Abstract
We conducted a screen for glossy-eye flies that fail to incorporate BrdU in the third larval instar eye disc but exhibit normal neuronal differentiation and isolated 23 complementation groups of mutants. These same phenotypes were previously seen in mutants for cytochrome c oxidase subunit Va. We have molecularly characterized six complementation groups and, surprisingly, each encodes a mitochondrial protein. Therefore, we believe our screen to be an efficient method for identifying genes with mitochondrial function.
MITOCHONDRIAL function is essential for a number of important cellular processes, such as the generation of ATP (reviewed in Ackerman and Tzagoloff 2005), apoptosis (reviewed in Newmeyer and Ferguson-Miller 2003), and the regulation of aging (reviewed in Ohta 2003). Recently, work from our laboratory has shown that the metabolic status of a cell, controlled by mitochondrial function, also regulates the G1–S checkpoint in mitosis (Mandal et al. 2005). This is evident in tenured (tend) mutants, which contain a null mutation in the gene encoding cytochrome c oxidase subunit Va of complex IV of the mitochondrial electron transport chain. This mutation causes a reduction in ATP generated in mutant cells to ∼40% of wild-type levels, triggering the activation of AMPK and p53, which leads to the eventual downregulation of cyclin E. A G1–S mitotic checkpoint is then enforced, preventing cells from reentering the cell cycle (Mandal et al. 2005).
Adult tend mutants have glossy eyes lacking lens material, normally secreted by cone and pigment cells of the eye. In earlier studies, the lens-depleted glossy-eye phenotype was described in lozenge mutants, which are defective in cone and pigment cell specification (Flores et al. 1998). However, in the case of tend, such defects are caused by the lack of a sufficient number of precursor cells from which cone and pigment cells arise (Mandal et al. 2005).
The events that give rise to the tend adult eye phenotype occur in larval eye development. The larval Drosophila eye imaginal disc undergoes two distinct phases of proliferation (Wolff and Ready 1991). During the early larval stages prior to differentiation, all cells divide frequently and asynchronously, apparently without regulation. In the third larval instar, the morphogenetic furrow sweeps across the eye disc to pattern the cells (Ready et al. 1976). Within the morphogenetic furrow, cells arrest in G1 and, following the passage of the furrow, either differentiate into neuronal photoreceptors or reenter the cell cycle and undergo a terminal round of division termed the second mitotic wave (SMW). This generates the precursors for cone and pigment cell lineages (Wolff and Ready 1991). Misregulation of the SMW will lead to a loss of these accessory cell types. In the third larval instar, cells mutant for tend anterior to the furrow slow down in cell cycle progression, while those posterior to the furrow fail to cross the G1–S checkpoint due to impaired ATP production (Mandal et al. 2005). Therefore, in addition to undergoing fewer rounds of mitosis prior to the passage of the furrow, tend mutants lack the SMW and, as a result, have a dramatically reduced number of accessory cells. Importantly, in tend mutant cells, cell divisions in second and early third instar eye discs are not affected. As a result, tend mutants give rise to large clones in the adult eye. This is different from the phenotype resulting from mutant clones in the basic cell cycle machinery, which would not be able to give rise to any mutant clones. Furthermore, early patterning and differentiation of neurons are also not affected in tend, as mutant clones express pan-neuronal, as well as cell-type-specific, markers and extend axons normally to the optic lobe (Mandal et al. 2005).
On the basis of the nature of the tend phenotype, we hypothesized that a simple screen that identifies glossy-eye flies, followed by a secondary screen for a subset of the mutants in which cells exhibit proliferation defects but have relatively normal patterning in the third larval instar, will potentially identify nuclear genes that encode mitochondrial proteins. Our hypothesis is supported by our earlier finding that, like tend, flies mutant in the mitochondrial components pdsw, mRpL4, and mRpL17 exhibit a glossy-eye phenotype and fail to incorporate BrdU in the larval eye disc but stain normally with the ELAV antibody (Mandal et al. 2005). In this article, we show that an unbiased screen for glossy-eye mutants that exhibit these larval phenotypes is remarkably effective in identifying genes with mitochondrial function.
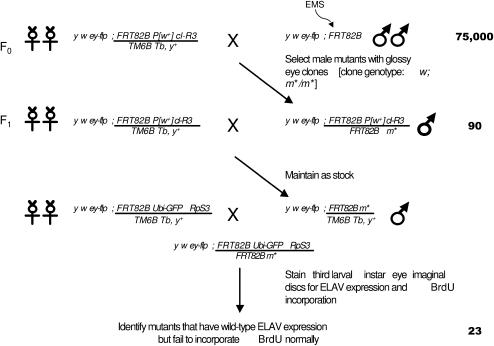
We conducted an eyeless-flp/FRT-based mitotic recombination screen (Newsome et al. 2000) on chromosome arm 3R (Figure 1). y w ey-flp; FRT 82B males were mutagenized with 25 mm ethyl methanesulfonate (EMS) and crossed to y w ey-flp; FRT82B P[w+] cl-R3/TM6B Tb, y+ females to generate adult eye clones. ey-flp drives mitotic recombination between the FRT-containing chromosomes under the control of the eyeless enhancer, specifically in the embryonic anlagen of the developing eye (Newsome et al. 2000). The resulting clones of cells will be homozygous for the newly induced mutation and are white due to the lack of the w+ gene essential for pigmentation. A total of 75,000 adult flies were screened for the mosaic glossy-eye phenotype. Ninety mutants were isolated. Complementation analysis was then performed by crossing each of the 90 mutants with one another and looking for lethality in the offspring. This yielded a total of 46 complementation groups. All mutants were balanced over a TM6B Tb, y+ chromosome and maintained as stocks.
Figure 1.—
Crossing scheme for genetic screen. A total of 75,000 chromosomes were mutagenized and screened for the adult glossy-eye phenotype. A total of 90 glossy-eye mutants were isolated, and larval eye-disc clones were induced to study their BrdU incorporation and ELAV expression. Twenty-three failed to incorporate BrdU but expressed ELAV normally and were studied further. m* represents the EMS-induced mutation. The cl-R3 mutation is cell lethal and is used to eliminate cells that are homozygous for the chromosome that does not contain m*. Similarly, for larval clones, cells homozygous for the RpS3 (Minute) mutation are eliminated. Cells heterozygous for the cl-R3 and RpS3 mutations grow more slowly, allowing m*/m* cells to form large clones.
Next, third larval instar eye imaginal discs from the identified glossy-eye mutants were assayed for BrdU incorporation and stained with the neuron-specific ELAV antibody (Yao et al. 1993). Larval eye-disc clones were generated by crossing the mutant stocks to y w ey-flp; FRT82B Ubi-GFP RpS3/TM6B Tb, y+ flies. Clones homozygous for the mutation are negatively marked by their lack of GFP. The 46 complementation groups isolated from the glossy-eye screen fall into four categories on the basis of this characterization. Ten mutant groups exhibit both wild-type neuronal patterning and BrdU incorporation, indicating that the glossy-eye phenotype is due to mutations in genes that function after the third instar and are required for cone and pigment cell specification. Three mutant groups display abnormal ELAV staining patterns while cells incorporate BrdU normally. Since the formation of cone cells requires a combination of signals from differentiated neuronal photoreceptors (Flores et al. 2000), this phenotype likely occurs because of defects in the patterning of specific photoreceptors. Ten mutant groups exhibit defective ELAV staining patterns in early clusters as well as defects in BrdU incorporation, which can be attributed to mutations in genes required for specification of multiple cell types in the eye. Finally, 23 mutant groups fail to incorporate BrdU properly but show essentially normal patterning of neuronal clusters emerging from the furrow. They were further analyzed as part of this study.
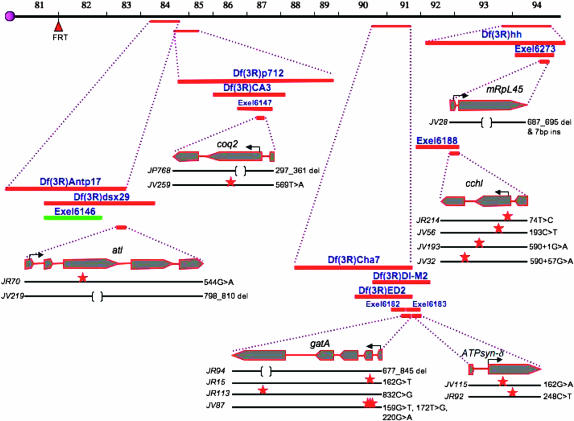
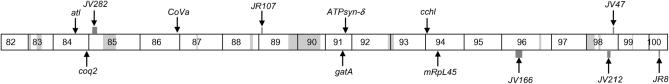
We have mapped all mutants from this last category to either available deficiencies on chromosome 3R from the Bloomington and Exelexis collections (Thibault et al. 2004) or attributed their map positions to gaps within the deficiency kit (Table 1A). We have cloned six mutant complementation groups, consisting of 24 alleles, and obtained sequence information on the molecular lesions of 15 of the alleles (Figure 2). As the guiding hypothesis of this screen was to identify mitochondrial mutations, we sequenced potentially mitochondrial genes within candidate regions as determined through BLAST analysis (Altschul et al. 1990) and MitoProt scores, which predict potential mitochondrial localization on the basis of the presence of a mitochondrial localization sequence (Claros and Vincens 1996). Using this method, we identified four mutants in CG6022, a homolog of cytochrome c heme lyase (cchl); two mutant alleles of CG9613, a homolog of coenzyme Q biosynthesis protein 2 (coq2); nine mutants in CG10092, a homolog of arginyl tRNA ligase (atl); three mutant alleles of ATP synthase-δ subunit (ATPsyn-δ); five mutants in glutamyl-tRNA amidotransferase A (gatA); and one mutant allele of mitochondrial ribosomal protein large subunit 45 (mRpL45). Combined with previously described mutants in pdsw, mRpL4, mRpL17, and tend, all mutations exhibiting the glossy-eye phenotype with defective BrdU incorporation and wild-type neuronal patterning cloned thus far could be rationalized as having roles in the mitochondrion (see below and Figure 3 and Table 1B). Note that this screen identified the first mutant alleles for CoVa, coq2, atl, ATPsyn-δ, gatA, and mRpL45. Single alleles of cchl, pdsw, mRpL4, and mRpL17 had been identified as lethal mutations in either the Exelixis collection or the Berkeley Drosophila Genome Gene Disruption Project, but were not previously genetically characterized (Spradling et al. 1999; Thibault et al. 2004) (Table 1B).
TABLE 1.
Mapping results and mutations in nuclear-encoded genes with mitochondrial function that exhibit a G1–S block in the third larval instar with normal neuronal differentiation
| A. Mapping results | |||
|---|---|---|---|
 | |||
| Cytological position | Gene name | Mitochondrial process | Alleles |
| B. Mitochondrial genes identified by the glossy eye–BrdU screen | |||
| 84E4 | CG10092 arginyl tRNA ligase (atl) | Protein biosynthesis–tRNA synthesis | JR70, JR77, JV83, JR101, JR203, JV219, JR247, JV250, JP782 |
| 84F11 | CG9613 coenzyme Q biosynthesis protein 2 (coq2) | Oxidative phosphorylation–complex III | JV259, JP768 |
| 91E4 | glutamyl-tRNA amidotransferase A (gatA) | Protein biosynthesis–tRNA synthesis | JR15, JV87, JR94, JR113, JR205 |
| 91F1 | ATP synthase subunit δ (ATPsyn-δ) | Oxidative phosphorylation–complex V of electron transport chain | JR92, JV115, JR238 |
| 94A1 | CG6022 cytochrome c heme lyase (cchl) | Oxidative phosphorylation–complex IV of electron transport chain | JV32, JV56, JV193, JR214; Exelixis c04553 (Thibault et al. 2004) |
| 94B6 | mitochondrial ribosomal protein large subunit 45 (mRpL45) | Protein biosynthesis–ribosome assembly | JV28 |
| Previously identified mitochondrial genes that cause the glossy-eye–defective BrdU incorporation phenotype | |||
| 23F3 | pdsw | Oxidative phosphorylation–complex I of electron transport chain | pdswk10101 (Spradling et al. 1999) |
| 35F11 | mitochondrial ribosomal protein large subunit 4 (mRpL4) | Protein biosynthesis–ribosome assembly | mRpL4k14608 (Spradling et al. 1999) |
| 61B3 | mitochondrial ribosomal protein large subunit 17 (mRpL17) | Protein biosynthesis–ribosome assembly | mRpL17KG06809 (Bloomington Drosophila Stock Center at Indiana University) |
| 86F9 | cytochrome c oxidase subunit Va (CoVa) | Oxidative phosphorylation–complex IV of electron transport chain | JP785 (Mandal et al. 2005) |
| Allele | Cytological position | No. of genes in region | Allele | Cytological position | No. of genes in region |
| Other mapping results | |||||
| JV282 | 85A6-B1 | 24 | JV212 | 98E1-E3 | 6 |
| JR107 | 89A5-A8 | 29 | JV47 | 98F5 | 9 |
| JV166 | 96A21-B4 | 34 | JR8 | 100B9 | 5 |
In A, dark shading indicates complementation groups that have mapped to a deficiency and light shading indicates gaps within the available deficiency kit. The candidate regions and potential mitochondrial genes for complementation groups that have not been cloned are also indicated.
Figure 2.—
Map positions and molecular lesions of identified mutations. Deficiency stock names and their approximate endpoints, as annotated by FlyBase, are indicated. Solid black lines, alleles identified from the screen; red stars, point mutations; brackets, deletions; red bars, deficiencies that fail to complement the mutant; green bars, deficiencies that complement the mutant.
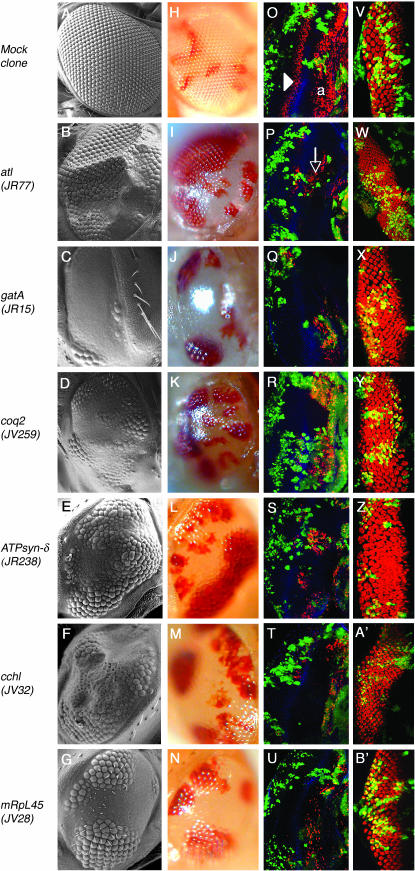
Figure 3.—
Representative adult and larval phenotypes of mutants from mapped complementation groups. (A–N) Eyes of adult flies examined by scanning electron microscopy (A–G) and bright-field microscopy (H–N). An eye containing mock clones is mosaic in color (H), but the entire eye is faceted as in wild-type tissue (A). In eyes mutant for the indicated mitochondrial genes, cells heterozygous for the mutation are faceted (B–G) and red (I–N) as in mock clones, while homozygous clones are glossy and white. In all images, the representative allele is indicated in parentheses and posterior is to the left. (O–U) 30-min BrdU incorporation (red) in third instar larval eye discs with mock (O) and mutant (P–U) clones. Armadillo (blue) marks the morphogenetic furrow. In an eye disc with mock clones (O), both green and nongreen tissue are wild type. BrdU is randomly incorporated anterior to the furrow (“a”); posterior to the furrow, a single band of BrdU incorporation (arrowhead) marks the SMW. In mutant eye discs, BrdU incorporation is lost in homozygous mutant tissue, marked by the lack of GFP, both anterior and posterior to the furrow. Incorporation remains normal in clones heterozygous for the mutation (green). For comments on the apparent local nonautonomy of this phenotype (arrow), see Mandal et al. (2005). (V–B′) ELAV antibody stainings of eye discs with mock (V) and mutant (W–B′) clones. Expression of ELAV is largely normal in all mutant clones.
CCHL:
Within the electron transport chain, CCHL participates in the function of complex IV by attaching a heme group to apocytochrome c, thereby converting it to a functional holocytochrome c and enabling its import through the inner membrane of the mitochondrion (reviewed in Kranz et al. 1998). In Saccharomyces cerevisiae, strains lacking or mutant in CCHL have increased levels of apocytochrome c in the cytoplasm (Dumont et al. 1991). From the screen, we have isolated four alleles of cchl (Figure 2) on the basis of their failure to complement Exelixis c04553 (Thibault et al. 2004), which harbors a lethal insertion in cchl. The first allele, JR214, harbors a T > C transition resulting in the amino acid substitution M25T. JV56 contains a C > T transition, resulting in a premature stop codon at Q65. The remaining two alleles, JV193 and JV32, have G > A transitions at the first and last base pair of intron 2, respectively, and thus may result in the insertion of 19 amino acids if splicing cannot occur normally. All of the mutations occur within the heme lyase domain of the protein. Drosophila CCHL bears 53% identity to the human homolog and 42% identity to the S. cerevisiae homolog. According to MitoProt predictions, CCHL has a 28% likelihood of localizing to the mitochondrion; this relatively low likelihood may be due to the fact that mitochondrial heme lyases lack an N-terminal targeting signal and instead are directed to the mitochondrion by an internal localization sequence in the third quarter of the protein (Diekert et al. 1999). This is further supported by the fact that MitoProt also predicts a very low (4.8%) chance of mitochondrial localization for the S. cerevisiae CCHL.
ATPsyn-δ:
Studies in Escherichia coli show that F1F0-ATP synthase, complex V of the electron transport chain, generates ATP through cooperation between a “rotor” and a “stator” (reviewed in Boyer 1997; Weber et al. 2004). The stator consists of two β-subunits and one δ-subunit (reviewed in Dunn et al. 2000), and loss of the δ-binding site on the α-subunit of F1 results in impaired growth and ATP synthesis in vivo (Weber et al. 2004). Three alleles of ATPsyn-δ were isolated from the screen (Figure 2). JV115 contains a G > A transition that results in a premature stop codon at W54, while JR92 has a C > T transition that prematurely terminates the protein at Q83. A molecular lesion for the third allele, JR238, has not been found; presumably, it is located within a regulatory region of the gene. The mutations all lie within the ATP synthase-δ domain of the protein. MitoProt analysis suggests that Drosophila ATPsyn-δ has a 92% likelihood of localizing to the mitochondria.
COQ2:
coq2 encodes the enzyme parahydroxybenzoate (PHB):polyprenyltransferase (Ashby et al. 1992). As part of a multi-step process involved in ubiquinone biosynthesis, PHB:polyprenyltransferase catalyzes the transfer of a polyprenyl group from polyprenyl diphosphate to parahydroxybenzoate to form 3-polyprenyl-4-hydroxybenzoate (Winrow and Rudney 1969). Once synthesized, ubiquinone functions as an electron transporter between complexes I, II, and III of the electron transport chain (Green 1966). Two alleles of coq2 were identified from the screen (Figure 2). The first allele, JP768, contains a 94-bp deletion mutation starting at A100 that causes a frameshift in the remaining sequence. All amino acids downstream of the deletion are altered, and a premature stop codon is introduced after the 232nd amino acid. JV259 has a T > A transversion that results in a premature stop codon at L189. Both mutations fall within the UbiA prenyltransferase domain of COQ2. Drosophila COQ2 has 45% identity to the S. cerevisiae homolog and a 97% likelihood of being imported into the mitochondrion according to MitoProt analysis.
GatA:
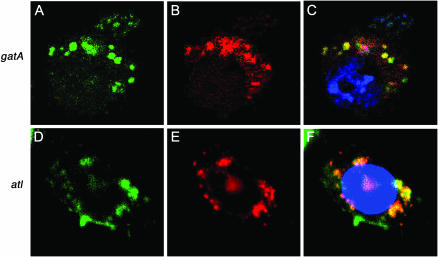
Aminoacylation studies have shown that there is no detectable glutaminyl–tRNA synthase activity in cyanobacteria, Gram-positive bacteria, and chloroplasts and mitochondria of plants and animals (Wilcox and Nirenberg 1968; Schon et al. 1988). Instead, these organisms and organelles synthesize glutamine by mischarging a tRNAGln with glutamate and then using glutamyl–tRNA amidotransferases to convert the glutamate to glutamine (Schon et al. 1988). According to MitoProt predictions, Drosophila GatA has a relatively low probability (56%) of localizing to the mitochondrion. To determine the exact subcellular location of GatA, we constructed a GatA–GFP fusion protein, transfected Drosophila S2 cells with the construct, and co-immunostained the cells with a GFP antibody and MitoTracker Orange CM-H2TMRos, a dye that localizes to the mitochondrion in response to its membrane potential. This colocalization assay shows that GatA is indeed a mitochondrial protein (Figure 4, A–C). Thus, both functional and localization studies and the nature of other mutants isolated by our screen establish that GatA has a mitochondrial function. Five alleles of gatA were isolated from our screen; we have located molecular lesions in four alleles (Figure 2). JR94 has a 168-bp deletion in exon 5. JR15 harbors a G > T transversion, resulting in the amino acid substitution Q54H. JR113 has a C > G transversion that causes the substitution H278D. Finally, JV87 has three point mutations, G > T, T > G, and G > A, which result in the following amino acid changes: Q53H, A58S, and V74I. All of the mutations lie within the putative amidase domain of the protein.
Figure 4.—
GatA and ATL localize to the mitochondrion. Co-immunostaining of GatA–GFP and ATL–GFP fusion proteins (A and D, green), respectively, with MitoTracker Orange CM-H2TMRos (Molecular Probes, Eugene, OR) (B and E) shows mitochondrial localization of the fusion proteins (C and F, yellow). Full-length cDNAs were cloned using the Gateway cloning system (Invitrogen, San Diego) into the pAWG vector containing the actin promoter and sequence coding for a C-terminal GFP protein. Blue, DAPI staining (C) and TOPRO3 (F) staining.
ATL:
It has been determined that S. cerevisiae has two genes encoding arginyl tRNA ligase (ATL), with one functioning in the cytoplasm and the other, MSR1, in the mitochondrion (Tzagoloff and Shtanko 1995). The Drosophila genome has three genes encoding proteins with potential arginyl tRNA ligase activity on the basis of Gene Ontology annotation in FlyBase (http://www.flybase.org): atl, Aats-arg, and CG8097 (Drysdale and Crosby 2005). BLAST analysis shows that ATL is 36% identical to Msr1p, while AATS-arg and CG8097 are 28 and 24% identical, respectively. According to MitoProt analysis, ATL is only 8.4% likely to localize to the mitochondrion. This low number did not support a mitochondrial function for ATL. To pinpoint its subcellular location, we constructed an ATL–GFP fusion protein and demonstrated that it colocalizes with MitoTracker Orange CM-H2TMRos to the mitochondrion by immunostaining S2 cells transfected with the fusion protein (Figure 4, D–F). Thus, ATL's subcellular localization, similarity to Msr1p, and its suite of phenotypic similarities to other known mitochondrial mutants, indicate that this protein functions in the mitochondrion. Our screen has identified nine alleles of atl, and we have located molecular lesions in two of these nine alleles (Figure 2). JR70 has a G > A transition resulting in the amino acid change G182R. JV219 contains a 12-bp in-frame deletion starting at V266. Both mutations occur within the catalytic arginyl tRNA core domain.
mRpL45:
mRpL45 is a protein of 599 amino acids with a molecular weight of 26 kDa (reviewed in Graack and Wittmann-Liebold 1998). At least 50 mitochondrial ribosomal proteins exist within the S. cerevisiae mitochondrial ribosome, among which at least 39 are classified as large subunit proteins. In humans, these ribosomal subunits help synthesize 13 mitochondrial proteins essential for oxidative phosphorylation, and mutations in the genes encoding them are associated with many disorders (reviewed in Sylvester et al. 2004). According to MitoProt, mRpL45 has an 88% chance of localizing to the mitochondrion. One mutant allele of mRpL45 was identified from the screen (Figure 2). JV28 harbors a 9-bp deletion and a 7-bp insertion in exon 2 that results in a frameshift starting at V230 and a premature stop codon at the 279th amino acid. The mutation occurs within the TIM44 domain, which is involved in transporting the protein across the inner membrane of the mitochondrion (reviewed in Pfanner and Geissler 2001).
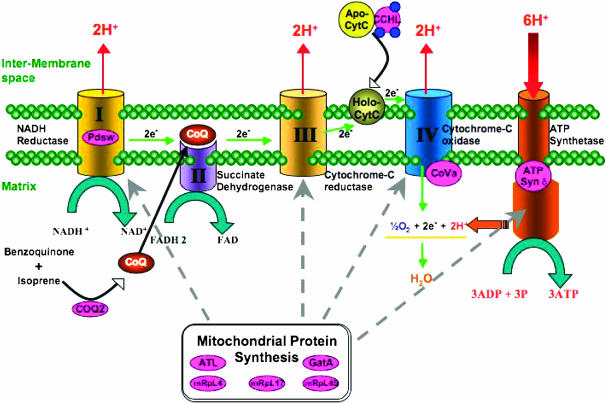
The generation of ATP in eukaryotic cells is largely dependent on oxidative phosphorylation in mitochondria (Figure 5). ATP originates from the oxidation of NADH and FADH2 via the electron transport chain, which consists of a series of four electron transporting complexes (I–IV) and the terminal complex V, which generates ATP. Complexes I and II transfer electrons, from NADH and FADH2, respectively, to ubiquinone (CoQ) via a series of iron–sulfur clusters. Ubiquinone, which is a mobile carrier, transfers these electrons to complex III. Cytochrome c, another mobile carrier protein, then delivers the electrons from complex III to complex IV where it reduces dioxygen to water. Concurrent with electron transport, complexes I, II, and IV pump protons from the matrix into the intermembrane space, forming an electrochemical proton gradient across the inner membrane, which is used to generate ATP by ATP synthase or complex V. Five mutants discussed in this study—tend, pdsw, coq2, cchl, and ATP syn-δ—function either within or between complexes of the electron transport chain. These mutants likely activate the G1–S checkpoint as described for tend (Mandal et al. 2005). The remaining mutants—atl, gatA, mRpL4, mRpL17, and mRpL45—function in diverse roles within the mitochondrial protein biosynthesis pathways. The mitochondrial genome encodes 13 proteins that are essential for oxidative phosphorylation, and defects within the protein translation system are likely to cause downstream effects in energy production and subsequently affect the G1–S transition of the cell.
Figure 5.—
Schematic of complexes involved in oxidative phosphorylation in the mitochondrion. Proteins highlighted in pink are encoded by mutant genes identified from this screen or described in Mandal et al. (2005).
The screen described here is the most efficient in any higher eukaryote for identifying nuclear-encoded genes that function in the mitochondrion. All six mutants mapped thus far are likely to have roles in mitochondrial processes. This is further supported by the fact that previously described P-element mutants in pdsw, mRpL4, and mRpL17 also exhibit the same phenotypes as mutants isolated from our screen (Mandal et al. 2005). This is an independent confirmation that this screen is uniquely capable of selecting for disruptions in cell cycle regulation in the developing eye disc, caused by the activation of a metabolic checkpoint initiated by the mitochondrion. However, not all of the mutants isolated in this screen are expected to be disruptions in mitochondrial proteins. In Mandal et al. (2005), we described a pathway linking mitochondrial processes to cell cycle regulation in the nucleus. Two key members of this pathway, AMPK and p53, are known cytosolic proteins. This pathway is not yet fully understood, and we expect that our screen will also provide missing cytosolic links, in addition to mitochondrial ones described here. Our mapping results show that the screen is very likely to have identified some nonmitochondrial genes. For example, JV212 maps to 98E1-98E3 (Table 1, A and B), a region encompassing six genes, none of which bears homology to other mitochondrial genes or contains a mitochondrial localization sequence. This is again true for JR8 (Table 1, A and B), which maps to 100B9, another region containing five genes that are not likely to be mitochondrial.
An additional advantage to our screen is that it may be a more accurate predictor of mitochondrial function than the currently available in silico databases. For example, MitoProt predicts relative low probabilities of mitochondrial localization for CCHL (28%), Pdsw (10%), GatA (56%), and ATL (8.4%). We have demonstrated through immunostaining in this study that both GatA and ATL localize to the mitochondrion. Pdsw, a NADH dehydrogenase, and CCHL have been shown in previous functional studies to play roles in complexes I and IV, respectively, of the electron transport chain. Although MitoProt will be able to reliably predict mitochondrial localization for the proteins that fit its algorithm, our screen provides the more relevant in vivo information.
This study identified several new mutations in genes that function in either oxidative phosphorylation or mitochondrial protein biosynthesis. Consistent with our earlier observation using tend (Mandal et al. 2005), these new mutants exhibit normal early cell divisions, a G1–S block at the third larval instar, and proper neuronal differentiation. The new mutants help to further establish that attenuated mitochondrial function, caused by a loss in one of its many individual components, will result in an enforcement of the G1–S checkpoint of the cell cycle. The fact that each of the characterized mutants with this suite of phenotypes encodes a protein with a mitochondrial function suggests that our method can serve as a general screen for identifying nuclear-encoded genes with mitochondrial function. This study represents yet another example of the power of Drosophila genetics and mutant screens in the discovery and refinement of important cellular processes.
Acknowledgments
We thank Julia Thompson, Kha Nguyen, and Ryan Skophammer for their help with the screen and the Bloomington Stock Center and the Exelixis Stock Center for fly stocks. This study was supported by National Institutes of Health grant R01-EY08152 to U.B. U.B. is a Howard Hughes Medical Institute (HHMI) professor and G.C. is an HHMI instructor. We acknowledge HHMI for supporting research efforts by our undergraduate students, six of whom (A.C., J.M., K.Y., Q.A.F., G.L.G., and W.K.) are included here as authors.
References
- Ackerman, S. H., and A. Tzagoloff, 2005. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 80: 95–133. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ashby, M. N., S. Y. Kutsunai, S. Ackerman, A. Tzagoloff and P. A. Edwards, 1992. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267: 4128–4136. [PubMed] [Google Scholar]
- Boyer, P. D., 1997. The ATP synthase: a splendid molecular machine. Annu. Rev. Biochem. 66: 717–749. [DOI] [PubMed] [Google Scholar]
- Claros, M. G., and P. Vincens, 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779–786. [DOI] [PubMed] [Google Scholar]
- Diekert, K., G. Kispal, B. Guiard and R. Lill, 1999. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. USA 96: 11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., and M. A. Crosby, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont, M. E., T. S. Cardillo, M. K. Hayes and F. Sherman, 1991. Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5487–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, S. D., D. T. McLachlin and M. Revington, 2000. The second stalk of Escherichia coli ATP synthase. Biochim. Biophys. Acta 1458: 356–363. [DOI] [PubMed] [Google Scholar]
- Flores, G. V., A. Daga, H. R. Kalhor and U. Banerjee, 1998. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development 125: 3681–3687. [DOI] [PubMed] [Google Scholar]
- Flores, G. V., H. Duan, H. Yan, R. Nagaraj, W. Fu et al., 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85. [DOI] [PubMed] [Google Scholar]
- Graack, H. R., and B. Wittmann-Liebold, 1998. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J. 329(Pt 3): 433–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D. E., 1966. Comprehensive Biochemistry, pp. 309–326. Elsevier, Amsterdam.
- Kranz, R., R. Lill, B. Goldman, G. Bonnard and S. Merchant, 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29: 383–396. [DOI] [PubMed] [Google Scholar]
- Mandal, S., P. Guptan, E. Owusu-Ansah and U. Banerjee, 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9: 843–854. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D. D., and S. Ferguson-Miller, 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112: 481–490. [DOI] [PubMed] [Google Scholar]
- Newsome, T. B., B. Asling and B. J. Dickson, 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Ohta, S., 2003. A multi-functional organelle mitochondrion is involved in cell death, proliferation and disease. Curr. Med. Chem. 10: 2485–2494. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., and A. Geissler, 2001. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2: 339–349. [DOI] [PubMed] [Google Scholar]
- Ready, D. F., T. E. Hanson and S. Benzer, 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53: 217–240. [DOI] [PubMed] [Google Scholar]
- Schon, A., C. G. Kannangara, S. Gough and D. Soll, 1988. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature 331: 187–190. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, J. E., N. Fischel-Ghodsian, E. B. Mougey and T. W. O'Brien, 2004. Mitochondrial ribosomal proteins: candidate genes for mitochondrial disease. Genet. Med. 6: 73–80. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Tzagoloff, A., and A. Shtanko, 1995. Mitochondrial and cytoplasmic isoleucyl-, glutamyl- and arginyl-tRNA synthetases of yeast are encoded by separate genes. Eur. J. Biochem. 230: 582–586. [DOI] [PubMed] [Google Scholar]
- Weber, J., A. Muharemagic, S. Wilke-Mounts and A. E. Senior, 2004. Analysis of sequence determinants of F1Fo-ATP synthase in the N-terminal region of alpha subunit for binding of delta subunit. J. Biol. Chem. 279: 25673–25679. [DOI] [PubMed] [Google Scholar]
- Wilcox, M., and M. Nirenberg, 1968. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl. Acad. Sci. USA 61: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winrow, M. J., and H. Rudney, 1969. The incorporation of p-hydroxybenzoic acid and isopentenyl pyrophphate into ubiquinone precursors by cell-free preparations of rat tissues. Biochem. Biophys. Res. Commun. 37: 833–840. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1991. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113: 841–850. [DOI] [PubMed] [Google Scholar]
- Yao, K. M., M. L. Samson, R. Reeves and K. White, 1993. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol. 24: 723–739. [DOI] [PubMed] [Google Scholar]