Abstract
Data from completely sequenced genomes are likely to open the way for novel studies of the genetics of nonmodel organisms, in particular when it comes to the identification and analysis of genes responsible for traits that are under selection in natural populations. Here we use the draft sequence of the chicken genome as a starting point for linkage mapping in a wild bird species, the collared flycatcher—one of the most well-studied avian species in ecological and evolutionary research. A pedigree of 365 flycatchers was established and genotyped for single nucleotide polymorphisms in 23 genes selected from (and spread over most of) the chicken Z chromosome. All genes were also found to be located on the Z chromosome in the collared flycatcher, confirming conserved synteny at the level of gene content across distantly related avian lineages. This high degree of conservation mimics the situation seen for the mammalian X chromosome and may thus be a general feature in sex chromosome evolution, irrespective of whether there is male or female heterogamety. Alternatively, such unprecedented chromosomal conservation may be characteristic of most chromosomes in avian genome evolution. However, several internal rearrangements were observed, meaning that the transfer of map information from chicken to nonmodel bird species cannot always assume conserved gene orders. Interestingly, the rate of recombination on the Z chromosome of collared flycatchers was only ∼50% that of chicken, challenging the widely held view that birds generally have high recombination rates.
THE possibility of identifying genes coding for traits that are under selection in natural populations and, given the availability of such knowledge, subsequently studying key aspects of evolutionary biology has only recently become a realistic goal (Feder and Mitchell-Olds 2003; Slate 2005; Vasemagi and Primmer 2005). Examples of questions that should be possible to address are how local adaptation relates to genotypic variation and how genetic variation can be maintained for fitness-related traits by, for instance, genotype–environment interaction (Stearns 1992). Various approaches may be taken to attack the missing link between genotypes and phenotypes in wild populations of nonmodel organisms, including candidate gene approaches (Tabor et al. 2002), transcriptome profiling through EST sequencing (Le Quere et al. 2004) or microarray-based hybridization (Drnevich et al. 2004), genome scans for regions subject to selective sweeps (Storz 2005), and linkage/quantitative trait loci (QTL) mapping (Slate 2005). To various extents, all these approaches require some prior knowledge of the genetics of the species under study, which until recently has been a major limiting factor in the case of most natural populations. This is particularly true for genetic mapping approaches in which a large number of polymorphic markers have to be developed and placed on a primary linkage map.
However, an increasing number of linkage maps are now being reported from natural populations of organisms, such as butterflies (Jiggins et al. 2004), fishes (Chistiakov et al. 2005), fungi (Marra et al. 2004), insects (Lorenzen et al. 2005), molluscs (Hubert and Hedgecock 2004), and amphibians (Smith et al. 2005). They are so far generally limited to species that can easily be bred in captivity or where sufficiently large litter sizes are being produced in natural settings and are accessible to sampling to allow the establishment of pedigrees necessary for linkage analysis. Unfortunately, this is not the case for most species of birds. In addition to the well-established linkage map of the chicken (Groenen et al. 2001), mapping is underway in some domestic galliforms of agricultural interest, including quail (Kikuchi et al. 2005) and turkey (Reed et al. 2005), but there are only preliminary attempts at linkage mapping in wild species, notably by the work of Hansson et al. (2005). Given the role birds play in evolutionary research, improved genetic resources, such as the development of detailed linkage maps, are likely to open the way for advancing our understanding of the role of natural selection in wild bird populations (Edwards et al. 2005).
In this study we focus on one of the most well-studied avian species in evolutionary research, the collared flycatcher (Ficedula albicollis). By the standards of the field, it has become a “role model” organism for studies of, for example, life history evolution, mate choice, sexual selection, and speciation (Gustafsson and Sutherland 1988; Gustafsson and Pärt 1990; Gustafsson et al. 1995; Ellegren et al. 1996; Saetre et al. 1997; Qvarnström et al. 2000, 2006, Merilä et al. 2001; Veen et al. 2001; Michl et al. 2002), and this is also valid for its closely related sister species, the pied flycatcher (F. hypoleuca) (Alatalo et al. 1986; Merino et al. 1996; Tapio and Lehikoinen 2000; Both and Visser 2001). For the former species, this work mainly originates from a long-term study of a Baltic Sea island population. This population, showing high philopatry, has been closely followed and monitored for fitness-related traits, including lifetime reproductive success, for >25 years. It forms an ideal setting for searching for genotype–phenotype–function connections, provided that at least some basic knowledge of the species' genome can be obtained. In addition, natural hybridization between the two flycatcher species makes this system highly suitable for studying the genetic basis of reproductive isolation barriers.
The recent report of a draft sequence of the chicken genome (International Chicken Genome Sequencing Consortium 2004) offers a most promising resource for the transfer of genetic information among bird species and for the study of avian evolutionary genomics (Ellegren 2005). Here, we make use of chicken genomic resources to make a targeted attempt at linkage mapping in collared flycatchers. We have generated a pedigree of >350 birds with parentage confirmed by genetic profiling; the pedigree consists of 27 half-sib families where each male has mated with several females. In this study we specifically focus on the Z sex chromosome, according to the following. First, we evaluate the usefulness of the chicken genome sequence for developing conserved sex-linked gene markers in a distantly related bird species. Chicken belongs to the order Galliformes and collared flycatcher to the order Passeriformes; these lineages split early in the radiation of neognath (non-ratite) birds, probably ∼100 million years ago (van Tuinen and Hedges 2001). Second, we use these gene markers for the identification of single nucleotide polymorphisms (SNPs) in the collared flycatcher. Third, by collecting data for some 20,000 SNP genotypes in the pedigree (53 SNPs typed in 365 individuals) we establish a high-density, gene-based linkage map of the collared flycatcher Z chromosome. Finally, we use the flycatcher Z chromosome linkage map to address the questions of conserved synteny in avian sex chromosome evolution and the evolution of avian recombination rates.
MATERIALS AND METHODS
Sampling and DNA extraction:
Blood samples were collected from collared flycatchers breeding in nest boxes on the islands Öland and Gotland in the Baltic Sea in 2001–2004. Approximately 10 μl blood was collected from adults and offspring by venipuncture of the cutaneus ulnar wing vein with a sterile syringe. Blood was stored frozen in EDTA buffer until usage. DNA was extracted by cell digestion with proteinase K at 37° overnight, followed by three rounds of phenol-chlorophorm DNA purification. Purified DNA was precipitated with NaAc and 95% EtOH, washed once in 70% EtOH, and the dried pellet was dissolved in water.
The structure of the pedigree with 365 birds is described in supplemental Table 1 (http://www.genetics.org/supplemental/). The final linkage analysis was made for 62 litters with a total of 280 offspring, fathered by 27 males, each of which had mated with 2–4 females; the pedigree thus had the two-generation half-sib design.
Identification of extra-pair offspring:
Extra-pair paternity is known to occur in collared flycatchers (Sheldon and Ellegren 1999) and illegitimate offspring must obviously be excluded from linkage analysis, at least for the study of male recombination fractions. Although extra-pair offspring (EPO) are most likely to be identified from non-Mendelian inheritance in large-scale SNP data sets, we sought to minimize the number of EPO being subjected to SNP typing by first genotyping all families for five microsatellite markers (FhU2, FhU4, Mcyμ4, Pdoμ5, and Pca3) (Ellegren 1992; Primmer et al. 1996; Double et al. 1997; Griffith et al. 1999; Dawson et al. 2000). These markers were analyzed essentially following the conditions described in the original reports, using fluorescently labeled primers. Fragment length analysis was performed on a MegaBACE 96 capillary instrument using ROX size standard (Applied Biosystems), and genotypes were scored using the software Genetic Profiler (Amersham Biosciences).
Offspring whose genotypes deviated from Mendelian expectations on the basis of the presumed parental genotypes at two or more microsatellite loci were excluded from further analyses. Although retaining offspring with an incompatible genotype combination at a single locus meant that some EPO were likely to remain after this first selection, it allowed for including offspring in the final analysis in which a microsatellite genotyping error or a microsatellite mutation had potentially occurred. Of the 344 offspring initially tested, 44 (12.8%) were identified as EPO according to microsatellite fingerprinting. From the subsequent SNP genotyping another 20 offspring were also classified as EPO, giving a total frequency of 18.6%. After removal of these 64 offspring, the final data set used in linkage analysis included 280 offspring.
Marker development and screening for SNP markers:
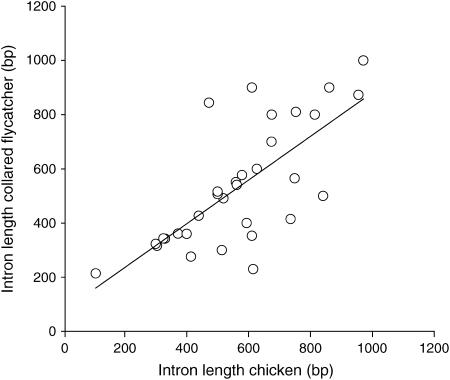
The protocol for identification of polymorphic markers to be used in flycatcher linkage analysis was to design PCR primers from exonic sequences flanking introns of Z-linked chicken genes, amplification of the orthologous loci in flycatchers, followed by SNP screening using resequencing of eight unrelated male birds. We initially selected 135 candidate markers, which in each case flanked a single intron of an equal number of Z-linked genes. The markers were chosen on basis of (i) being evenly distributed along the chicken Z chromosome in the draft assembly (http://www.ensembl.org/Gallus_gallus/mapview?chr=Z), (ii) flanking introns preferably in the size range of 200–1000 bp in chicken (which, assuming that intron length in chicken and flycatcher would be correlated, was expected to facilitate amplification and direct sequencing in flycatcher; the assumed correlation was subsequently confirmed within the range of intron sizes analyzed: r2 = 0.52, P < 0.01, Figure 1), and (iii) showing some degree of exonic sequence conservation when BLASTed (http://www.ncbi.nih.gov/BLAST/) to other vertebrate genome sequences (mainly human, mouse, rat, and zebrafish). Importantly, particularly conserved regions were targeted for primer design. Most of the markers come from Ensembl predicted genes (http://www.ensembl.org/info/data/docs/genome_annotation.html). In addition, we specifically searched for chicken introns containing microsatellite sequences using the default setting of the Sputnik program (http://espressosoftware.com/pages/sputnik.jsp); 42 such introns were identified, included in the total of 135. The position of genes on the chicken Z chromosome as indicated in Table 1 is according to build 1.1 of the chicken genome; these positions are likely to change when new builds are constructed and given that errors in the draft assembly seem to occur (see below).
Figure 1.—
Correlation between intron size (in base pairs) in chicken and collared flycatcher (y = 0.81x + 74.7).
TABLE 1.
All gene markers placed on the collared flycatcher linkage map
| Position on chicken Zc | Lengthd
|
BLASTN
|
|||||
|---|---|---|---|---|---|---|---|
| Gene | Gene symbola | Gene IDb | Exon | Intron | Best hit | Score | |
| Vasculin | GPBP1 | 23746 | 1005436 | 120 | 844 | Z 1005351 | 428E-31 |
| Importin-11 | IPO11 | 23770 | 2866719 | 0 | 744 | Z 1784654 | 0,082 |
| ADAMTS6 variant 2 | ADAMTS6 | 23786 | 3783173 | 56 | 491 | Z 3783135 | 150E-122 |
| Peptidylprolyl isomerase domain | PPWD1 | 23799 | 3862227 | 107 | 577 | Z 3862780 | 630E-43 |
| Zinc transporter 5 | SLC30A5 | 23871 | 5128095 | 93 | 361 | Z 5128443 | 130E-33 |
| Growth hormone receptor precursor | GHR | 23973 | 6621490 | 13 | 507 | Z 6622618 | 0,016 |
| NAD(P) transhydrogenase | NNT | 23990 | 6824865 | 0 | 540 | — | — |
| Poly (ADP-ribose) polymerase, member 8 | PARP8 | 24009 | 7878620 | 35 | 895 | Z 7878950 | 0,00011 |
| Similar to TGF β-inducible protein 1 | 24105 | 24105 | 9914755 | 166 | 360 | Z 9915877 | 120E-61 |
| Ras GTPase-activating-like protein | IQGAP2 | 24162 | 10609097 | 150 | 343 | Z 10609027 | 500E-57 |
| Glycine dehydrogenase | GLDC | 24274 | 12149860 | 99 | 452 | Z 12150318 | 140E-43 |
| N-acylsphingosine amidohydrolase 3 like | ASAH3L | 24328 | 13516515 | 0 | 573 | Z 13516956 | 0,0043 |
| Amyloid β A4 precursor | APBA1 | 24379 | 15556988 | 0 | 632 | Z 15557080 | 0,0051 |
| Transient receptor cation channel 3 | TRPM3 | 24411 | 15902290 | 108 | 541 | Z 15902836 | 330E-81 |
| Transient receptor cation channel 6 | TRPM6 | 24457 | 17282846 | 77 | 733 | Z 17283040 | 120E-18 |
| Hypothetical protein | 24555 | 24555 | 19651339 | 75 | 427 | Z 19651751 | 230E-09 |
| Hypothetical protein | 24638 | 24638 | 22459475 | 83 | 332 | Z 22454175 | 130E-26 |
| Phosphodiesterase 6 β subunit | GAF | 24804 | 25383953 | 83 | 487 | Z 25384564 | 660E-33 |
| Neuregulin-1 | NRG1 | 24876 | 25804434 | 59 | 506 | Z 25803889 | 270E-25 |
| ATP-binding cassette A member 1 | ABCA1 | 24891 | 26519192 | 164 | 230 | Z 26519097 | 160E-62 |
| Fructose-bisphosphate aldolase B | ALDOB | 25056 | 27566818 | 62 | 460 | Z 27541710 | 640E-13 |
| Zinc finger FYVE domain 16 | ZFYVE16 | 25117 | 28483373 | 85 | 379 | Z 28483805 | 260E-31 |
| Hypothetical protein | 25189 | 25189 | 29865246 | 31 | 878 | — | — |
For hypothetical proteins, gene symbols were given according to the ENSEMBL transcript ID.
Gene IDs are given as the last five numbers of the ENSEMBL transcript ID (ENSGALT000000xxxxx).
Position of the first nucleotide in the exon immediately upstream of the intron amplified in the collared flycatcher on the chicken Z chromosome, according to the draft genome assembly (build 1.1).
Length of the obtained sequence in collared flycatchers.
PCR reactions were performed in 20-μl reactions in a PTC-100 thermal cycler (MJ Research) with 0.5 pmol of each primer, 50 μm of each dNTP, 2.5 mm MgCl2, 0.025 units AmpliTaq Gold (Applied Biosystems), and 50 ng of template DNA. The general temperature profile was 5 min at 95°, 30–40 cycles of 30 sec at 95°, 30 sec at annealing temperature (45–60°) and 30–60 sec at 72°, followed by a final extension step at 72° for 5–10 min. Primer sequences are provided in supplemental Table 2 (http://www.genetics.org/supplemental/). Amplification success was initially examined using three male samples. For markers revealing a specific amplification product, a total of eight male samples (16 chromosomes) were amplified and these products were purified with Exo-SAP (USB). Four μl of purified product was subject to sequencing using 1 pmol of forward or reverse primer and 5 μl of DYEnamic ET Terminator cycle sequencing mix (Amersham Biosciences) for 29 cycles of 20 sec at 95°, 15 sec at 50°, and 30 sec at 72°. Sequencing reactions were purified with the 96-well Autoseq system (Amersham Biosciences) and run on a MegaBACE 96 capillary instrument.
Sequences were edited and aligned in Sequencher (Gene Codes Corporation) and SNPs, typically identified from overlapping forward and reverse sequencing, were scored by hand. The obtained collared flycatcher sequences were used in BLASTN searches at Ensembl (http://www.ensembl.org/Gallus_gallus) to confirm orthology to a single region on chicken Z chromosome. SNPs subsequently used for pedigree genotyping were selected with preference for high frequency alleles (>20% minor allele frequency, MAF).
SNP genotyping:
The 365 individuals were genotyped at each of 53 selected SNP loci using the 12-plex GenomeLab SNPStream system (Beckman Coulter; Bell et al. 2002). The primers for PCR and minisequencing were designed using the Autoprimer software (http://www.autoprimer.com, Beckman Coulter) and are available upon request; note that these were internal primers and thus different from the exonic, chicken-derived primers (supplemental Table 2 at http://www.genetics.org/supplemental/) initially used for obtaining sequence information from the collared flycatcher. The overall genotype call rate was 96%, and the accuracy was 99.94% according to duplicate analysis of 22% of the total number of genotypes (4694/21,479). When possible, missing data were inferred from the genotypes of parents/offspring.
Linkage analysis:
Genotype data were used to infer intra-intronic haplotypes of all individuals assuming no recombination between SNPs within introns in the single generation scored. All linkage analyses were done in CRIMAP (Green et al. 1990) using an LOD score of 3.0 as threshold for significant linkage. Pairwise linkage analyses between all marker pairs were done using the option two point and a framework marker map was created with the option build. To include all linked loci in an extended build, unordered loci from the first build were tested with recurrent runs of the option flips4 until no better order could be obtained. For comparison of gene orders and recombination rates of collared flycatcher and chicken, data from chicken were primarily taken from http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9031&chr=Z. However, since the draft assembly contains errors in order of contigs on the Z, we used information from the most recent version (H. Cheng, unpublished data) of the East Lansing chicken linkage map (available at http://poultry.mph.msu.edu/) to get as accurate data as possible on gene order and recombination rates in chicken. The East Lansing genetic map was developed by multipoint analysis using 88 backcross progeny and >2500 markers, including 1207 SNPs based on the chicken draft genome assembly (WASHUC1). Throughout the article we use the term synteny to refer to conserved gene content of orthologous chromosomes, irrespective of gene order.
RESULTS
Development of polymorphic gene markers in collared flycatchers:
Of 135 primer pairs (corresponding to 135 different genes) designed from chicken exon sequence, 75 (55%) amplified a specific product in flycatchers and 39 (29%) of these could be readily sequenced. We used the retrieved flycatcher sequences in BLAST searches against the chicken genome. In the great majority of cases there was a single best hit to the expected gene on the chicken Z chromosome, indicating orthology (Table 1). All cases of weakly supported orthology concerned loci for which there was no or only limited exon sequence in collared flycatcher retrieved.
From resequencing of 18,450-bp intronic sequence in each of eight unrelated males, a total of 126 SNPs were identified, yielding an average SNP density of 1 every 146 bp of Z-chromosome sequence. Thirty-one introns showed at least one SNP with an MAF of >20% in the 16 chromosomes screened. Fifty-three such high-frequency SNPs from the 31 introns were subsequently genotyped in the pedigree. For 22 introns initially identified to contain microsatellite sequences in chicken, none showed length polymorphism in collared flycatcher. Generally, these introns showed little sign of remaining simple repetitive sequence in the collared flycatcher.
Linkage mapping:
We collected ∼20,000 SNP genotypes (53 sites × 365 individuals) in a mini-sequencing platform for SNP typing. Seven SNP markers turned out to be monomorphic in the pedigree and for another three markers only one to two families were informative for segregation analysis; these markers were excluded from further analyses. The remaining 43 SNPs were from 23 different introns (Table 1) and since no recombination event could be detected between SNPs within introns in an initial two-point analysis, genotype data from markers with >1 SNP were converted to haplotype data, which were used in subsequent analysis. The 23 genes are rather evenly spread from position 0.1–29.9 Mb in the draft assembly of the chicken Z chromosome sequence (which has a total length, excluding gaps, of 31 Mb).
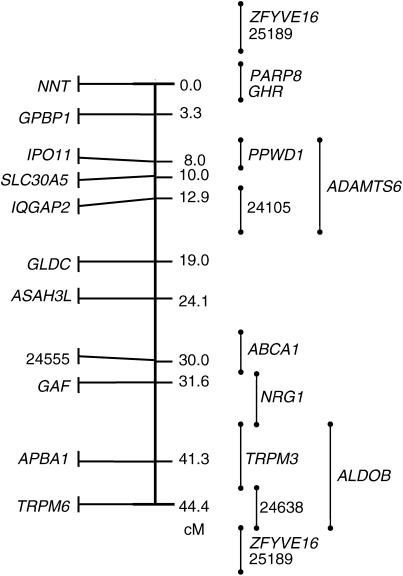
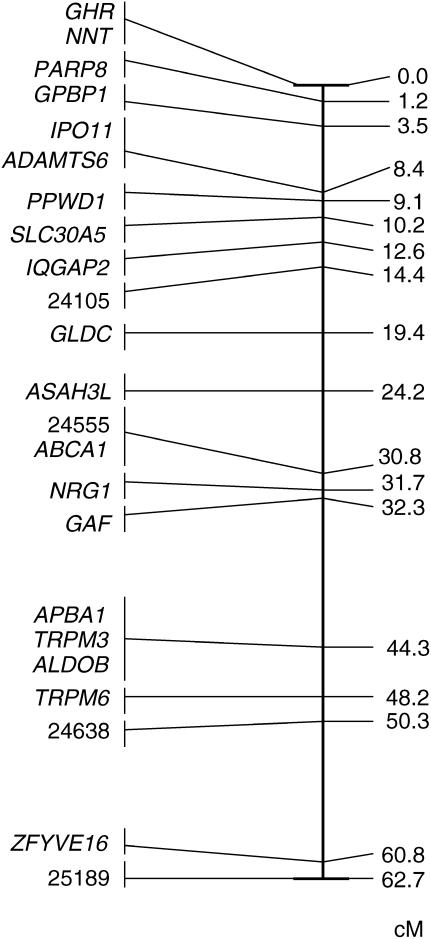
Two-point analysis revealed that all gene markers were significantly (LOD score >3.0) linked to at least one other locus in a way consistent with these 23 genes being part of a single linkage group. Since all parent and offspring females showed just one allele at all marker loci, we concluded that this linkage group is located on the collared flycatcher Z chromosome. The build procedure of CRIMAP resulted in a 44.4-cM linkage group with 11 loci significantly ordered (LOD score >3.0) in a framework map (Figure 2). Most of the remaining loci can be placed with significant support in either of two alternative intervals. A best-order linkage group for all 23 loci was subsequently obtained by evaluation using the flips4 option; this group spanned 62.7 cM (Figure 3).
Figure 2.—
A framework linkage map of the collared flycatcher Z chromosome. Positions are given as the cumulative genetic position of gene markers in the linkage map. Unordered markers are shown to the right with alternative locations indicated by vertical bars. Note that most unordered markers can be placed with statistical support in either of two locations.
Figure 3.—
A best-order linkage map of the collared flycatcher chromosome based on all gene markers genotyped in the pedigree. Positions are given as the cumulative genetic position of gene markers in the linkage map.
Gene order rearrangements during avian sex chromosome evolution:
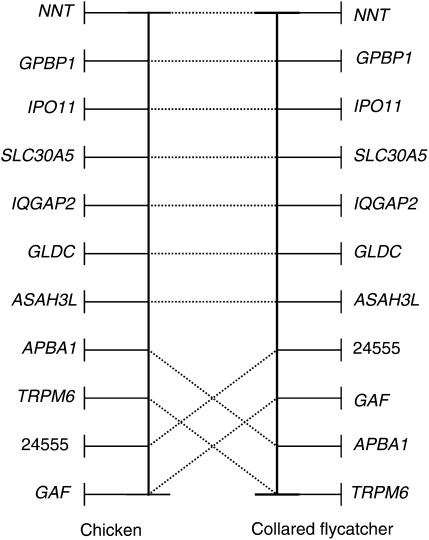
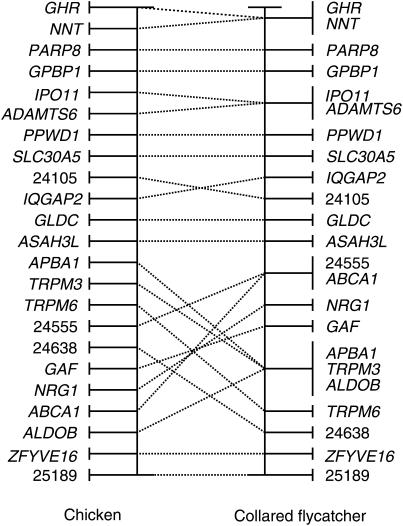
A comparison of the order of loci on the flycatcher framework map to the order of orthologous loci on the chicken physical map reveals two major rearrangements close to the respective ends of the flycatcher linkage group. However, one of these (Figure 4), NNT, being distal to GPBP1, IPO11, and SLC30A5 in the flycatcher map but proximal in the chicken physical map, is likely to represent an error in the draft assembly of the chicken genome since the same order as we find in flycatcher has recently also been observed in chicken linkage analysis (H. Cheng, unpublished data). The other rearrangement (for which chicken physical and genetic data are concordant) concerns a 14-cM flycatcher/10-Mb chicken interval tagged by four loci, APBA1, TRPM6, 24555, and GAF (Figure 4; note while two of these loci had relatively low BLAST scores, two were highly supported). The order of loci in the two species cannot straightforwardly be accounted for by a single inversion event and hence point at more complex structural changes. Moreover, the best-order map indicates additional rearrangements of complex nature in the vicinity of APBA1-TRPM6-24555-GAF (Figure 5). An inversion is indicated for a segment harboring 24105 and IQGAP2 but here the distance between loci is small, within map error. It can be concluded that while Z-chromosome synteny is perfectly conserved among the 23 genes investigated in the chicken–flycatcher comparison, a number of internal rearrangements have taken place during avian sex chromosome evolution.
Figure 4.—
A comparison of the relative location of orthologous genes on the chicken and collared flycatcher Z chromosome maps. Gene order in collared flycatcher is from the framework linkage map presented in Figure 2. In chicken, data are from the draft genome assembly updated with information from linkage mapping.
Figure 5.—
A comparison of the relative location of orthologous genes on the chicken and collared flycatcher Z chromosome maps. Gene order in collared flycatcher is from the best-order linkage map presented in Figure 3. In chicken, data are from the draft genome assembly updated with information from linkage mapping.
Comparison of recombination rates:
Given that the most distal markers in the best-order flycatcher map do not seem to be involved in intrachromosomal rearrangements, we can tentatively compare chicken and flycatcher recombination rates by comparing the length of our linkage group with the length of the part of the chicken Z chromosome genetic map flanked by the corresponding markers. This syntenic segment has a genetic length of 63 and 135 cM in collared flycatcher and chicken, respectively, indicating that the recombination rate in flycatchers is only ∼50% of that in chicken. Analyses of shorter segments within the maps give similar differences (data not shown).
DISCUSSION
Although cytogenetic approaches, mainly cross-species fluorescence in situ hybridization with chromosome-specific probes (ZOO-FISH), have been used to study the patterns of synteny conservation in birds (Shetty et al. 1999; Guttenbach et al. 2003; Shibusawa et al. 2004a,b; Itoh and Arnold 2005), genome evolution at the level of gene content and gene order across phylogenetically divergent bird lineages has not been investigated so far. A main goal of this study was therefore to analyze chicken and collared flycatcher gene maps in a comparative genomic context. An evaluation of the extent to which information on the organization of the chicken genome can be transferred to nonmodel species is of significance both for future genomic studies of natural populations and for our general understanding of avian genome evolution. Moreover, by focusing on the organization of the Z chromosome, we can address the degree of sex chromosome conservation in a system of female heterogamety. The latter is motivated by the fact that while there have been numerous interchromosomal rearrangements among autosomes during mammalian evolution (Murphy et al. 2005), the gene content of the X chromosome is largely conserved across all eutherian lineages (Murphy et al. 1999; Raudsepp et al. 2004). Is this a consequence of the mode of dosage compensation in mammals or is it a general feature of sex chromosome evolution?
Conserved synteny on the avian Z chromosome:
We found that 23 of 23 genes more or less evenly distributed along the Z chromosome of chicken also map to the Z chromosome of the collared flycatcher. This result shows that there is a very high degree of sex chromosome conservation at the level of gene content across distantly related bird lineages, which mimics the situation seen for the mammalian X chromosome. A possible explanation is that a particularly high degree of conservation is inherently associated with heteromorphic sex chromosomes. Ohno (1973) first hypothesized that there is selection to maintain the gene content of the X chromosome, because if dosage compensation occurs on a gene-by-gene basis, the transfer of a gene from an autosome to X would initially imply an imbalance in expression levels between the two sexes that cause problems in development and other life processes. Also, the translocation of an X-linked and dosage-compensated gene to an autosome may be associated with deleterious effects on expression levels. Potentially, the observed high degree of conserved synteny on the avian Z chromosome might be a consequence of similar selective constraints.
However, the case for, and the mode of, dosage compensation in birds is still a matter of uncertainty (Ellegren 2002). It seems clear that Z chromosome inactivation does not occur (Schmid et al. 1989). There are reports of equalized expression levels in males and females for a limited number of Z-linked genes (Kuroda et al. 2001; McQueen et al. 2001), possibly regulated at the level of transcription since both Z chromosomes of male chicken are transcribed (Kuroda et al. 2001; Kuroiwa et al. 2002). The observations of a heterogeneous, noncoding RNA that accumulates on the Z chromosome in the female nucleus but is heavily methylated and silent in males (Teranishi et al. 2001) and Z chromatin being enriched with hyperacetylated histones in females but not in males (Bisoni et al. 2005) point to epigenetic mechanisms similar to that suggested to be involved with dosage compensation in other organisms. While more work is clearly needed for avian dosage compensation to be understood, the absence of Z chromosome inactivation and the difference in genetic basis for sex determination between birds and mammals indicate that conserved synteny of sex chromosomes may be a general feature of heterogametic organisms, irrespective of whether there is male or female heterogamety or how dosage compensation is mediated.
An alternative explanation for our findings is that the high degree of conservation at the level of gene content is typical for most avian chromosomes, i.e., not being restricted to the Z chromosome. There are several lines of evidence indicating that, overall, genomic stability is higher in birds than in mammals. Comparative map data suggest that chromosomal rearrangements have occurred at a significantly lower rate in the avian than in the mammalian lineage (Burt et al. 1999; Bourque et al. 2005). Moreover, studies of karyotype evolution in birds using cross-species chromosome painting probes (ZOO-FISH) have revealed an extensive degree of chromosomal conservation across phylogenetically divergent lineages (Shetty et al. 1999; Guttenbach et al. 2003; Shibusawa et al. 2004a,b; Itoh and Arnold 2005), although there are exceptions, as in birds of prey (Bed'Hom et al. 2003; de Oliveira et al. 2005). In most cases, chicken-derived probes of macrochromosomes and of the Z chromosome each hybridize to a single chromosome in paleognathous as well as in other neognathous birds. Furthermore, a karyotype of 2n = 78 is widely conserved across birds (Burt 2002). Although none of these approaches has the resolution of addressing genome conservation at the level of gene content, together they show that avian genomes are unusually stable at a gross cytogenetic level. Additional gene mapping data, or genome sequence information, from avian lineages distant from the chicken will be needed for unveiling whether a similarly high conservation of gene content that we observed for the avian Z chromosome is also found in autosomes.
Intrachromosomal rearrangements:
In contrast to the observation of conserved synteny, several changes in gene order during avian sex chromosome evolution were evident from the collared flycatcher linkage map. Intrachromosomal rearrangements of the Z chromosome have also been indicated by chromosome painting among Galliform species (Shibusawa et al. 2004a,b), i.e., among species that are much more closely related than in the Galliformes–Passeriformes comparison. The finding of internal rearrangements has important practical consequences for the exploitation of data from the chicken genome in studies of less well-characterized bird species. If such rearrangements tend to occur on a broad scale across the genome, then map information will often not be directly transferable between species. This caveat is important to point out because a high genomic stability at the level of conserved synteny may incorrectly be taken to suggest that gene orders are also conserved. Moreover, the complex nature of the rearrangements inferred in Figures 4 and 5 means that it can be difficult to predict the location of homologous genes unless detailed comparative map information is available. We finally note that conserved synteny but frequent internal rearrangements are also a hallmark of mammalian X chromosome evolution (Blair et al. 1994; Millwood et al. 1997; Bourque et al. 2004).
Recombination rate evolution:
An important observation in this study was that the recombination frequency on the collared flycatcher Z chromosome seems lower than that in chicken. On the basis of early linkage mapping studies in chicken, subsequently confirmed by genome analysis, it has generally been thought that birds have high recombination rates, at least when compared to most mammals. To some extent this can be explained by the effect of the number of chromosomes; with an obligate chiasma per chromosome or chromosome armper meiosis needed for proper segregation, the total genetic distance of a genome should be expected to correlate with the number of chromosomes. However, recombination rates (in centimorgans per megabase) of individual chicken chromosomes are also high when compared to the rate in similarly sized mammalian chromosomes (International Chicken Genome Sequencing Consortium 2004). Our data now suggest that high recombination rates may not be a ubiquitous feature of bird genomes, at least not the sex chromosomes. Preliminary data from microsatellite-based linkage groups in the great reed warbler (Acrocephalus arundinaceus) point in a similar direction (Hansson et al. 2005).
There is a body of literature on the evolution of recombination rates (see, e.g., reviews by Otto and Lenormand 2002; Rice 2002). It would be premature at this point to speculate on how, for example, life history differences between chicken and flycatcher could relate to differences in recombination rates between the two lineages. However, one more principal characteristic is worth mentioning. First, strong selection for particular traits is likely to be coupled with selection for high recombination rate, since the highest selective response is expected when beneficial alleles are inherited together and unfavorable combinations removed (Rodell et al. 2004). In other words, the avoidance of Hill–Robertson interference is dependent on the rate of recombination. Empirical data from plants indicate that strong artificial selection during domestication has selected for elevated recombination rates (Ross-Ibarra 2004) and there are also observations in the same direction among domestic animals (Burt and Bell 1987). Thus, it is possible that chicken domestication may have led to the evolution of a higher rate of recombination compared to other birds.
Conserved gene markers:
The idea of systematic use of evolutionarily conserved exonic primers spanning introns (Lyons et al. 1997) has proven useful for polymorphism ascertainment in a variety of organisms (Friesen et al. 1999; Hellborg and Ellegren 2003; Aitken et al. 2004), including birds (Primmer et al. 2002). The availability of the recently obtained chicken genome sequence (International Chicken Genome Sequencing Consortium 2004) now offers a huge resource of potential targets for conserved anchors in the avian genome. Since Galliformes, together with Anseriformes, likely form the most basal clade (Galloanserae) among neognath (non-ratite) birds (Edwards et al. 2005), the use of chicken-derived primer sequences for amplification of orthologous sequence in a passerine bird constitutes a critical test of the applicability of chicken gene sequences in distantly related birds. The observed success ratio of 29% (39/135; to the point of obtaining flycatcher intron sequence data) is similar to that reported by Primmer et al. (2002) (20%; 8/41). Clearly, however, this ratio can be increased substantially by including sequence information from another bird species in primer design.
Linkage maps are most often constructed using anonymous markers like microsatellites or, to some extent, amplified length polymorphisms and this applies in particular to less well-characterized nonmodel organisms. However, while the polymorphism content of microsatellites is clearly higher than that of SNPs, it might be anticipated that SNP-based maps will come to find an increased application in the future. First, the increasing number of completely sequenced genomes will facilitate the development of PCR-based markers in nonmodel organisms, as demonstrated in this study. Second, SNPs in genes have a clear advantage in comparative mapping, in that they make identification of synteny and conserved chromosomal segments straightforward, as also demonstrated here. Third, there is rapid progress in technology for large-scale SNP genotyping with increased throughput at decreased costs.
Conclusions:
Obtaining the genome sequence of the chicken was an important step toward increased understanding of avian biology and this study has clearly revealed the usefulness of chicken as a model organism for genetic studies of other birds. Specifically, we have demonstrated an approach by which polymorphic markers in a focal species can be developed on the basis of sequence information from chicken and how such markers subsequently can be used for genetic mapping in a nonmodel organism. Moreover, our study reveals highly conserved synteny between the sex chromosomes of chicken and a passerine bird. Importantly, it should be possible to extend this approach to whole-genome coverage, selecting markers evenly distributed along all chicken chromosomes. We anticipate that this will pave the way for studies of other bird species, with the ultimate goal of using genetic maps for the identification of genes underlying phenotypic traits and the subsequent study of selection directly on genotypes instead of phenotypes. Such work is now in progress in collared flycatchers.
Acknowledgments
We thank Tomas Axelsson, Dan Fredriksson, Ulrika Liljedahl, and Ann-Christine Syvänen for excellent technical assistance in SNP genotyping, performed at the SNP Technology Platform at Uppsala University. Financial support was obtained from the Swedish Research Council to H.E.
References
- Aitken, N., S. Smith, C. Schwarz and P. A. Morin, 2004. Single nucleotide polymorphism (SNP) discovery in mammals: a targeted-gene approach. Mol. Ecol. 13: 1423–1431. [DOI] [PubMed] [Google Scholar]
- Alatalo, R. V., A. Lundberg and C. Glynn, 1986. Female pied flycatchers choose territory quality and not male characteristics. Nature 323: 152–153. [Google Scholar]
- Bed'Hom, B., P. Coullin, Z. Guillier-Gencik, S. Moulin, A. Bernheim, et al. 2003. Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res. 11: 335–343. [DOI] [PubMed] [Google Scholar]
- Bell, P. A., S. Chaturvedi, C. A. Gelfand, C. Y. Huang, M. Kochersperger et al., 2002. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. BioTechniques (Suppl.): 70–72, 74, 76–77. [PubMed]
- Bisoni, L., L. Batlle-Morera, A. P. Bird, M. Suzuki and H. A. McQueen, 2005. Female-specific hyperacetylation of histone H4 in the chicken Z chromosome. Chromosome Res. 13: 205–214. [DOI] [PubMed] [Google Scholar]
- Blair, H. J., V. Reed, S. H. Laval and Y. Boyd, 1994. New insights into the man-mouse comparative map of the X chromosome. Genomics 19: 212–220. [DOI] [PubMed] [Google Scholar]
- Both, C., and M. E. Visser, 2001. Adjustment to climate change is constrained by arrival date in a long-distance migrant bird. Nature 411: 296–298. [DOI] [PubMed] [Google Scholar]
- Bourque, G., P. A. Pevzner and G. Tesler, 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 14: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., E. M. Zdobnov, P. Bork, P. A. Pevzner and G. Tesler, 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res. 15: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A., and G. Bell, 1987. Mammalian chiasma frequencies as a test of two theories of recombination. Nature 326: 803–805. [DOI] [PubMed] [Google Scholar]
- Burt, D. W., 2002. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96: 97–112. [DOI] [PubMed] [Google Scholar]
- Burt, D. W., C. Bruley, I. C. Dunn, C. T. Jones, A Ramage et al., 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402: 411–413. [DOI] [PubMed] [Google Scholar]
- Chistiakov, D. A., B. Hellemans, C. S. Haley, A. S. Law, C. S. Tsigenopoulos et al., 2005. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics 170: 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, D. A., O. Hanotte, C. Greig, I. R. K. Stewart and T. Burke, 2000. Polymorphic microsatellites in the blue tit Parus caeruleus and their cross-species utility in 20 songbird families. Mol. Ecol. 9: 1941–1944. [DOI] [PubMed] [Google Scholar]
- de Oliveira, E. H., F. A. Habermann, O. Lacerda, I. J. Sbalqueiro, J. Wienberg et al., 2005. Chromosome reshuffling in birds of prey: the karyotype of the world's largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus). Chromosoma 114: 338–343. [DOI] [PubMed] [Google Scholar]
- Double, M. C., D. Dawson, T. Burke and A. Cockburn, 1997. Finding the fathers in the least faithful bird: a microsatellite-based genotyping system for the superb fairy-wren Malurus cyaneus. Mol. Ecol. 6: 691–693. [Google Scholar]
- Drnevich, J. M., M. M. Reedy, E. A. Ruedi, S. Rodriguez-Zas and K. A. Hughes, 2004. Quantitative evolutionary genomics: differential gene expression and male reproductive success in Drosophila melanogaster. Proc. R. Soc. Lond. B 271: 2267–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S. V., W. Bryan Jennings and A. M. Shedlock, 2005. Phylogenetics of modern birds in the era of genomics. Proc. R. Soc. Lond. B 272: 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H., 1992. Polymerase-chain-reaction (PCR) analysis of microsatellites—A new approach to studies of genetic relationships in birds. Auk 109: 886–895. [Google Scholar]
- Ellegren, H., 2002. Dosage compensation: Do birds do it as well? Trends Genet. 18: 25–28. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., 2005. The avian genome uncovered. Trends Ecol. Evol. 20: 180–186. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., L. Gustafsson and B. C. Sheldon, 1996. Sex ratio adjustment in relation to paternal attractiveness in a wild bird population. Proc. Natl. Acad. Sci. USA 93: 11723–11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder, M. E., and T. Mitchell-Olds, 2003. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Friesen, V. L., B. C. Congdon, M. G. Kidd and T. P. Birt, 1999. Polymerase chain reaction (PCR) primers for the amplification of five nuclear introns in vertebrates. Mol. Ecol. 8: 2147–2149. [DOI] [PubMed] [Google Scholar]
- Green, P., K. A. Falls and S. Crooks, 1990. Documentation for CRI-MAP, Version 2.4. Washington University School of Medicine, St. Louis.
- Griffith, S. C., I. R. K. Stewart, D. A. Dawson, I. P. F. Owens and T. Burke, 1999. Contrasting levels of extra-pair paternity in mainland and island populations of the house sparrow (Passer domesticus): Is there an “island effect”? Biol. J. Linn. Soc. 68: 303–316. [Google Scholar]
- Groenen, M. A., H. H. Cheng, N. Bumstead, B. F. Benkel, W. E. Briles et al., 2001. A consensus linkage map of the chicken genome. Genome Res. 10: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, L., and T. Pärt, 1990. Acceleration of senescence in the collared flycatcher Ficedula albicollis by reproductive costs. Nature 347: 279–281. [Google Scholar]
- Gustafsson, L., and W. J. Sutherland, 1988. The costs of reproduction in the collared flycatcher Ficedula albicollis. Nature 335: 813–815. [Google Scholar]
- Gustafsson, L., A. Quarnström and B. C. Sheldon, 1995. A trade-off between a life-history and a secondary sexual trait. Nature 375: 311–313. [Google Scholar]
- Guttenbach, M., I. Nanda, W. Feichtinger, J. S. Masabanda, D. K. Griffin et al., 2003. Comparative chromosome painting of chicken autosomal paints 1–9 in nine different bird species. Cytogenet. Genome Res. 103: 173–184. [DOI] [PubMed] [Google Scholar]
- Hansson, B., M. Akesson, J. Slate and J. M. Pemberton, 2005. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc. R. Soc. Lond. B 272: 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg, L., and H. Ellegren, 2003. Y chromosome conserved anchored tagged sequences (YCATS) for the analysis of mammalian male-specific DNA. Mol. Ecol. 12: 283–291. [DOI] [PubMed] [Google Scholar]
- Hubert, S., and D. Hedgecock, 2004. Linkage maps of microsatellite DNA markers for the Pacific Oyster Crassostrea gigas. Genetics 168: 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium, 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- Itoh, Y., and A. P. Arnold, 2005. Chromosomal polymorphism and comparative painting analysis in the zebra finch. Chromosome Res. 13: 47–56. [DOI] [PubMed] [Google Scholar]
- Jiggins, C. D., J. Mavarez, M. Beltran, W. O. McMillan, J. S. Johnston et al., 2004. A genetic linkage map of the mimetic butterfly, Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, S., D. Fujima, S. Sasazaki, S. Tsuji, M. Mizutani, et al., 2005. Construction of a genetic linkage map of Japanese quail (Coturnix japonica) based on AFLP and microsatellite markers. Anim. Genet. 36: 227–231. [DOI] [PubMed] [Google Scholar]
- Kuroda, Y., N. Arai, M. Arita, M. Teranishi, T. Hori et al., 2001. Absence of Z-chromosome inactivation for five genes in male chickens. Chromosome Res. 9: 457–468. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, A., T. Yokomine, H. A. Sasaki, M. Tsudzuki, K. Tanaka et al., 2002. Biallelic expression of Z-linked genes in male chickens. Cytogenet. Genome Res. 99: 310–314. [DOI] [PubMed] [Google Scholar]
- Le Quere, A., A. Schutzendubel, B. Rajashekar, B. Canback, J. Hedh et al., 2004. Divergence in gene expression related to variation in host specificity of an ectomycorrhizal fungus. Mol. Ecol. 13: 3809–3819. [DOI] [PubMed] [Google Scholar]
- Lorenzen, M. D., Z. Doyungan, J. Savard, K. Snow, L. R. Crumly et al., 2005. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics 170: 741–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, L. A., T. F. Laughlin, N. G. Copeland, N. A. Jenkins, J. E. Womack et al., 1997. Comparative anchor tagged sequences (CATS) for integrative mapping of Mamm. Genomes. Nat. Genet. 15: 47–56. [DOI] [PubMed] [Google Scholar]
- Marra, R. E., J. C. Huang, E. Fung, K. Nielsen, J. Heitman et al., 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen, H. A., D. McBride, G. Miele, A. P. Bird and M. Clinton, 2001. Dosage compensation in birds. Curr. Biol. 11: 253–257. [DOI] [PubMed] [Google Scholar]
- Merilä, J., L. E. B. Kruuk and B. C. Sheldon, 2001. Cryptic evolution in a wild bird population. Nature 412: 76–79. [DOI] [PubMed] [Google Scholar]
- Merino, S., J. Potti and J. Moreno, 1996. Maternal effort mediates the prevalence of trypanosomes in the offspring of a passerine bird. Proc. Natl. Acad. Sci. USA 193: 5726–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl, G., J. Török, S. C. Griffith and B. C. Sheldon, 2002. Experimental analysis of sperm competition mechanisms in a wild bird population. Proc. Natl. Acad. Sci. USA 99: 5466–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millwood, I. Y., M. T. Bihoreau, D. Gauguier, G. Hyne, E. R. Levy et al., 1997. A gene-based genetic linkage and comparative map of the rat X chromosome. Genomics 40: 253–261. [DOI] [PubMed] [Google Scholar]
- Murphy, W. J., S. Sun, Z. Q. Chen, J. Pecon-Slattery and S. J. O'Brien, 1999. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 9: 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W. J., D. M. Larkin, A. Everts-van der Wind, G. Bourque, G. Tesler et al., 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1973. Ancient linkage groups and frozen accidents. Nature 244: 259–262. [DOI] [PubMed] [Google Scholar]
- Otto, S., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Primmer, C. R., A. P. Møller and H. Ellegren, 1996. A wide-range survey of cross-species microsatellite amplification in birds. Mol. Ecol. 5: 365–378. [DOI] [PubMed] [Google Scholar]
- Primmer, C. R., T. Borge, J. Lindell and G. P. Saetre, 2002. Single-nucleotide polymorphism characterization in species with limited available sequence information: high nucleotide diversity revealed in the avian genome. Mol. Ecol. 11: 603–612. [DOI] [PubMed] [Google Scholar]
- Qvarnström, A., T. Pärt and B. C. Sheldon, 2000. Adaptive plasticity in mate preference linked to differences in reproductive effort. Nature 405: 344–347. [DOI] [PubMed] [Google Scholar]
- Qvarnström, A., J. E. Brommer and L. Gustafsson, 2006. Testing the genetics underlying the co-evolution of mate choice and ornament in the wild. Nature 441: 84–86. [DOI] [PubMed] [Google Scholar]
- Raudsepp, T., E. J. Lee, S. R. Kata, C. Brinkmeyer, J. R. Mickelson et al., 2004. Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc. Natl. Acad. Sci. USA 101: 2386–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, K. M., L. D. Chaves, M. K. Hall, T. P. Knutson and D. E. Harry, 2005. A comparative genetic map of the turkey genome. Cytogenet. Genome Res. 111: 118–127. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 2002. Experimental tests of the adaptive significance of sexual recombination. Nat. Rev. Genet. 3: 241–251. [DOI] [PubMed] [Google Scholar]
- Rodell, C. F., M. R. Schippe and D. K. Keenan, 2004. Modes of selection and recombination response in Drosophila melanogaster. J. Hered. 95: 70–75. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra, J., 2004. The evolution of recombination under domestication: a test of two hypotheses. Am. Nat. 163: 105–112. [DOI] [PubMed] [Google Scholar]
- Saetre, G.-P., T. Moum, S. Bure, M. Král, M. Adamjan et al., 1997. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature 387: 589–592. [Google Scholar]
- Schmid, M., E. Enderle, D. Schindler and W. Schempp, 1989. Chromosome banding and DNA replication patterns in bird karyotypes. Cytogenet. Cell Genet. 52: 139–146. [DOI] [PubMed] [Google Scholar]
- Sheldon, B. C., and H. Ellegren, 1999. Sexual selection resulting from extrapair paternity in collared flycatchers. Anim. Behav. 57: 285–298. [DOI] [PubMed] [Google Scholar]
- Shetty, S., D. K. Griffin and J. A. Graves, 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7: 289–295. [DOI] [PubMed] [Google Scholar]
- Shibusawa, M., M. Nishibori, C. Nishida-Umehara, M. Tsudzuki, J. Masabanda et al., 2004. a Karyotypic evolution in the Galliformes: An examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet. Genome Res. 106: 111–119. [DOI] [PubMed] [Google Scholar]
- Shibusawa, M., C. Nishida-Umehara, M. Tsudzuki, J. Masabanda, D. K. Griffin et al., 2004. b A comparative karyological study of the blue-breasted quail (Coturnix chinensis) and California quail (Callipepla californica, Odontophoridae). Cytogenet. Genome Res. 106: 82–90. [DOI] [PubMed] [Google Scholar]
- Slate, J., 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14: 363–379. [DOI] [PubMed] [Google Scholar]
- Smith, J. J., D. K. Kump, J. A. Walker, D. M. Parichy and S. R. Voss, 2005. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics 171: 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, S. C., 1992. The Evolution of Life Histories. Oxford University Press, Oxford.
- Storz, J. F., 2005. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 14: 671–688. [DOI] [PubMed] [Google Scholar]
- Tabor, H. K., N. J. Risch and R. M. Myers, 2002. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat. Rev. Genet. 3: 391–397. [DOI] [PubMed] [Google Scholar]
- Tapio E., and E. Lehikoinen, 2000. Pollution: Recovery of breeding success in wild birds. Nature 403: 851–852. [DOI] [PubMed] [Google Scholar]
- Teranishi, M., Y. Shimada, T. Hori, O. Nakabayashi, T. Kikuchi et al., 2001. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res. 9: 147–165. [DOI] [PubMed] [Google Scholar]
- van Tuinen, M., and S. B. Hedges, 2001. Calibration of avian molecular clocks. Mol. Biol. Evol. 18: 206–213. [DOI] [PubMed] [Google Scholar]
- Vasemagi, A., and C. R. Primmer, 2005. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol. Ecol. 14: 3623–3642. [DOI] [PubMed] [Google Scholar]
- Veen, T., T. Borge, S. C. Griffith, G.-P. Saetre, S. Bures et al., 2001. Hybridization and adaptive mate choice in flycatchers. Nature 411: 45–50. [DOI] [PubMed] [Google Scholar]