole-genome or small-scale duplication events (Maere et al. 2005; Moore and Purugganan 2005). In addition, selective retention of redundancy, as in the case of developmental genes, has often been related to genetic robustness (Wagner 2005).
Germins constitute a group of homologous proteins found only in “true” cereals (Hill 1937), including barley, maize, oat, rice, rye, and wheat (Lane 2002). All contain a characteristic sequence, PHIHPRATEI, known as the germin box (Lane et al. 1991). The first described germin was detected as a marker of the onset of growth during germination of isolated wheat embryos (Thompson and Lane 1980) and was later found to have oxalate oxidase activity (Lane et al. 1993). This oxidase activity generates two molecules of carbon dioxide and one molecule of hydrogen peroxide for every molecule of oxalate and dioxygen. The generation of hydrogen peroxide by germins is consistent with their proposed roles in defense and development (Lane 1994, 2002). Germins of uncharacterized enzymatic activity, such as barley GerB and GerF, are referred to as germin-like proteins (GLPs).
One hundred twenty-four germin and germin-like barley cDNAs have been grouped according to their amino acid homology into five subfamilies, designated HvGER-I to HvGER-V (Druka et al. 2002). GerB and GerF are two closely linked loci on barley chromosome 1 (7H) bin 8 and belong to the HvGER-III subfamily (Druka et al. 2002). Both genes share high nucleotide sequence identity in coding and noncoding regions. These constitute an excellent pair of paralogous genes to study the evolution and fate of recently duplicated genes in a diploid cereal species such as barley.
A previous report localized high levels of oxalate oxidase activity to the epicarp of the developing barley grain, but it could not link this activity to GerB or GerF expression in this tissue using transient expression analysis (Wu et al. 2000). Also, sequence analysis of barley EST libraries did not strictly discriminate the expression patterns of these two genes, due to their highly conserved nature (Druka et al. 2002). In this report, the use of single-stranded conformation polymorphism (SSCP) analysis of RT–PCR products allowed us to elucidate the developmental and tissue-specific expression patterns of each gene, providing evidence of both overlapping redundancy and early subfunctionalization. The ability of both promoter sequences to direct reporter gene expression in various tissues is discussed in relation to the evolutionary fate of recently duplicated genes, the putative developmental function of these GLPs, and the potential utility of these promoters for targeted transgene expression in barley.
MATERIALS AND METHODS
Materials:
Seeds of barley (Hordeum vulgare L.) cultivars Morex, Steptoe, and Golden Promise and of wild barley (H. vulgare ssp. spontaneum, PI 391093) were obtained from the USDA–ARS National Small Grains Collection (Aberdeen, ID). Plants were grown in 3.8-liter pots (1:1 mixture of peat moss and vermiculite) in a greenhouse with 26° days and 13–26° nights. Supplemental 400 W sodium arc vapor lights were used during a 16-hr photoperiod. Growth chambers were maintained at 16–18° under the same photoperiod. Fusarium graminearum strain NRRL 29169 was obtained from Kerry O'Donnell (USDA, ARS, National Center for Agricultural Utilization Research, Peoria, IL). The fungus was cultured as per Skadsen and Hohn (2004).
RNA extraction and differential display:
Total RNA from all tissues, except ovaries, anthers, and seeds, was extracted with guanidinium thiocyanate (Chirgwin et al. 1979). Ovary and anther RNAs were extracted using an RNeasy kit (QIAGEN). Seed RNAs were extracted as described (Skadsen 1993). Pericarp RNA for differential display was extracted from Morex seeds at the early dough stage of development. Morex seeds were imbibed for 8 hr, and developing shoots were harvested 1–6 days from the beginning of imbibition (dpi). Developing spikes were staged as described in Skadsen et al. (2002). Pericarp/testa from developing seed were staged as follows: (1) approximate pollination, (2) elongating, (3) gelatinous, and (4) dough; seed developmental stages were as described in Skadsen et al. (2000). Pericarp and flag leaf poly(A)+ mRNAs were purified on an oligo(dT) cellulose column (type III, Collaborative Biomedical Products). Syntheses of first strand pericarp and leaf cDNAs were carried out according to Krug and Berger (1987). Routine molecular procedures were performed as described (Sambrook and Russell 2001). Differential display of cDNAs was performed as Liang and Pardee (1992) with modifications (Federico et al. 2005).
Cloning and sequencing of cDNA:
A radiolabeled DNA band (designated GerB) prominent among pericarp differential display products, but absent in flag leaf products, was excised from the gel and used as a template in unlabeled tertiary PCRs, performed as above. A 680-bp product was cloned into the pCR2.1 vector and used to transform Escherichia coli INVαF′ cells (Invitrogen). The cloned insert was sequenced using Big Dye fluorescent terminators (Applied Biosystems). Sequences were determined by the University of Wisconsin Biotechnology Center.
The 5′ sequences of the GerB and GerF mRNAs were determined by rapid amplification of cDNA ends (RACE) using the GeneRacer kit, as described by Invitrogen. A GeneRacer RNA oligonucleotide (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGGAAGGAGUAGAAA) was ligated to the 5′ ends of pericarp, coleoptile, and lemma/palea mRNAs. A GeneRacer oligo(dT) primer [5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18] was used to prime transcription with AMV reverse transcriptase. One gene-specific downstream primer (GSP1) was designed using the sequence from the 680-bp GerB fragment: GSP1 (5′-GTGCCAGGGAGATGCCGAGGGTGTTGA). GSP1 was used to amplify the 5′ cDNA end of GerB and GerF from pericarp, coleoptile, and lemma/palea RACE-ready cDNAs using GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACACTGA) as the upstream primer, in separate PCR reactions.
Sequence analyses were performed using the BCM Search Launcher interface (Smith et al. 1996), unless stated otherwise. Homology searches were performed to all sequences in GenBank and TIGR barley EST databases using the BLAST algorithm (Altschul et al. 1990). Nucleotide and protein sequence alignments were performed using ClustalW 1.8 (Thompson et al. 1994). Nucleotide replacement (Ka) and synonymous (Ks) substitutions were estimated using K-estimator v6.0 (Comeron 1999).
RNA gel and Southern blot analyses:
Preparation of gel blots, preparation of 32P-dCTP radiolabeled probe (Feinberg and Vogelstein 1983), hybridization and washing conditions, and autoradiography were conducted as previously described (Skadsen et al. 1995). Following hybridization with 3′ 246-bp GerB, PR-1, and ribosomal cDNA probes, RNA gel blots were rinsed at 50°. Blots contained 10 μg total RNA per lane. Total RNA cDNA probe was prepared as previously described (Federico et al. 2005). Transcript levels were quantified by electronic autoradiography using an Instant Imager (Packard, Prospect, CT). Counts per minute obtained with the 246-bp GerB 3′-UTR probe were normalized relative to counts per minute values obtained from 18S ribosomal RNA hybridizations. PCR primers were designed to obtain a 454-bp barley cDNA PR-1 (GenBank accession Z21494) probe. This served as a positive control for host response to infection. Southern blots contained 10 μg of genomic DNA per lane. DNA was digested to completion with EcoRV, BamHI, or HindIII and electrophoretically separated on 0.8% agarose gels. Two GerB cDNA probes (680 bp and 246 bp) and a Stowaway-like miniature inverted-repeat transposable element (MITE) (160 bp) probe were used to hybridize Southern blots.
Cloning and analysis of GerB and GerF genomic sequences:
Different PCR primers were designed to amplify and clone overlapping regions of the GerF and GerB genes from Morex genomic DNA to corroborate that the authentic nuclear genes had been cloned. To examine gene sequence conservation between GerB and GerF, we used the main VISTA (mVISTA) program (Mayor et al. 2000). To locate repetitive sequences in GerB and GerF genes, homology searches to all sequences in TIGR Hordeum repeat database were performed using the BLAST algorithm (Altschul et al. 1990). Homology search of putative cis-acting DNA elements was performed with the PLACE (plant cis-acting regulatory DNA elements) database (Higo et al. 1999).
SSCP analysis of GerB and GerF RT–PCR products:
A pair of primers (GerSSCP1 5′-ACTCTCTTTCATCTGGGCAT and GerSSCP2 5′-GAGCACCTCTGTATTCCA) was designed to amplify GerB and GerF 3′-UTRs. GerB and GerF RT–PCR products were 213 bp and differed by only four substitutions, which yielded a pairwise nucleotide identity of 98.1%. Total RNAs (5 μg) were treated with RQ10 DNAse (Promega, Madison, WI). cDNA syntheses were performed using Superscript III (Invitrogen, Carlsbad, CA) in 20-μl reactions. Aliquots (5 μl) of one-tenth cDNA dilutions were used as templates for each RT–PCR reaction. RT–PCR reactions (10 μl) also included 1 unit Taq polymerase (Perkin-Elmer, Norwalk, CT), 1× buffer (Perkin-Elmer), 10% (v/v) DMSO, 0.4 mm dNTPs, 1 μCi [α32P]-dCTP (3000 Ci/mmol; Amersham Biosciences, Piscataway, NJ), and 2 μm of each primer. Amplification started with a 94° denaturation step (3 min), followed by 24 cycles of 15 sec at 94°, 30 sec at 50°, and 30 sec at 72°, with a final 72° extension of 7 min. Fifty microliters of SSCP loading buffer [95% formamide, 10 mm NaOH, 0.25% (w/v) xylene cyanol, 0.25% (w/v) bromophenol blue] were added to each RT–PCR reaction. RT–PCR products were then heated for 10 min at 96° and quenched on ice for 10 min. A total of 4 μl of each sample was loaded on a 0.75 X mutation detection enhancement gel (MDE; Cambrex BioScience, Walkersville, MD) and electrophoresed at 7 W constant power for 20–22 hr at 21–25° in a Bio-Rad sequencing apparatus. Electrophoresis and gel buffers used were 0.6× and 0.63× TBE, respectively. Following electrophoresis, the gels were dried on Whatman G3 paper and exposed to film between Kodak intensifying screens for 24–48 hr.
Transcript levels were quantified by electronic autoradiography, as above. Counts per minute values obtained from individual GerB and GerF-specific SSCP fragments were used to estimate the contribution of each paralog to each tissue's Ger transcript pool. This cDNA–SSCP analysis yields quantitative estimates of transcript ratios in template pools (Cronn and Adams 2003). A calibration curve spanning different GerB:GerF template pools (100/0, 75/25, 50/50, 25/75, 0/100) showed that ratios could be estimated on a reaction tube basis (supplemental Figure 1 at http://www.genetics.org/supplemental/). Transcript levels were considered approximately equal when ratios for the two paralogs ranged from 50/50 to 60/40; preferential expression of one paralog was assumed when ratios were 61/39 or greater.
F. graminearum infection of detached spikes:
Barley spikes (cv. Morex) were excised when they reached the elongating stage (Figure 3H, stage 2; according to Skadsen et al. 2000) and placed in 50-ml plastic test tubes. Spikes were either mock-inoculated (water) or inoculated with F. graminearum (2030 spores/ml). Tubes were slowly rotated for 1 hr at 22°. After this inoculation, spikes were placed on 1% water agar and incubated at 22° for 24 and 48 hr. Spikes were rinsed prior to dissecting lemmas. Two independent experiments were conducted.
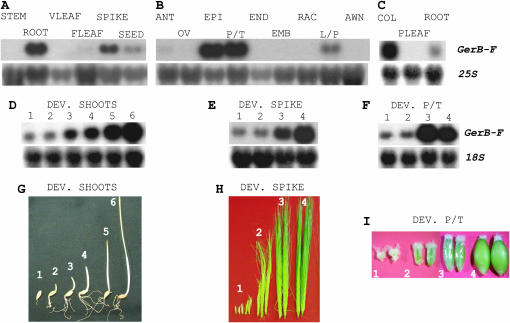
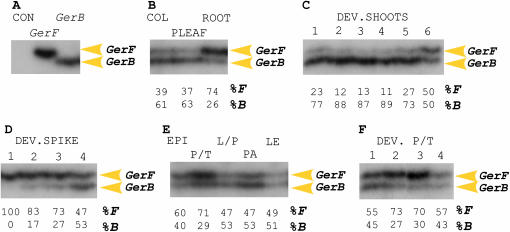
Figure 3.—
RNA gel blot analysis of GerB-F expression in barley organs. Blots were hybridized with the GerB 3′-UTR probe and rehybridized with total RNA probe. (A) GerB-F is expressed in roots, developing spikes, and seeds but not in stems, vegetative (VLEAF), or flag leaves (FLEAF). (B) GerB-F expression in spike tissues. GerB-F is highly expressed in the epicarp (EPI) and pericarp/testa (P/T). Low expression was found in lemma/palea (L/P) and at trace levels in anthers (ANT). No expression is associated with ovary (OV), endosperm (END), embryo (EMB), rachis (RAC), or awn tissues. (C) In 4-dpi seedling organs, GerB-F is highly expressed in coleoptiles (COL) and at low levels in roots. (D) GerB-F expression in developing shoots (1–6 dpi). (E) GerB-F expression in developing spikes. Stages of development were as in H. (F) GerB-F expression in developing pericarp/testa (P/T). Stages of seed development were as in I. (G) Stages of shoot development (1–6 dpi). (H) Stages of spike development were as follows: (1) pre-lemma, (2) elongating, (3) awn extension, and (4) emerging from boot (Skadsen et al. 2002). (I) Stages of seed development are described in materials and methods.
Promoter deletion analyses:
PCR primers were designed to produce 5′ deletions of the Ger promoters (Figure 6A). Morex genomic DNA was used as a template in PCR reactions using Pfx polymerase (Invitrogen). Products were translationally fused to the gfp reporter gene by replacing the Ubi1 promoter contained in the HindIII–NcoI fragment of the pAHCSGFP vector (Kaeppler et al. 2000). All expression vectors contained the Ger 5′-UTR region, which is identical in GerB and GerF. Constructs were sequenced to ensure correct synthesis. For analysis of promoter deletions, developing barley spikes, seeds, and leaves were harvested from greenhouse-grown plants on the day of transformation. Lemmas and paleas were removed from the seeds to expose the pericarp epidermis (epicarp). Coleoptiles were harvested from 1-day-old seedlings grown at 21°. Transient expression experiments were performed as previously described (Federico et al. 2005).
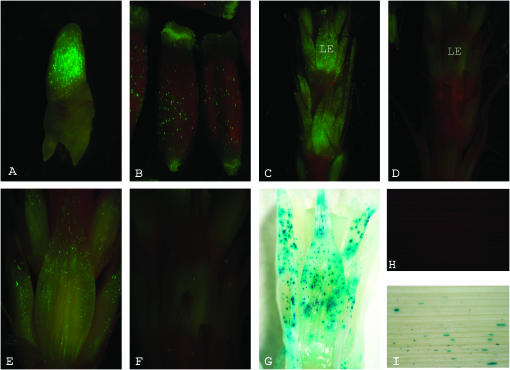
Figure 6.—
Different GerB and GerF promoter deletions drive transient gfp reporter expression in various barley organs. All constructs were co-bombarded with Ubi∷uidA expression vector, pAHC25, as an internal control. (A) Coleoptile (1 dpi). Expression is strong (+++) under GerB (−1308/+15 bp) promoter. (B) Epicarp (gelatinous). Expression is strong (+++) under GerF (−1156/+15 bp) promoter. (C) Developing spike (elongating stage). Expression is strong (+++) under GerF (−1156/+15 bp) promoter. LE, lemma. (D) Developing spike (elongating stage). Expression is weak (+) under the GerF (−97/+15) promoter. (E) Lemma (elongating stage). Expression is strong (+++) under GerF (−356/+15 bp) promoter. (F) Lemma (elongating stage). Expression is weak (+) under the GerF (−97/+15) promoter, showing the effect of the −356/−97 fragment deletion. (G) Ubiquitin-driven uidA gene expression in lemma, same as in F. (H) No expression is detected in mature leaves under GerB (−1308/+15 bp) promoter. (I) Ubiquitin-driven uidA gene expression in mature leaves.
RESULTS
GerB and GerF encode two paralogous germin-like proteins:
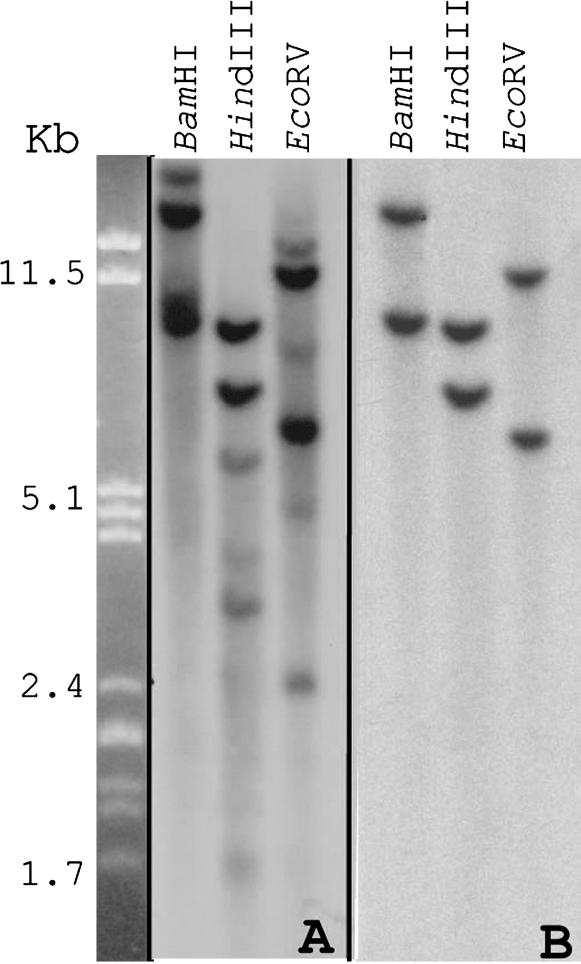
Differential display was used to produce cDNAs corresponding to the 3′ end of mRNAs found in the pericarp but not in flag leaves of barley cultivar Morex. One of these cDNAs was selected for further analysis due to its signal strength and specificity. Sequencing of this 680-bp cDNA revealed 100 and 99% nucleotide identity with the barley GerB and GerF genes, respectively. These two paralogs are part of a gene family in barley that encodes GLPs (Wu et al. 2000; Druka et al. 2002). Southern blot analysis of Morex genomic DNA, hybridized with the 680-bp GerB cDNA, confirmed that GerB belongs to a relatively small gene family (Figure 1A). This probe hybridized strongly to two and weakly to four HindIII and EcoRV fragments. Hybridization with a GerB 3′-UTR fragment (246 bp), which differs only by 5 bases with the corresponding GerF 3′-UTR fragment, revealed that this probe is specific for both GerB and GerF and does not cross hybridize with other barley GLP genes (Figure 1B). The same restriction fragment length polymorphism was observed when genomic DNA from wild barley (H. vulgare ssp. spontaneum) and two other barley cultivars (Steptoe and Golden Promise) were hybridized (supplemental Figure 2 at http://www.genetics.org/supplemental/).
Figure 1.—
Southern blots of Morex genomic DNA digested with BamHI, HindIII, or EcoRV. (A) The presence of a small germin-like gene family in barley is detected when a probe comprising the 3′-UTR and part of GerB coding sequence (680 bp) is used for hybridization. (B) GerB and GerF each appear to be single-copy genes when a 246-bp fragment composing part of the GerB 3′-UTR was used as a probe.
The transcription start sites of GerB and GerF were mapped to 15 bp upstream of the start of translation by sequencing 5′ RACE products. The deduced amino acid sequences revealed that both proteins contain 226 amino acids and differ only in amino acid number 19 (R in GERB, W in GERF). A likely signal peptide cleavage site was predicted to occur between amino acids 23 and 24 for both proteins (Wu et al. 2000). This agrees with the notion that GLPs might be associated with cell walls, as in the case of wheat germin (Lane 1994). Interestingly, GERB and GERF mature peptides are indistinguishable from each other since amino acid 19 is part of the signal peptide.
The level of replacement and synonymous site nucleotide divergence ratio (Ka/Ks = 0.00182/0.01409 = 0.129) indicates that this gene pair is likely undergoing purifying selection. Confounding effects of gene conversion events cannot be dismissed since GerB and GerF are two closely linked loci (Druka et al. 2002). However, high protein homogeneity has been maintained in this barley gene family; Ka/Ks ratios between unlinked gene family members are <0.3 for all pairs tested (supplemental Table 1 at http://www.genetics.org/supplemental/), which strongly indicates high function constraint of protein evolution. This is not surprising; structural and functional analyses revealed that barley and wheat germins have enzymatic activity only as hexamers (Lane et al. 1993; Woo et al. 2000).
Nucleotide sequence identity between these two paralogs averages 94.8% over 2329 bp (Figure 2A). Interestingly, 276 bp of proximal promoter have been 100% conserved in these two paralogs. The level of 5′ noncoding sequence conservation remains high upstream of this region but shows signs of divergence (nucleotide substitutions, deletion, and insertions). Exon I and II exhibit 98.3 and 99.6% sequence identity, respectively. Intron and 3′ noncoding regions have diverged faster than coding regions, exhibiting identities of 94 and 98%, respectively.
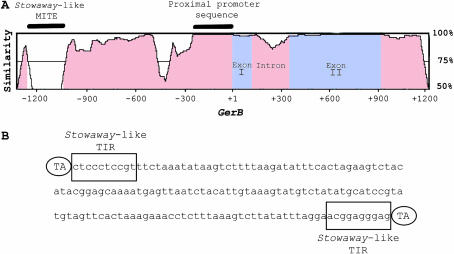
Figure 2.—
GerB and GerF are paralogous genes. (A) Barley cv. Morex GerB and GerF genomic sequence similarity is depicted in a VISTA plot. GerB sequence is plotted on the x-axis. Percentage similarity between GerF and GerB is plotted on the y-axis. The window size is 100 bp. Insertion of a Stowaway-like MITE in the GerB 5′ noncoding region (0% similarity) and 276 bp of highly conserved proximal promoter region (100% similarity) are denoted by solid bars. (B) Stowaway-like MITE sequence insertion (160 bp) present in GerB 5′ noncoding region. The first 10 bp of the terminal inverted repeats (TIRs) are conserved and match the maize Stowaway family of transposable elements consensus 5′-CTCCCTCCRT-3′ (boxed). The ovals depict the dinucleotide TA as target sequence duplications (TSDs).
A 160-bp MITE occurs 1074 bp upstream of the start of translation in GerB (Figure 2A). Analysis of its sequence revealed the duplication of the dinucleotide (TA) insertion site, the presence of 10-bp terminal inverted repeats (TIRs) matching the consensus TIR (5′-CTCCCTCCRT-3′) of maize Stowaway MITEs (Figure 2B ) (Wessler et al. 1995), and the potential to form stable secondary structures. In GerF, the target insertion site (TA) is present but not duplicated, which suggests that the insertion of this Stowaway-like MITE occurred after the duplication event. Southern blot analysis showed that this MITE belongs to a family present in high copy numbers in barley (supplemental Figure 3 at http://www.genetics.org/supplemental/).
Combined Ger B-F expression in vegetative and reproductive organs:
We determined the combined GerB-F expression pattern during development, since GerB and GerF are indistinguishable on RNA gel blots. GerB-F mRNA occurred in roots, developing spikes, and seeds, but no expression was observed in stems or leaves of mature plants (Figure 3A). In spike tissues, GerB-F mRNA levels were high in the pericarp epidermis (epicarp) and the pericarp/testa fraction (containing the epicarp) but low in lemma/palea. Trace levels were found in anthers. No expression was observed in ovary, endosperm, rachis, or awns (Figure 3B). In 4-dpi seedlings, GerB-F mRNA levels were high in coleoptiles and low in roots (Figure 3C). Trace levels of expression in seedling leaves are observed only after long exposure times (not shown).
To determine developmental expression, 1- to 6-dpi shoots, developing spikes, and developing pericarp/testa were analyzed. GerB-F expression steadily increased after germination, during growth and elongation of the shoot (coleoptile plus primary leaf; Figure 3 D and G). Similarly, GerB-F mRNA levels increased during spike development; the highest expression level was observed at stage 4, when spikes emerge from the boot sheath (Figure 3 E and H). During seed development, highest GerB-F mRNA levels were observed at the gelatinous stage (stage 3, Figure 3F). This stage precedes the beginning of epicarp desiccation (Figure 3I).
cDNA–SSCP analysis reveals diverging expression patterns of GerB and GerF genes:
Due to the highly conserved nature of GerB and GerF mRNA sequences, we used SSCP analysis of RT–PCR products to characterize their individual tissue-specific and developmental patterns of expression. This analysis clearly distinguished GerB from GerF transcripts (Figure 4A) and revealed that even though the promoter sequences of these two paralogs remain highly similar (Figure 2A), their expression patterns have diverged both in a tissue-specific and temporal manner. GerB and GerF are expressed in seedling and spike tissues, but their relative mRNA levels differ. GerB is more highly expressed in coleoptiles and primary leaves, whereas GerF is mainly expressed in roots (Figure 4B). GerB is expressed at higher levels (73–89%) in developing shoots at 1–5 dpi. At 6 dpi, however, when the combined GerB-F signal is the highest in RNA gel blots (Figure 3D) both genes contribute equally to the transcript pool (Figure 4C).
Figure 4.—
cDNA–SSCP analysis of GerB and GerF expression in barley organs. Transcript levels are shown as percentages determined by Instant Imager analysis. (A) RT–PCR controls. CON, no DNA template control; GerF and GerB, plasmid controls. (B) Seedlings (4 dpi). COL, coleoptile. PLEAF, primary leaf. (C) Developing shoots (1–6 dpi). (D) Developing spikes, staged as in Figure 3H. (E) Spike tissues. EPI, epicarp; P/T, pericarp/testa; L/P, lemma/palea; PA, palea; LE, lemma. (F) Pericarp/testa at different seed developmental stages as in Figure 3I. Results from only one strand are shown for simplicity (RT–PCR products were labeled on both strands).
GerF predominates throughout barley spike development (Figure 4D), accounting for 100% of the mRNA levels observed in RNA gel blots (Figure 3E) during pre-lemma (stage 1), 83% during elongating (stage 2), and 73% during awn extension (stage 3). Similarly to what was observed in shoots of developing seedlings, GerB and GerF contribute equally to the transcript pool (stage 4; Figure 4D) at the stage exhibiting the highest levels of GerB-F mRNA (Figure 3E), spike emergence. Independent sampling of lemmas and paleas (approximate pollination) also showed equal contribution of GerB and GerF to the transcript pool (Figure 4E). In the combined pericarp/testa fraction, GerF is preferentially expressed in the middle stages of seed development, elongating (stage 2), and gelatinous (stage 3; Figure 4F). Independent sampling of combined pericarp/testa fraction at stage 3 (Figure 4E) also revealed preferential expression of GerF.
Ger B-F expression is upregulated under biotic stress:
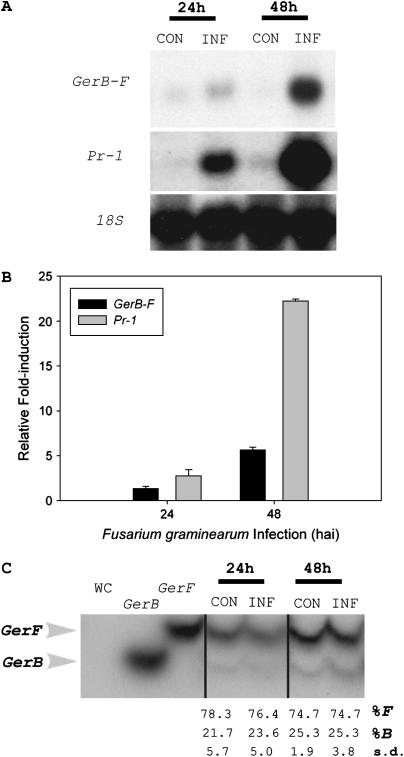
Reinforcement of cell walls is a common plant defense mechanism against fungal invasion (Agrios 1997). In wheat spikes, formation of large papillae has been observed after inoculation with F. culmorum (Kang and Buchenauer 2000). In addition, accumulation of a barley GLP (HvOxOLP) transcript correlates with papillae formation in powdery mildew-infected leaves (Wei et al. 1998). To test whether GerB and/or GerF respond to fungal infection, we inoculated detached spikes with F. graminearum (see materials and methods).
Lemma GerB-F mRNA levels in inoculated spikes increased slightly 24 hr after infection (hai) but were sixfold higher than controls by 48 hai (Figure 5, A and B). In wheat spikes, F. graminearum inoculation caused the upregulation of several defense response genes including peroxidase, PR-1, PR-2, PR-3, PR-4, and PR-5 (Pritsch et al. 2000). Thus, the induction of PR-1 mRNA levels in lemmas of detached barley spikes after F. graminearum inoculation served as a positive control for host response to infection. PR-1 mRNA levels were clearly detectable in F. graminearum-inoculated spikes, 3-fold higher than controls by 24 hai and 22-fold higher by 48 hai (Figure 5, A and B). cDNA–SSCP analysis showed that GerF is preferentially expressed in lemmas before and after F. graminearum inoculation (Figure 5C), accounting for ∼75% of the expression levels observed in RNA gel blots (Figure 5A). Both genes respond equally to infection, since their relative contribution to the transcript pool did not change during infection.
Figure 5.—
Biotic stress response in Fusarium graminearum-inoculated barley spikes. (A) RNA gel blot analysis of combined GerB and GerF (GerB-F) expression levels in lemmas. Detached spikes were either mock-inoculated (CON) or inoculated with F. graminearum (INF) for 24 or 48 hr. Blots were rehybridized with a barley PR-1 cDNA probe (positive control) and a barley 18S probe (loading control). (B) Fold induction after F. graminearum inoculation. Bars represent induction levels observed for GerB-F (solid) and PR-1 (shaded) in inoculated spikes relative to control spikes. GerB-F and PR-1 signals (counts per minute) were normalized using the corresponding counts per minute of 18S. Normalized signals for inoculated spikes were divided by the normalized signals of control spikes to obtain fold inductions. (C) cDNA–SSCP analysis of GerB and GerF expression in lemmas of mock-inoculated (CON) and F. graminearum-inoculated (INF) spikes. WC, no DNA template control; GerB and GerF, plasmid controls. Relative transcript levels (%) and corresponding standard deviations (s.d.) were determined by Instant Imager analysis.
Determination of active promoter sequence:
Wu et al. (2000) reported that β-glucuronidase expression driven by the barley GerF promoter occurs specifically in the testa and epicarp of the developing seed, while expression driven by the barley GerB promoter occurs only in the testa. However, our data indicate that both promoters should drive reporter gene expression not only in the epicarp but also in coleoptile, lemma, palea, and root tissues (Figures 3 and 4). As expected, both full-length promoters drove gfp expression in coleoptiles (Figure 6A), epicarps (Figure 6B), and lemma/palea of developing spikes (Figure 6C). No visible differences were observed between equivalent GerB and GerF promoters (dashed lines, Figure 7A).
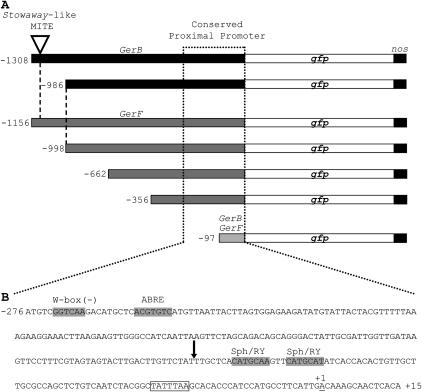
Figure 7.—
GerB and GerF promoter deletion series. (A) All constructs in the 5′ deletion series contained the 5′-UTR translationally fused to gfp followed by the nos 3′ polyadenylation signal. The arrowhead denotes the insertion of a Stowaway-like MITE in GerB 5′ noncoding region (−1234/−1074). The dashed line aligns GerB and GerF promoter sequences for comparison. The dotted line indicates the proximal promoter region conserved in both paralogs. (B) Conserved proximal upstream promoter (276 bp) and 5′-UTR (15 bp) sequences. Putative cis-acting DNA elements are indicated by shaded boxes. The putative TATA box at position −31 is boxed. The start of transcription (+1) is underlined. The start of the −97/+15 deletion is denoted by an arrow.
Series of 5′ GerB and GerF promoter deletions were constructed to locate regions conferring tissue specificity and promoter strength (Figure 7A). The deletion containing only 97 bp of a conserved proximal promoter region (Figure 7, A and B) was sufficient to drive gfp reporter gene expression in epicarps, coleoptiles, and spikes (lemma/palea), but with greatly reduced strength. This indicated that the −356/−97 GerF region was necessary for high promoter activity. When this region was deleted, expression was greatly decreased in spikes (Figure 6, C and D), lemmas (Figure 6, E and F), and epicarps (data not shown). This decline suggests that important cis-acting DNA elements must be located in this region. β-glucuronidase activity from the co-bombarded pAHC25 vector (Ubi∷uidA) served as an internal control in all bombardments (Figure 6G).
Insertion of MITEs within genes can modify promoter sequences (Yang et al. 2001; El Amrani et al. 2002). In addition, they are capable of generating an inverted repeat hairpin loop, which makes them preferential targets of the novo DNA methylation (Bender 1998). No visual differences in tissue specificity or gfp intensity were observed between GerB promoters containing or lacking the Stowaway-like MITE insertion, −1308/+15 and −986/+15, respectively (data not shown). Expression changes at the epigenetic level, however, cannot be discarded in the native gene since transient particle bombardment experiments are conducted using naked DNA.
As expected, no gfp expression was observed in mature leaves following bombardment with either GerB (Figure 6H) or GerF promoters. Expression of β-glucuronidase demonstrated that leaves were competent to express transgenes (Figure 6I). Expression was not observed in any tissue following bombardments with a promoterless gfp vector (data not shown). No transient expression experiments were conducted on roots due to their inherent green autofluorescence under blue light.
DISCUSSION
GerB and GerF are two highly conserved paralogous genes:
Differential display screening for genes that are expressed in the pericarp epidermis (epicarp) of barley, but not in leaves, detected a highly specific GLP-encoding gene, GerB. Sequence and Southern blot analyses (Figure 1) confirmed that GerB belongs to a small gene family (Wu et al. 2000; Druka et al. 2002), that GerF is its closest paralog, and that the duplication event occurred before barley domestication (supplemental Figure 3 at http://www.genetics.org/supplemental/). GerB and GerF genomic sequences are highly conserved (Figure 2A) and encode apoplastic proteins that differ only by one amino acid in the signal peptide. Thus, GERB and GERF mature proteins are identical. The rate of nucleotide synonymous substitution (Ks) between these two coding sequences is 0.01409, suggesting that these duplicates originated recently (Ks < 0.02) (Moore and Purugganan 2003). This finding represented an opportunity to evaluate the individual expression patterns of recently duplicated paralogs in barley.
GerB and GerF exhibit overlapping redundancy and early signs of subfunctionalization:
The duplication–degeneration–complementation model, or subfunctionalization, suggests that a pair of paralogs accumulate degenerative yet complementary mutations that fractionate the ancestral gene's subfunctions (Force et al., 1999). As a result, paralog expression patterns diverge, yet they complement to reproduce the ancestral expression pattern of the single gene progenitor. Due to the highly conserved nature of GerB and GerF mRNAs, we characterized their individual tissue-specific and developmental patterns of expression using RNA gel blots (Figure 3) coupled with cDNA–SSCP analysis (Figure 4). Both paralogs continue to be expressed and exhibit overlapping redundancy and signs of early subfunctionalization. GerB is predominantly expressed in coleoptiles and primary leaves (developing shoots 1–5 dpi; Figure 4C), while GerF is predominantly expressed in roots (Figure 4B), developing spikes (stages 1–3; Figure 4D), and pericarp/testa (Figure 4, E and F).
Adams et al. (2003) used cDNA–SSCP analysis to follow the expression of homeologs of 40 different genes in allotetraploid cotton, describing the occurrence of developmentally regulated, organ-specific reciprocal gene silencing (one duplicate predominantly expressed in one organ and its counterpart in another) in several gene homeologs including one oxalate oxidase gene (B5). They explained these changes in gene expression invoking epigenetic mechanisms, probably involving organ-specific chromatin states and position effects (Adams et al. 2003; Liu and Wendel 2003). Similarly, our cDNA–SSCP analysis revealed that GerB expression predominates over GerF expression during most of shoot development (Figure 4C), while the opposite is true during spike development (Figure 4D). Interestingly, both GLP genes are expressed equally at the final stage in both developmental series, at which the highest GerB-F expression levels were observed in RNA gel blots (Figure 3, D and E; Figure 4, C and D). The reciprocal silencing of genes that encode nearly identical proteins, as in the case of GerB and GerF or B5 (Adams et al. 2003), could be functionally and selectively important for dosage effect reasons. Dosage effects have been observed for many genes, including key regulators of developmental processes, in both diploid and polyploid species (reviewed by Osborn et al. 2003). Alternatively, they may only represent two duplicated genes at an early and progressive stage of subfunctionalization.
GerB and GerF retain a conserved proximal promoter:
GerB and GerF spatial, temporal, and inducible patterns of expression make the promoters of these genes potential candidates for targeting Fusarium head blight resistance in barley. Analysis of GerB and GerF genomic sequences (Figure 2A) revealed that these paralogs retain an identical 276-bp 5′ proximal promoter region (dotted line; Figure 6, A and B). Previously, we showed that a 247-bp Ltp6 promoter sequence drove the expression of gfp in transgenic barley plants, reproducing the expression pattern of the native Ltp6 gene (Federico et al. 2005). This demonstrated that most of the tissue-specific and developmental determinants were present in the proximal promoter. Similarly, several putative cis-acting DNA elements located in GerB and GerF proximal promoters (Figure 6B) may explain the expression patterns exhibited by this pair of duplicated genes. Two Sph/RY-like elements (Bäumlein et al. 1992) at positions −80 and −90, one ABA-responsive element (ABRE; Hattori et al. 2002) at position −256, and a W-box (Eulgem et al. 1999) at position −265 are found within the 276 bp of the conserved proximal promoter region (Figure 6B).
Arabidopsis ABI3, which is orthologous to maize Vp1, has been postulated to function as a general regulator for the timing of developmental transitions throughout the life cycle of plants (Rohde et al. 2000). ABI3/Vp1 encodes a transcription factor capable of acting on at least two distinct types of cis-elements, ABRE and Sph/RY. While its action on Sph/RY involves direct binding to this element, its effect on ABREs is accomplished indirectly via the interaction with other transcription factors, such as TRAB1 and ABI5 (Hobo et al. 1999; Nakamura et al. 2001). One ABRE in particular, ACGTGTC, was found to be significantly enriched in ABI3/Vp1- and ABA-inducible gene promoters (Suzuki et al. 2003). Interestingly, the putative ABRE motif located at position −256 has the same sequence (Figure 6B). If this motif is functional in GerB and GerF promoters, the decreased GerF promoter activity observed after deleting the −356/−97 region (Figure 7, C–F) could be the result of disrupting important protein–protein interactions. These interactions might involve transcription factors bound to this ABRE and the two Sph/RY-like motifs (RY/ABRE complex) located at positions −80 and −90 (Figure 6B).
GerB and GerF expression patterns are consistent with proposed roles in cell wall modification during plant development and defense:
Germin isoforms are discrete markers of wheat development and are associated with cell walls during seed germination and maturation (Thompson and Lane 1980; Lane et al. 1992). Their hydrogen peroxide (H2O2)-generating capacity together with their mRNA accumulation patterns and apoplastic location are consistent with a role in cell wall extensibility, restriction of organ growth, and lignification during plant development and defense (Lane 1994). Cell wall modification during development (e.g., germination) or pathogen infection often requires the cross-linking of cell wall polymers (Carpita and Gibeaut 1993; Dumas et al. 1995; Lane 1994, 2002). Germins might mediate cell wall modifications by locally generating the H2O2 required for such reactions (Lane 1994).
Wu et al. (2000) located prominent oxalate oxidase activity in root tips and root vascular tissues of barley seedlings and in the epicarp and root primordia of developing seeds but could not link this activity to GerB or GerF using transient expression assays. In wheat, oxalate oxidase activity has been localized to cell walls of the coleoptiles in elongating shoots (Caliskan and Cuming 1998) and to cell walls of epicarps, lemma/palea, and glumes in developing seeds (Lane 2000). In this report, we have shown that both GerB and GerF are expressed in roots, coleoptiles, epicarp, and lemma/palea tissues (Figures 3 and 4). In addition, we have shown that GerB and GerF promoters are capable of driving the expression of a reporter gene in coleoptiles, epicarps, and lemma/palea (Figure 7, A–C). Thus, GerB and GerF expression patterns correlate with the observed oxalate oxidase activity in barley.
GerB and GerF response to F. graminearum infection suggests that these genes might provide the H2O2 required for cell wall strengthening and/or signaling under pathogen attack (Figure 5). The putative W-box, TTGACC, present in GerB and GerF conserved proximal promoter sequence (Figure 6B) could mediate this pathogen response, as it has been shown in the Pathogenesis-related Class 10 (PR-10) genes in parsley (Eulgem et al. 1999).
GLPs can catalyze a variety of enzymatic reactions. GLPs from tobacco (NectarinI) and a moss (BuGLP) are superoxide dismutases (SODs) (Carter and Thornburg 2000; Yamahara et al. 1999); a GLP from barley (HvGLP1) is an ADP-glucose pyrophosphatase/phosphodiesterase (Rodriguez-Lopez et al. 2001) and a GLP from pear (PpABP20) is an auxin-binding protein (Ohmiya 2002). Interestingly, a protein sequence comparison between GERB and GERF and a group of germins and GLPs of known enzymatic activity relates them to those exhibiting SOD activity (supplemental Figure 4 at http://www.genetics.org/supplemental/). This activity, as in the case of the oxalate oxidases, could also provide the H2O2 required for cell wall cross-linking during development and/or pathogen infection.
Retention of GerB and GerF expression after duplication correlates with putative function:
Loss is the most likely fate of a duplicated gene (Walsh 1995; Lynch and Connery 2003). Fewer than 27% of A. thaliana (Blanc et al. 2003) and 16% of Saccharomyces cerevisiae (Wong et al. 2002) genes have been retained as duplicates after evolution through polyploidization. Retention has often been explained by functional divergence occurring by either neo- or subfunctionalization (Force et al. 1999; Blanc and Wolfe 2004b). GerB and GerF spatial and temporal divergence of expression suggests that this pair of paralogs is likely undergoing the early stages of subfunctionalization. However, retention can also be explained by a selective advantage acquired through the functionally redundant activity of duplicated genes (Osborn et al. 2003). Loss and retention of duplicated genes has recently been found to correlate with gene function (Moore and Purugganan 2003; Maere et al. 2005). We hypothesize that GerB and GerF have been retained as duplicates in the barley genome because they play important roles in plant development and defense. During development, changes in the relative timing of events can result in extensive morphological changes (Hunter and Poethig 2004). Events such as cessation of coleoptile elongation and lignification of lemma, palea, and epicarp need to occur at specific developmental stages. Correct timing may be controlled in part by the RY/ABRE complex present in the highly conserved proximal promoter region (Figure 6B). Thus, retention of redundancy could have been selectively maintained due to the fitness cost of developmental error (Gu et al. 2003; Moore et al. 2005). Additionally, functional retention of these duplicated genes could be explained by the selective advantage provided by increased levels of gene product (Force et al. 1999; Gu et al. 2003; Osborn et al. 2003) during defense responses against pathogen infection.
Acknowledgments
We thank John Herbst, Laura Oesterle and Yim Candace Chan for technical assistance and Chris Pyres for critically reviewing the manuscript. This work was supported in part by the American Malting Barley Association, the United States Department of Agriculture, and the Fulbright Program (fellowship to M.L.F). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture.
References
- Adams, K., R. Cronn, R. Percifield and J. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios, G. (Editor), 1997. Plant Pathology, pp. 93–114. Academic Press, San Diego.
- Altschul, S., W. Gish, W. Miller, E. Myers and D. Lipman, 1990. Basic local alignment research tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bäumlein, H., I. Nagy, R. Bassüner, R. Villaroel, D. Inze et al., 1992. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 2: 233–239. [PubMed] [Google Scholar]
- Bender, J., 1998. Cytosine methylation of repeated sequences in eukaryotes: the role of DNA pairing. Trends Biochem. Sci. 23: 252–256. [DOI] [PubMed] [Google Scholar]
- Birchler, J., U. Bhadra, M. Bhadra and D. Auger, 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and K. Wolfe, 2004. a Widespread paleoploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and K. Wolfe, 2004. b Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., K. Hokamp and K. Wolfe, 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan, M., and A. Cuming, 1998. Spatial specificity of H2O2-generating oxalate oxidase gene expression during wheat embryo germination. Plant J. 15: 165–171. [DOI] [PubMed] [Google Scholar]
- Carpita, N., and D. Gibeaut, 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1–30. [DOI] [PubMed] [Google Scholar]
- Carter, C, and R. W. Thornburg, 2000. Tobacco nectarin I: purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J. Biol. Chem. 275: 36726–36733. [DOI] [PubMed] [Google Scholar]
- Chirgwin, J. M., A. Prybyla, R. J. Mac Donald and W. J Rutter, 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18: 5294–5299. [DOI] [PubMed] [Google Scholar]
- Comeron, J., 1999. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15: 763–764. [DOI] [PubMed] [Google Scholar]
- Cronn, R., and K. Adams, 2003. Quantitative analysis of transcript accumulation from genes duplicated by polyploidy using cDNA-SSCP. BioTechniques 34: 726–734. [DOI] [PubMed] [Google Scholar]
- Druka, A., D. Kudrna, C. Kannangara, D. von Wettstein and A. Kleinhofs, 2002. Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc. Natl. Acad. Sci. USA 99: 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, B., G. Freyssinet and K. Pallet, 1995. Tissue-specific expression of germin-like oxalate oxidase during development and fungal infection of barley seedlings. Plant Physiol. 108: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amrani, A., L. Marie, A. Aïnouche, J. Nicolas and I. Couée, 2002. Genome-wide distribution and potential regulatory functions of AtATE, a novel family of miniature inverted-repeat transposable elements in Arabidopsis thaliana. Mol. Genet. Gen. 267: 459–471. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., P. Rushton, E. Schmelzer, K. Hahlbrock and I. Somssich, 1999. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18: 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico, M., H. Kaeppler and R. Skadsen, 2005. The complex developmental expression of a novel-stress responsive barley Ltp gene is determined by a shortened promoter sequence. Plant Mol. Biol. 57: 35–51. [DOI] [PubMed] [Google Scholar]
- Feinberg, A., and B. Vogelstein, 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Analytical Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. Pickett, A. Amores, Y. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., L. Steinmetz, X. Gu, C. Scharfe, R. Davis et al., 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66. [DOI] [PubMed] [Google Scholar]
- Hattori, T., M. Totsuka, T. Hobo, Y. Kagaya and A. Yamamoto-Toyoda, 2002. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 43: 136–140. [DOI] [PubMed] [Google Scholar]
- Higo, K., Y. Ugawa, M. Iwamoto and T. Korenaga, 1999. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acid Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A., 1937. Economic Botany, pp. 309–333. McGraw Hill, New York.
- Hobo, T., Y. Kowyama and T. Hattori, 1999. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic–induced transcription. Proc. Natl. Acad. Sci. USA 96: 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C., and S. Poethig, 2004. ReFUSing to grow up. Dev. Cell 7: 288–289. [DOI] [PubMed] [Google Scholar]
- Kaeppler, H., G. Menon, R. Skadsen, A. Nuutila and A. Carlson, 2000. Transgenic oat plants via visual selection of cells expressing green fluorescent protein. Plant Cell Rep. 19: 661–666. [DOI] [PubMed] [Google Scholar]
- Kang, Z., and H. Buchenauer, 2000. Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiol. Mol. Plant Pathol. 57: 255–268. [Google Scholar]
- Krug, M. S., and S. L. Berger, 1987. First-strand cDNA synthesis primed with oligo(dT). Methods Enzymol. 152: 316–325. [DOI] [PubMed] [Google Scholar]
- Lane, B., 1994. Oxalate, germin, and the extracellular matrix of higher plants. FASEB J 8: 294–301. [DOI] [PubMed] [Google Scholar]
- Lane, B., 2000. Oxalate oxidases and differentiating surface structure in wheat: germins. Biochem. J. 349: 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, B., 2002. Oxalate, germins, and higher plant-pathogens. IUBMB Life 53: 67–75. [DOI] [PubMed] [Google Scholar]
- Lane, B., F. Bernier, E. Dratewka-Kos, R. Shafai, T. Kennedy et al., 1991. Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. J. Biol. Chem. 266: 10461–10469. [PubMed] [Google Scholar]
- Lane, B., A. Cuming, Y. Fregeau, N. Carpita, W. Hurkman et al., 1992. Germin isoforms are discrete temporal markers of wheat development. Pseudogermin is a uniquely thermostable water-soluble oligomeric protein in ungerminated embryos and like germin in germinated embryos, it is incorporated to cell walls. Eur. J. Biochem. 209: 961–969. [DOI] [PubMed] [Google Scholar]
- Lane, B., J. Dunwell, J. Ray, M. Schmitt and A. Cuming, 1993. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268: 12239–12242. [PubMed] [Google Scholar]
- Liang, P., and A. Pardee, 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971. [DOI] [PubMed] [Google Scholar]
- Liu, B., and J. Wendel, 2003. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29: 365–379. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. Connery, 2003. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3: 489–505. [PubMed] [Google Scholar]
- Lynch, M, and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S., S. De Bodt, J. Raes, T. Casneuf, M. Van Montagu et al., 2005. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., M. Brudno, J. Schwartz, A. Poliakov, E.M Rubin et al., 2000. Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046. [DOI] [PubMed] [Google Scholar]
- Moore, R., and M. Purugganan, 2003. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 100: 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R., and M. Purugganan, 2005. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128. [DOI] [PubMed] [Google Scholar]
- Moore, R., S. Grant and M. Purugganan, 2005. Molecular population genetics of redundant floral-regulatory genes in Arabidopsis thaliana. Mol. Biol. Evol. 22: 91–103. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., T. Lynch and R. Finkelstein, 2001. Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635. [DOI] [PubMed] [Google Scholar]
- Ohmiya, A., 2002. Characterization of ABP19/20, sequence homologues of germin-like protein in Prunus persica L. Plant Sci. 163: 683–689. [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Osborn, T., J. Pires, J. Birchler, D. Auger, Z. Chen et al., 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Paterson, A., 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsch, C., G. Muehlbauer, W. Bushnell, D. Somers and C. Vance, 2000. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant-Microbe Interact. 13: 159–169. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez, M., E. Baroja-Fernandez, A. Zandueta-Criado, B. Moreno-Bruna, F. J. Muñoz et al., 2001. Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett. 490: 44–48. [DOI] [PubMed] [Google Scholar]
- Rohde, A., S. Kurup and M. Holdsworth, 2000. ABI3 emerges from the seed. Trends Plant Sci. 5: 418–419. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Skadsen, R., 1993. Aleurones from a barley with low α-amylase activity become highly responsive to gibberellin when detached from the starchy endosperm. Plant Physiol. 102: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skadsen, R., and T. Hohn, 2004. Use of Fusarium graminearum transformed with gfp to follow infection patterns in barley and Arabidopsis. Physiol. Mol. Plant Pathol. 64: 45–53. [Google Scholar]
- Skadsen, R., P. Schulze-Lefert and J. Herbst, 1995. Molecular cloning, characterization and expression analysis of two catalase isozyme genes in barley. Plant Mol. Biol. 29: 1005–1014. [DOI] [PubMed] [Google Scholar]
- Skadsen, R., P. Sathish and H. Kaeppler, 2000. Expression of thaumatin-like permatin PR-5 genes switches from the ovary wall to the aleurone in developing barley and oat seeds. Plant Sci. 156: 11–22. [DOI] [PubMed] [Google Scholar]
- Skadsen, R. W., P. Sathish, M. L. Federico, T. Abebe, J. Fu et al., 2002. Cloning of the promoter for a novel barley gene, Lem1, and its organ-specific promotion of Gfp expression in lemma and palea. Plant Mol. Biol. 49: 545–555. [DOI] [PubMed] [Google Scholar]
- Smith, R., B. Wiese, M. Wojzynski, D. Davison and K. Worley, 1996. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6: 454–462. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., M. Ketterling, Q.-B. Li and D. McCarty, 2003. Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Phys. 132: 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, E., and B. Lane, 1980. Relation of protein synthesis in imbibing wheat embryos to the cell-free translational capacities of bulk mRNA from dry and imbibing embryos. J. Biol. Chem. 255: 5965–5970. [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A., 2005. Distributed robustness versus redundancy as causes of mutational robustness. BioEssays 27: 176–188. [DOI] [PubMed] [Google Scholar]
- Walsh, J., 1995. How often do duplicated genes evolve new functions? Genetics 139: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Z. Zhang, C. Andersen, E. Schmelzer, P. Gregersen et al., 1998. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol. Biol. 36: 101–112. [DOI] [PubMed] [Google Scholar]
- Wessler, S., T. Bureau and S. White, 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5: 814–821. [DOI] [PubMed] [Google Scholar]
- Wong, S., G. Butler and K. Wolfe, 2002. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. USA 99: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, E-J., J. Dunwell, P. Goodenough, A. Marvier and R. Pickersgill, 2000. Germin is a manganese containing homohexamer with oxalate and superoxide dismutase activities. Nat. Struct. Biol. 7: 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wu, S., A. Druka, H. Horvath, A. Kleinhofs, G. Kannangara et al., 2000. Functional characterization of seed coat-specific members of the barley germin gene family. Plant Physiol. Biochem. 38: 685–698. [Google Scholar]
- Yamahara, T., T. Shiono, T. Suzuki, K Tanaka, S. Takio et al., 1999. Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J. Biol. Chem. 274: 33274–33278. [DOI] [PubMed] [Google Scholar]
- Yang, G., J. Dong, M. Chandrasekharan and T. Hall, 2001. Kiddo, a new transposable element family closely associated with rice genes. Mol. Genet. Genomics 266: 417–424. [DOI] [PubMed] [Google Scholar]