Abstract
Although heritable microorganisms are increasingly recognized as widespread in insects, no systematic screens for such symbionts have been conducted in Drosophila species (the primary insect genetic models for studies of evolution, development, and innate immunity). Previous efforts screened relatively few Drosophila lineages, mainly for Wolbachia. We conducted an extensive survey of potentially heritable endosymbionts from any bacterial lineage via PCR screens of mature ovaries in 181 recently collected fly strains representing 35 species from 11 species groups. Due to our fly sampling methods, however, we are likely to have missed fly strains infected with sex ratio-distorting endosymbionts. Only Wolbachia and Spiroplasma, both widespread in insects, were confirmed as symbionts. These findings indicate that in contrast to some other insect groups, other heritable symbionts are uncommon in Drosophila species, possibly reflecting a robust innate immune response that eliminates many bacteria. A more extensive survey targeted these two symbiont types through diagnostic PCR in 1225 strains representing 225 species from 32 species groups. Of these, 19 species were infected by Wolbachia while only 3 species had Spiroplasma. Several new strains of Wolbachia and Spiroplasma were discovered, including ones divergent from any reported to date. The phylogenetic distribution of Wolbachia and Spiroplasma in Drosophila is discussed.
THE extent of symbiotic associations in animals is prompting a new evaluation of the role of microorganisms, particularly bacteria, in animal development, ecology, and evolution (McFall-Ngai 2002; Backhed et al. 2005). Although pathogenic infections are more intensively studied, recent studies of divergent groups, including mollusks, nematodes, annelids, insects, and mammals, reveal that chronic, noninvasive associations with particular bacterial lineages are common and are often beneficial or even required for the development and reproduction of hosts (Nelson and Fisher 1995; McFall-Ngai 2002; Brummel et al. 2004; Backhed et al. 2005; Baumann 2005; Taylor et al. 2005). Beneficial effects include dietary supplementation through biosynthesis of needed nutrients, developmental interactions that prime the immune system, improved tolerance to thermal stress, and defenses against natural enemies. At the same time, many chronic infectious agents have subtle deleterious effects on hosts, blurring the distinction between pathogenic and mutualistic associations.
Of particular interest are heritable microorganisms, which are especially widespread in insects (see Buchner 1965; Werren et al. 1995a ; Jeyaprakash and Hoy 2000; Moran et al. 2005b). Many of these are mutualistic, but some exert distinctive effects on host reproduction, such as biasing sex ratio, effecting parthenogenesis, or causing incompatibility in crosses with uninfected strains of the same host species (Werren et al. 1995b).
The genus Drosophila provides the primary insect genetic model system for studies of evolution and diversification (Powell 1997) and for studies of infectious processes and immunity (Mylonakis and Aballay 2005). Drosophila species lack so-called “primary symbionts” (ancient obligate associations in which symbionts occupy specialized host organs, Buchner 1965) but they do form facultative associations with maternally transmitted symbionts that undergo occasional horizontal transfer into naïve hosts.
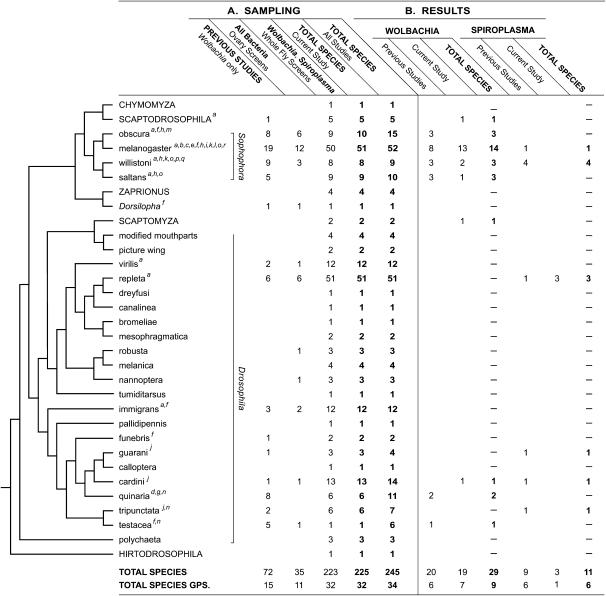
Despite the broad interest in Drosophila for ecological, evolutionary, and genetic studies, and the recent investigations of heritable symbionts in insects generally, few Drosophila species have been screened for the presence of heritable endosymbionts. Indeed, some associations have been discovered in the course of recent genomic sequencing projects in which symbiont DNA has been intermixed with that of the hosts (Salzberg et al. 2005a,b). The little deliberate screening that has been performed has been restricted mostly to Wolbachia pipientis, estimated to infect up to 70% of insect species, from many insect orders (Jeyaprakash and Hoy 2000). In Drosophila, most screening for Wolbachia has concentrated on long-term laboratory cultures (Giordano et al. 1995; Werren and Jaenike 1995; Werren et al. 1995a,b; Bourtzis et al. 1996; Zhou et al. 1998; Clark et al. 2005; Salzberg et al. 2005a,b; Miller and Riegler 2006) as opposed to natural populations (Vavre et al. 1999; Jaenike et al. 2003; Charlat et al. 2004; Veneti et al. 2004; Dyer and Jaenike 2005; Haine et al. 2005; Montenegro et al. 2006). Furthermore, there has been considerable phylogenetic bias in the species screened. Of the 69 species of the family Drosophilidae for which Wolbachia screening results have been published, 68 belong to the genus Drosophila (Figure 1). Of these, 41 belong to the subgenus Sophophora, which has ∼500 species, including Drosophila melanogaster, while only 26 belong to the larger subgenus Drosophila, which has ∼1500 species, excluding the Hawaiian Drosophila and the Scaptomyza (Markow and O'Grady 2006). Of the 20 species infected with Wolbachia, 17 belong to the subgenus Sophophora while only three belong to the subgenus Drosophila. Thus, uneven taxonomic sampling could underlie the observation that 17 of the 20 genus Drosophila species reported to harbor Wolbachia are from the subgenus Sophophora.
Figure 1.—
(A) Phylogenetic distribution of drosophilid species screened for endosymbiotic bacteria. Phylogenetic relationships among most of the species groups and genera in the subfamily Drosophilinae are based on Markow and O'Grady (2006). Capitalized taxon names represent genera other than the genus Drosophila. Noncapitalized names are species groups within the genus Drosophila; the subgenus to which they belong is also indicated. For each taxon, numbers represent (from left to right, respectively) number of species screened mainly for Wolbachia in previous studies, number of species screened for all groups of bacteria (i.e., ovary extracts) in this study, number of species screened specifically for Wolbachia and Spiroplasma (i.e., whole flies) in this study, the combined total number of species screened in this study, the total species screened for Wolbachia on the basis of previous and this study. (B) Species found to be positive for Wolbachia or Spiroplasma from (left to right) previous studies, this study, the combined total, first for Wolbachia and then for Spiroplasma. Previous studies: aBourtzis et al. (1996); bCharlat et al. (2004); cClark et al. (2005); dDyer and Jaenike (2005); eGiordano et al. (1995); fHaine et al. (2005); gJaenike et al. (2003); hMiller and Riegler (2006); iMontenegro et al. (2005); jMontenegro et al. (2006); kSalzberg et al. (2005a,b); lVavre et al. (1999); mVeneti et al. (2004); nWerren and Jaenike (1995); oWerren et al. (1995a,b); pWilliamson and Poulson (1979); qWilliamson et al. (1999); rZhou et al. (1998).
The only other heritable symbiont group reported for Drosophila species is Spiroplasma (Williamson and Poulson 1979; Williamson et al. 1999; Montenegro et al. 2005, 2006), which, along with related bacteria in the phylum Mollicutes, is widespread in insect hosts (Gasparich 2002) and which sometimes causes son killing in infected females (Anbutsu and Fukatsu 2003; Montenegro et al. 2005; Veneti et al. 2005). In Drosophila, Spiroplasma infections are currently documented in five species of the subgenus Sophophora—D. melanogaster (Montenegro et al. 2005); D. willistoni, D. nebulosa, D. paulistorum, and D. equinoxialis (Williamson and Poulson 1979; Williamson et al. 1999); and four species of the subgenus Drosophila—D. hydei (Ota et al. 1979); and D. neocardini, D. paraguayensis, and D. ornatifrons (Montenegro et al. 2006).
Examples of sex ratio bias or male killing have been reported for a few other Drosophila species (reviewed in Anbutsu and Fukatsu 2003), but the causative agents have not been identified. Other bacterial groups, including the Gammaproteobacteria (e.g., Moran et al. 2005a) and the phylum Bacteroidetes (Zchori-Fein and Perlman 2004), are also common as opportunistic heritable symbionts of insects, having major effects on reproduction, but their extent in Drosophila is not known. However, these bacterial groups also include transient colonizers of external surfaces and/or guts of insects; as a result, PCR amplification of whole insect DNA extracts with primers diagnostic to these groups would give little information regarding heritable symbiotic associations. One way to circumvent this problem is to screen extracts from dissected ovaries, the tissue most likely to harbor any heritable endosymbionts, but this procedure is time consuming. Wolbachia lacks closely related free-living or facultative counterparts, making its detection feasible by screening whole insect extracts with Wolbachia-specific PCR primers.
Given the array of heritable endosymbionts reported in other insects and the limited information about Drosophila, we have addressed the following questions: (1) What are the frequency and diversity of heritable bacterial endosymbionts in natural populations of Drosophila species? and (2) Are there any detectable phylogenetic patterns affecting endosymbiont infection within the genus Drosophila? Due to our fly sampling procedure, we likely would have missed sex ratio-distorting bacteria but should have detected bacteria that cause other effects on their hosts. Our initial PCR screen, using several sets of both “universal” and taxonomically restricted bacterial primers with DNA from dissected ovarioles, focused upon newly established isofemale strains from natural populations of 35 species (from 11 species groups) representing both major Drosophila subgenera (Figure 1). Because this initial screening revealed only the presence of Wolbachia and Spiroplasma, symbionts that can be definitively diagnosed using DNA extracted from whole flies, we then examined the distribution of these two symbionts across a total of 223 species from the Tucson Drosophila Species Stock Center.
MATERIALS AND METHODS
Drosophila species and strains screened:
Over 4700 individual flies from >1500 strains were screened, with usable results obtained for 1401 strains. The complete list of strains, their origin, and screening procedures used are provided as supplemental material (Appendix 1at http://www.genetics.org/supplemental/) and in an online database (http://amadeus.biosci.arizona.edu/∼bjn/flyendo/index.php).
Fly strains for exhaustive screening of ovarian tissues:
For the screening of heritable endosymbionts from any lineage of bacteria, we used primarily recently collected (<2 year) isofemale lines. Our rationale was based on our aim of detecting heritable symbionts present in wild fly populations (but possibly lost or acquired in long-term lab stocks). At the same time, isofemale lines were retained with the aim of having their progeny for further studies if endosymbionts were encountered and to validate fly species identification by a combination of examination of male genitalia and molecular approaches. For most species, identification could be confirmed by sequencing of fragments of the mitochondrial genes cytochrome oxidase I (Folmer et al. 1994) and II (Liu and Beckenbach 1992), while 1 kb of sequence flanking the X-linked microsatellite locus X008 was required to discriminate D. pseudoobscura from D. persimilis (Machado et al. 2002), and a fragment of xanthine dehydrogenase (xdh) (MJ-Xdh-798 5′-GAGCCAGACATTGGTGGAG-3′ and MJ-Xdh-1496 5′-AAGTAGGACTTGTGCTCGATGG-3′; L. Matzkin, unpublished data) was sequenced to distinguish among certain members of the repleta species group.
Fly strains for targeted screens:
For the screening of Wolbachia and Spiroplasma only, we used both recent and older collections, mostly derived from the Tucson Stock Center. We screened 223 species from the Tucson Stock Center (including most of the species used in the exhaustive screens). At the time of screening, this represented effectively all species in the collection that were neither in quarantine nor in the critical care unit. For the majority of the species, more than one strain was available and screened.
DNA extraction:
Ovarian dissection and DNA extraction:
We examined three to four females per fly strain. To ensure that ovaries contained large numbers of mature oocytes, mature females were placed in freshly yeasted vials for 2–3 days prior to dissection. Each female was anesthetized with CO2, surface sterilized in 95% ethanol, and dissected under sterile phosphate buffer. Ovaries were extracted carefully with sterile forceps (making sure the gut was not broken during the dissection), rinsed briefly in 0.5% bleach (0.03% sodium hypochlorite final concentration) and in sterile water, placed in a sterile microtube, frozen immediately in liquid nitrogen, and kept at −80° until DNA extraction. DNA was extracted with the DNEAsy Kit (QIAGEN, Valencia, CA) following the protocol for Gram-positive bacteria (i.e., lysozyme extraction). Two separate elutions in AE buffer of 30 μl were performed for each sample.
Whole-fly DNA extraction for targeted screens:
We examined three females per fly strain. Females were anesthetized with CO2; each female was placed individually into the well of a 96-well PCR plate on dry ice and kept at −80° until extraction. Each fly was crushed on ice with a sterile pestle in 48 μl of squish buffer (10 mm Tris–HCl; 1 mm EDTA; 25 mm NaCl) and 2 μl proteinase K (20 mg/ml) (Gloor et al. 1993). This was then incubated 30 min at 37° and 5 min at 95°.
PCR screening/sequencing:
For ovary extracts, we conducted PCR screens with each of the seven primer pairs listed in Table 1. The first two should amplify ∼1400 bp of the 16S ribosomal RNA gene of most Eubacteria. The 559F–35R pair should amplify ∼1000 bp of the 3′ end on the 16S rRNA gene, the intergenic spacer (ITS or IGS), which varies in size, and the first ∼35 bp of the 23S gene. However, it should not amplify the relatively few bacteria for which the 16S and 23S rRNA genes are not in the same operon, such as Buchnera (Tamas et al. 2002) and W. pipientis (Wu et al. 2004; Foster et al. 2005). To increase our chances of detecting endosymbionts, we also used four additional primer pairs, each of which amplifies a specific group of bacteria known to include heritable endosymbionts of insects (i.e., Bacteroidetes, Spiroplasma, and several Gram-positive Wolbachia, Cytophaga-like organisms; see Table 1).
TABLE 1.
Primer pairs used for PCR screening
| Primer pair (5′ to 3′) | Target gene (fragment size) | Target group | Annealing temp. [Mg+] |
|---|---|---|---|
| 10F AGTTTGATCATGGCTCAGATTGa | 16S rRNA (∼1500 bp) | Most bacteria | 60° [1.5 mm] |
| 1507R TACCTTGTTACGACTTCACCCCAGa | |||
| 27F GAGAGTTTGATCCTGGCTCAGb | 16S rRNA (∼1470 bp) | Most bacteria | 55° [1.5 mm] |
| 1492R GGTTACCTTGTTACGACTTb | |||
| 559F CGTGCCAGCAGCCGCGGTAATACc | 16S-ITS-35R (>1000 bp) | Most bacteria (not Wolbachia) | 58° [1.5 mm] |
| 35R CCTTCATCGCCTCTGACTGCd | |||
| 10FF AGAGTTTGATCATGGCTCAGGATGc | 16S rRNA (∼1300 bp) | Cytophaga–Flavobacterium–Bacteroidetes | 58° [4.5 mm] |
| 1370R CGTATTCACCGGATCATGGCe | |||
| 63F GCCTAATACATGCAAGTCGAACd | 16S rRNA (∼450 bp) | Spiroplasma and several Gram-positive | 55° [1.5 mm] |
| TKSSsp TAGCCGTGGCTTTCTGGTAAf | |||
| WspF TGGTCCAATAAGTGATGAAGAAACTAGCTAg | wsp (∼600 bp) | Wolbachia | Touchdown 65–55° [1.5 mm] |
| wspR AAAAATTAAACGCTACTCCAGCTTCTGCACg | |||
| CLOf GCGGTGTAAAATGAGCGTGh | 16S rRNA (∼450 bp) | Cytophaga-like organism | 57° [1.5 mm] |
| CLOr1 ACCTMTTCTTAACTCAAGCCTh | |||
| LCO-1490 GGTCAACAAATCATAAAGATATTGGi | Mitochondrial COI | Most invertebrates | 45° [5 mm] |
| HCO-2198 TAAACTTCAGGGTGACCAAAAAATCAi |
We included positive and negative controls for every PCR run. PCR runs with failed positive controls or with positive negative controls were excluded from the results. The quality of each ovary DNA extract was assessed by amplification of the fly's mitochondrial Cytochrome Oxidase I (mtCOI) gene. Templates that were negative for this PCR were excluded from the results. To assess presence of endosymbionts, we conducted an initial PCR screening (12.5 μl PCR reaction) for all samples. Samples that were scored as positive in the first PCR were then subjected to a second PCR reaction (50 μl total volume) for confirmation and sequencing. Extremely weak amplifications that did not yield enough template for sequencing were regarded as negative.
Both strands of each PCR product were directly sequenced with an ABI 3700 at the University of Arizona's Genomics Analysis and Technology Core Facility. If sequence results were unclear, suggesting that more than one sequence type or multiple PCR fragments were present, then PCR products were cloned and then sequenced (∼3 clones/PCR fragment/individual). Sequences were assembled and edited with Sequencher 4.5 (Gene Codes, Ann Arbor, MI).
Identification of bacteria:
We used Blastn (Altschul et al. 1997) and/or Classifier (Cole et al. 2005) to determine the identity of bacterial sequences. If a sequence was ≥98% identical to a sequence found in GenBank, it was assigned to that bacterial species or group.
Interpretation of screening results:
Flies were scored as infected by an endosymbiont if at least one individual of that line yielded a positive PCR result that was confirmed with a second PCR and, in most cases, a sequence. However, due to the possibility of contamination by free-living or facultative bacteria not known to be heritable symbionts of arthropods (e.g., Escherichia coli, Pseudomonas), flies that yielded positive PCR and sequences for these bacteria were not scored as infected. In the majority of cases, three individual flies from an infected line gave PCR results for a given primer pair, and a sequence was obtained from at least one individual.
Alignment and phylogenetic analyses:
To investigate the phylogenetic affinities of endosymbionts found in this study, we conducted phylogenetic analyses of the 16S rRNA gene. For Wolbachia analyses, we included published sequences (at least 1340-bp long) representing the highest Blastn hits to new haplotypes and representatives of most Wolbachia supergroups. For Spiroplasma analyses, we included the highest Blastn hits and published sequences of related lineages and outgroups on the basis of Gasparich et al. (2004). Sequences were aligned by eye in MacClade 4.06 (Maddison and Maddison 2003). Unalignable characters were excluded from phylogenetic analyses. Our alignments have been deposited in TreeBase (http://www.treebase.org/treebase) under accession numbers SN2737-10782 and SN2737-10783. We used PAUP*4.0b10 (Swofford 1998) to construct a neighbor-joining (NJ) tree under the Kimura-2-parameter model (Kimura 1980) of Drosophila mtCOI sequences for verification of species identity.
Given the evidence for widespread recombination in Wolbachia (Baldo et al. 2006), particularly within the wsp gene (Werren and Bartos 2001; Reuter and Keller 2003; Baldo et al. 2005), we did not attempt to construct phylogenetic relationships using this gene, as they would probably not reflect the true phylogenetic history of the strains. We used PAUP* and Modeltest 3.7 (Posada and Crandall 1998) to infer the most appropriate model of sequence evolution for 16S rRNA gene of Wolbachia and Spiroplasma haplotypes. We conducted maximum likelihood heuristic searches assuming the models selected above. As a measure of support for our phylogenetic inferences, we used MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001) to obtain Bayesian posterior probabilities. Four simultaneous Monte Carlo Markov chains were run for 10,000,000 cycles, and sampled every 100 cycles, under a model that included a substitution rate for each type of transition and transversion (general time reversible), a proportion of invariable sites, and a gamma distribution of rates across sites. Posterior probabilities for each node were obtained from a consensus of trees excluding the initial set of cycles preceding convergence on stable likelihood values (i.e., the “burn in”).
Nomenclature:
All the sequences obtained were compared to GenBank sequences by Blastn. If the most similar sequence in the database was not identical to our sequence, then our sequence was regarded as a new haplotype. For Wolbachia sequences, we named new haplotypes according to the host species. If a haplotype was not new (i.e., 100% identical to a sequence in GenBank), it was labeled with a previously assigned name.
RESULTS
Exhaustive screening of flies from natural populations for heritable symbionts:
We examined ovaries of 181 fly strains from 35 species in 11 species groups (Appendix 2 at http://www.genetics.org/supplemental), aiming to discover all possible maternally transmitted bacterial symbionts. According to our scoring criteria, three of the seven primer pairs used for screening produced no positive results (Table 2). Because all samples included in the results gave positive reactions for the DNA isolations (on the basis of the PCR of mtCOI), and positive template controls were run for every primer pair, these negative results show that these bacteria, including Cardinium (Cytophaga like) and other Bacteroidetes, were absent from all samples. Furthermore, in cases in which the more universal primers produced products, indicating presence of some bacterial type in the sample, the sequenced products almost always corresponded to either Wolbachia or Spiroplasma, which were also revealed by the corresponding diagnostic primer screens. Thus, screening with the 10F–1507R and/or 27F–1492R universal primer pairs demonstrated the presence of Wolbachia in eight species (Table 3) from the melanogaster and willistoni species groups (subgenus Sophophora). We also detected the presence of Spiroplasma in one species (D. hydei) with the 27F–1492R primer pair (Table 4).
TABLE 2.
Number of species and strains scored as positive for each of the primer sets
| Primer pair | Tissue examined | No. of species that were positive | Total no. of species examined | No. of strains that were positive | Total no. of strains examined |
|---|---|---|---|---|---|
| Universal 16S (10F–1507R) | Ovaries | 4W | 35 | 24W | 181 |
| Universal 16S (27F–1492R) | Ovaries | 8W 1S | 35 | 36W 5S | 181 |
| Universal 16S–23S (559F–35R) | Ovaries | 0 | 35 | 0 | 181 |
| Bacteroidetes 16S (10FF–1370R) | Ovaries | 0 | 35 | 0 | 181 |
| Cardinium and near relatives (CLOf1–CLOr1) | Ovaries | 0 | 35 | 0 | 181 |
| Spiroplasma 16S (63F–TKSSsp) | Ovaries and whole flies | 3 | 225 | 18 | 1401 |
| Wolbachia wsp | Ovaries and whole flies | 19 | 225 | 271 | 1401 |
See materials and methods. W, sequence corresponded to Wolbachia 16S gene; S, sequence corresponded to Spiroplasma 16S gene.
TABLE 3.
Species found positive for infection, with Wolbachia and Wolbachia haplotypes found
| Species group or genus | Strains examined
|
Wolbachia infected
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Ovaries | Whole flies | Total | Ovaries | Whole flies | Total | % strains infected | wsp haplotypes | 16S haplotypes | ||
| cardini | D. arawakana | 71 | 71 | 63 | 63 | 89 | 2 wWil; 56 wSpt | ||||
| melanogaster | D. ananassae | 3 | 17 | 20 | 3 | 7 | 10 | 50 | 7 wRi; 3 wSpt | 3 wRia,b | |
| melanogaster | D. baimaii | 2 | 2 | 1 | 1 | 50 | 1 wBai (new) | ||||
| melanogaster | D. bicornuta | 2 | 2 | 2 | 2 | 100 | 2 wBic (new) | ||||
| melanogaster | D. mauritiana | 3 | 3 | 2 | 2 | 67 | 2 wNo | ||||
| melanogaster | D. melanogaster | 11 | 51 | 60 | 10 | 37 | 45 | 75 | 37 wMel | 8 wMelc | |
| melanogaster | D. nikananu | 1 | 2 | 2 | 1 | 1 | 1 | 50 | 1 wNik (new) | 1 wNika (new) | |
| melanogaster | D. pseudoananassae | 8 | 8 | 3 | 3 | 38 | 3 wPana | 3 wPanaa | |||
| melanogaster | D. pseudotakahashii | 2 | 2 | 1 | 1 | 50 | 1 wPse (new) | ||||
| melanogaster | D. quadraria | 1 | 1 | 1 | 1 | 100 | 1 wRi | ||||
| melanogaster | D. sechellia | 3 | 3 | 1 | 1 | 33 | 1 wHa | ||||
| melanogaster | D. simulans | 18 | 30 | 48 | 18 | 13 | 31 | 65 | 2 wMel; 27 wRi | 16 wRia,b | |
| melanogaster | takahashii subgroup | 1 | 1 | 1 | 1 | 100 | 1 wTak (new) | 1 wTakc (new) | |||
| melanogaster | D. teissieri | 1 | 2 | 2 | 1 | 2 | 2 | 100 | 2 wSpt | 1 wMelc | |
| saltans | D. sturtevanti | 7 | 7 | 2 | 2 | 29 | 2 wStv MI | ||||
| willistoni | D. tropicalis | 1 | 3 | 3 | 1 | 2 | 2 | 67 | 2 wWil | ||
| willistoni | D. willistoni | 1 | 195 | 196 | 1 | 100 | 101 | 52 | 78 wWil | 1 wWilc (new) | |
| Scaptodrosophila | Scaptodrosophila stonei | 2 | 2 | 1 | 1 | 50 | 1 wSto (new) | ||||
| Scaptomyza | Scaptomyza pallida | 1 | 1 | 1 | 1 | 100 | 1 wSpt | ||||
| Total: | 434 | 271 | 62 | ||||||||
PCR product obtained with both 27F–1492R and 10F–1507R primer pairs.
Sequence is 1 bp different from another wRi strain (AY833061).
PCR product obtained with 27F–1492R primer pair only.
TABLE 4.
Species found positive for infection with Spiroplasma and Spiroplasma haplotypes found
| Species group | Strains examined
|
Spiroplasma infected
|
% strains infected | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Ovaries | Whole flies | Total | Ovaries | Whole flies | Total | Spiroplasma 16S haplotype | |||
| repleta | D. aldrichi | 12 | 12 | 1 | 1 | 8 | 2 haplotype_3a | |||
| repleta | D. hydei | 9 | 32 | 33 | 7 | 9 | 9 | 27 | 8 haplotype_1b; 1 haplotype_2b | |
| repleta | D. mojavensis | 7 | 114 | 121 | 3 | 5 | 8 | 7 | 8 haplotype_4a | |
| Total: | 166 | 18 | 11 | |||||||
PCR product obtained with 63F–TksspR primer pair only.
PCR product obtained with both 63F–TksspR and 27F–1492R primer pairs.
Several known heritable endosymbionts of insects are within the Enterobacteriaceae (Gammaproteobacteria), and some screens were designed to detect members of this group. Screening with the 559F–35R universal primer set, which spans the intergenic spacer of the rRNA operon and thus excludes Wolbachia, which lacks an intact operon (Wu et al. 2004), revealed the presence of proteobacterial sequences in several ovary extracts. Some of these were identified with Blastn and Classifier as particular species or genera (e.g., E. coli, Pseudomonas, Sphingomonas). These sequences probably reflect contamination or opportunistic pathogenic infection of the tissue or extract. A few others were identified as Enterobacteriaceae, but the genus could not be identified on the basis of the DNA sequence; these sequences could represent heritable symbionts. However, in most cases we found these sequences in only one individual per line, and these bacteria were not detected with either of the two other universal primer pairs for the 16S rRNA gene. We adopted a conservative criterion and disregarded them as heritable endosymbionts. Screening with group-specific primers revealed the presence of Wolbachia in the same eight species as with the universal primers as well as in one additional species (D. tropicalis; Table 3) and the presence of Spiroplasma in D. hydei and D. mojavensis (Table 4); the Spiroplasma strains from D. mojavensis were not detected with any of the universal primer pairs. We found no evidence of other bacterial groups with the other group-specific primers (Table 2).
Targeted screens for Wolbachia and Spiroplasma:
We screened for the presence of Wolbachia and Spiroplasma with group-specific primers in whole-fly DNA extracts from 1255 strains from ∼223 species representing 32 species groups of the family Drosophilidae, most within the genus Drosophila (Appendix 2 at http://www.genetics.org/supplemental; Figure 1A). We found evidence of Wolbachia in 16 species from the genus Drosophila (representing 4 species groups) and in 1 species each from the genus Scaptomyza and the genus Scaptodrosophila (Table 3; Figure 1B). We found evidence for Spiroplasma in D. hydei, D. mojavensis, and D. aldrichi, all within the repleta species group (Table 4; Figure 1B). All positive findings were confirmed with sequencing, and all positives were found in more than one independently extracted fly from the line.
Frequency of endosymbionts:
Wolbachia was much more common than Spiroplasma. Wolbachia was detected in 8% of all species examined, whereas Spiroplasma occurred in only 1.3% of species. Within Wolbachia-infected species, 62% of strains examined were infected (Table 3). In contrast, within Spiroplasma-infected species, only 11% of strains were infected (Table 4). Spiroplasma was most frequent in D. hydei where it occurred in 27% of the strains examined.
Diversity of Wolbachia:
On the basis of a data set of 1374 bp of the 16S rRNA sequences, we found six haplotypes of Wolbachia, three of which had not been reported before in any organism. Most of our phylogenetic analyses of the Wolbachia 16S rRNA gene included only taxa for which at least 1340 bp were available, but a subset of the analyses were conducted on a shorter data set (820 bp) to allow for inclusion of other lineages. Similarly, in a subset of the analyses, we removed the Wolbachia lineages that appeared most divergent to the Drosophila-associated Wolbachia strains to reduce the effects of mutational saturation on our phylogenetic inferences. Most of the substitution models used in the maximum likelihood and Bayesian analyses included a different rate for almost every type of substitution, unequal base frequencies, as well as a specific proportion of invariable sites and a specific gamma shape parameter (discrete approximation; four categories) for rate differences among sites.
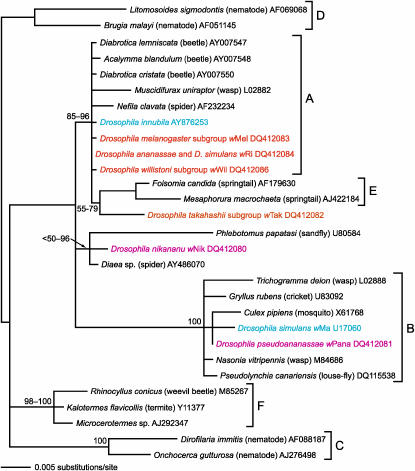
A consensus of our phylogenetic analyses (Figure 2) shows wPana (from D. pseudoananassae) within what is typically regarded as the B supergroup of Wolbachia, along with wNo (on the basis of a shorter sequence; not shown) and wMau strains of D. simulans. wNik (from D. nikananu) appeared to be most closely related to the Wolbachia strains from the sandfly Phlebotomus and the spider Diaea. However, in the absence of the Diaea strain (a shorter ∼820-bp sequence that was only included in a subset of the analyses) there was little or no support for the relationship between the wNik and the haplotype from Phlebotomus. Nevertheless, inclusion of the Diaea sequence resulted in very high support for this relationship (96% Bayesian posterior probability). wTak, the haplotype from an unidentified Drosophila species of takahashii subgroup, appeared to be related to the E supergroup found in springtails, but support for this relationship was low (55–79%). In our analyses, the E supergroup fell within what is traditionally recognized as the A supergroup with 85–96% support, and thus the A supergroup was not monophyletic. The strain from D. willistoni (wWil) was identical to the consensus of the trace archives of the D. willistoni whole genome shotgun sequencing project that contains fragments of the 16S rRNA gene but was otherwise unknown and unnamed. wWil was very similar to other Drosophila-associated Wolbachia: wRi and wMel (also found in our screenings), as well as to wHa and wAu, all of which were previously assigned to the A supergroup (Mercot and Charlat 2004; Baldo et al. 2005;).
Figure 2.—
Consensus of trees based on 16S rRNA gene of Wolbachia lineages inferred by maximum likelihood and Bayesian analyses. Numbers indicate the range of Bayesian posterior probabilities obtained under different sets of taxa and characters. Recognized Wolbachia supergroups are indicated by letters A–F. Taxon names indicate the host species and are followed by the GenBank accession number. Orange labels are haplotypes observed in this study but reported before, red labels had never been reported before, and blue labels are other Wolbachia strains associated with Drosophila reported in previous studies. Most analyses were based on a 1374-bp data set that included only taxa for which at least 1340 bp were available, but a subset of analyses included a taxon with a shorter sequence. Lineages from the D and C supergroups were removed sequentially in a subset of the analyses.
On the basis of the highly variable wsp gene, we found 14 haplotypes (GenBank accession nos. DQ412091–DQ412111; Table 3). Of these, 7 had been reported in Drosophila before and 7 were new haplotypes for Drosophila (i.e., wBai, wBic, wNik, wPana, wPse, wTak, and wSto). Although wsp is known to undergo widespread recombination in Wolbachia (Baldo et al. 2005), near-identical sequences likely reflect close relationship. Of these new haplotypes, 3 were very similar to other Drosophila-associated Wolbachia: the haplotype of Scaptodrosophila stonei (wSto) was 99% identical to that from D. septentriosaltans (wSpt; AY620209); the haplotype from D. bicornuta (wBic) was 99% identical to that from D. bifasciata (AJ27112; a male-killer) as well as to other non-Drosophila insects; and in agreement with the phylogenetic analyses of the 16S rRNA gene the haplotype from D. pseudonananassae (wPana) was 99% identical to B-clade sequences from D. simulans (wMa-AF020069 and wNo-AF020074). Four strains were quite different from anything reported from Drosophila before: that from D. baimaii (wBai) was 99% similar to Wolbachia from another dipteran (Pseudacteon curvatus; family Phoridae; AY878108); that from D. pseudotakahashii (wPse) was 99% similar to Wolbachia from fig and gall wasps and a heteropteran (AY567677, AY095154, AB109568); that from a species in the takahashii subgroup (wTak) was 99% similar to haplotypes from lice (AY331130); and that from D. nikananu (wNik) was very distinct from any Wolbachia reported to date, showing only 83% similarity to the closest sequence in GenBank, from a scarabid beetle host (Onthophagus vaulogeri; AY157683).
Diversity of Spiroplasma:
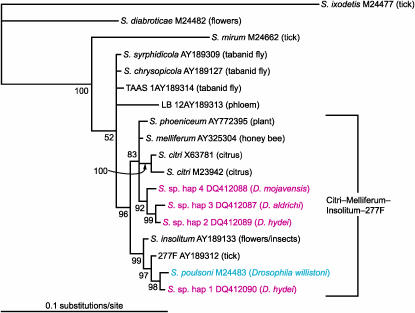
On the basis of a 439-bp fragment of the 16S rRNA gene of Spiroplasma, we found four Spiroplasma haplotypes in the lines examined (Table 4; Figure 3). Our phylogenetic analyses were restricted to this portion of the 16S rRNA gene because we could not obtain PCR products of the D. mojavensis strains with the universal primer pairs, which targeted a longer fragment (∼1500 bp), and because D. aldrichi was only included in the targeted surveys for Wolbachia and Spiroplasma, which examined a shorter fragment of the 16S rRNA gene of Spiroplasma. Each of the three species of Drosophila had a different haplotype, and two different haplotypes were observed in D. hydei: haplotype 1 was the most common in D. hydei while haplotype 2 only occurred in one line. Haplotype 1 was sister to S. poulsonii, from D. willistoni (subgenus Sophophora). These in turn were closely related to Spiroplasma from a tick and to S. insolitum, which infects flowers and insects. Haplotype 2 of D. hydei was closely related to the Spiroplasma found in D. mojavensis and D. aldrichi (also members of the repleta species group; subgenus Drosophila). These in turn were most closely related (83% Bayesian posterior probability) to S. citri, S. phoeniceum, and S. melliferum, which occur in plants and insects.
Figure 3.—
Maximum likelihood phylogeny of Spiroplasma 16S rRNA gene based on 439 characters. Haplotypes associated with Drosophila found in this study are indicated in red (hap 1–4), while the one found in a previous study is indicated in blue. Tree was rooted with Spiroplasma ixodetis. Numbers next to nodes indicate Bayesian posterior probabilities (>50%). Numbers next to taxon labels correspond to GenBank accessions, and host organism is indicated in parentheses.
DISCUSSION
Prior to this work there had been no systematic survey of heritable endosymbionts, other than Wolbachia, in Drosophila species. By examining 181 fly strains (from 35 species, 11 species groups) in the genus Drosophila for presence in ovarioles of endosymbionts from any lineage of bacteria (Figure 1), we have gained a more complete picture of the nature and scope of heritable endosymbiotic infections in this group of organisms. The most striking and unexpected result is that only two kinds of heritable endosymbionts were detected in these samples: Wolbachia and Spiroplasma. We contrast this to some other insects that possess a variety of bacterial symbionts, with high representation of Gammaproteobacteria (e.g., Baumann 2005). While our results imply a relatively low incidence of other heritable symbionts that do not cause sex-ratio distortion in Drosophila, such infections may occur in some populations or species. A previous study based on PCR screenings of different tissues of D. paulistorum, including ovaries, reports presence of a Proteus-like bacterium (Enterobacteriaceae; Gammaproteobacteria) (Miller et al. 1995). As mentioned above, we did detect presence of Enterobacteriaceae in some of our ovary extracts, but disregarded them as heritable endosymbionts because these occurrences were very sporadic and their DNA sequences did not allow a more specific identification. Although it is possible that some of these are truly heritable endosymbionts, our results suggest that if bacterial groups other than Wolbachia and Spiroplasma indeed associate with Drosophila, they do not appear to be widespread.
Absence of other heritable endosymbionts may reflect a robust innate immune response that eliminates infections by most bacterial groups. Drosophila species are saprophytic, utilizing necrotic plant material as feeding and breeding sites. Their niche is filled with a high diversity of microorganisms, many of which they consume along with the necrotic plant tissue. Exposure to microorganisms at all stages of their life cycle could have shaped the Drosophila immune system to resist infection by most bacteria. Indeed, Drosophila uses efficient mechanisms to prevent microbial infection (Tzou et al. 2002; Hoffmann 2003). Insect groups that are more prone to associations with heritable bacteria, such as sap-feeding insect families (Baumann 2005), may have less robust immune mechanisms, particularly against Gram-negative bacteria that replicate in the hemocoel. Currently, little is known about the immune system of sap-feeding insects such as aphids and relatives, although aphids are reported to have an attenuated encapsulation response (Mackauer 1986).
Despite the apparently robust immune system of Drosophila, Wolbachia and Spiroplasma have managed to invade a few species. One possible reason is that both Spiroplasma and Wolbachia appear to avoid recognition by innate immune systems (Bourtzis et al. 2000; Hurst et al. 2003). Previous work reported that 20 of 69 drosophilid species examined were infected with Wolbachia. Our study more than triples the number of drosophilid species screened for Wolbachia and Spiroplasma. Our results revealed infections in 9 additional species of the family Drosophilidae, including members of three groups previously unknown to have Wolbachia: cardini group (D. arawakana) in the genus Drosophila; and members of two other genera, Scaptomyza (S. pallida), and Scaptodrosophila (S. stonei). In some cases, infection by Wolbachia has persisted through decades (up to ∼60 years) of laboratory culture in the stock center, as reported by previous studies (Clark et al. 2005; Riegler et al. 2005).
While previous studies had examined several drosophilid species for Wolbachia, very few species had been surveyed for Spiroplasma, revealing nine Drosophila species infected by this bacterium: D. willistoni, D. paulistorum, D. nebulosa, and D. equinoxialis (willistoni species group, subgenus Sophophora); D. melanogaster (melanogaster species group, subgenus Sophophora); D. paraguayensis, D. ornatifrons, D. neocardini (Montenegro et al. 2006), and D. hydei (Ota et al. 1979) (tripunctata, guarani, cardini, and repleta groups, respectively; subgenus Drosophila), but this last one had not been confirmed by DNA sequencing. Our results revealed infections in D. hydei and in two additional members of the repleta group in which Spiroplasma had not been reported previously (D. aldrichi and D. mojavensis), but not in any of the other species groups reported before. This is probably due to the fact that most of the Drosophila-associated Spiroplasma strains reported to date are male killers, which were not likely to be found in our fly samples for reasons discussed above.
Frequency of Wolbachia and Spiroplasma:
Wolbachia was more common than Spiroplasma. Overall infection rates with Wolbachia were found to be low (8 and 12% of examined species; this study and all studies, respectively) compared to Wolbachia infection rates of insect species in general: 16.9–22% (using standard PCR; Werren et al. 1995a; Werren and Windsor 2000) and 70% (using long PCR; Jeyaprakash and Hoy 2000). Observation of low infection rates in our study could reflect a sampling bias. Many of our samples were derived from isofemale lines or old lab strains, which were unlikely to include male killers (including some Wolbachia, Spiroplasma, and other heritable bacteria). Indeed, we did not detect endosymbionts in several species in which male killers had been reported before (i.e., D. bifasciata, D. prosaltans, D. paulistorum, D. equinoxialis, D. nebulosa, and D. robusta; Magni 1953; Cavalcanti et al. 1957; Poulson 1966; Ikeda 1970; Williamson and Poulson 1979). However, our procedures would have enabled detection of symbionts causing cytoplasmic incompatibility (the most widely documented Wolbachia phenotype) or mutualistic phenotypes.
Phylogenetic distribution of Wolbachia and Spiroplasma:
Wolbachia and Spiroplasma appear to be concentrated in certain drosophilid groups. For example, on the basis of previous work and this study, 242 species have been screened for Wolbachia (Figure 1), most of which (229) belong to the genus Drosophila. Of these, 86 species belong to the subgenus Sophophora and 143 to the larger subgenus Drosophila. However, Wolbachia infections have been detected in 23 species of the subgenus Sophophora (out of 86; 27%) compared to only 4 species of the subgenus Drosophila (out of 143; 3%). Thus, Wolbachia is much more common in the subgenus Sophophora than in the subgenus Drosophila (G-test = 29.7; P = 4.9 × 10−8; d.f. = 1). This difference remains highly significant if only species with at least three tested strains are included. The proportion of Wolbachia-infected species in the subgenus Sophophora more closely reflects the overall proportion of Wolbachia-infected insect species (16.9–70%, depending on the screening method), while the proportion of infected species in the subgenus Drosophila is much lower. Whether or not this reflects resistance to Wolbachia in some Drosophila groups remains to be determined.
Far fewer species (11 of 228, on the basis of the present and past studies) are infected with Spiroplasma and these fall into six species groups within the genus Drosophila. In contrast to Wolbachia, no significant difference was observed between the subgenus Sophophora and subgenus Drosophila in the distribution of Spiroplasma (G-test = 0.17; P = 0.7; d.f. = 1). Interestingly, one of the groups that harbored Spiroplasma (the repleta group, subgenus Drosophila), which is well represented in our study (51 species), did not harbor any Wolbachia, but 3 of its species harbored Spiroplasma.
Diversity of Wolbachia:
Our screening revealed new haplotypes of Wolbachia and Spiroplasma, including some similar to previously reported haplotypes from Drosophila. Other Wolbachia strains were not similar to any previously reported in Drosophila but similar to ones reported from divergent taxa such as other dipterans, hymenopterans, heteropterans, and lice, providing further evidence that Wolbachia has been horizontally transmitted among very divergent taxa (Heath et al. 1999; Stevens et al. 2001). One of the haplotypes (wNik) was distinct from any reported to date, both in 16S rRNA and in wsp sequence. Another (wTak) appeared closely related to the E clade (from springtails) on the basis of the 16S rRNA gene, but its wsp sequence was 99% identical to a haplotype from lice regarded as a member of the B clade (Kyei-Poku et al. 2005). This disagreement between genes is expected due to the widespread recombination reported among and within several Wolbachia genes (Baldo et al. 2006). wNik and wTak were divergent from any haplotypes reported from Drosophila, but examination of multiple loci (e.g., Bordenstein and Rosengaus 2005; Casiraghi et al. 2005) may be necessary to accurately infer their phylogenetic affinities. In this regard, not recovering a monophyletic A supergroup may be the result of lack of phylogenetic signal in the 16S rRNA gene.
Occurrence of wWil (A supergroup) in both D. tropicalis and D. willistoni may reflect a recent horizontal transfer between these closely related species. The two infected D. tropicalis strains were collected at the same locality as one of the infected D. willistoni strains, suggesting horizontal transmission due to habitat sharing, as reported for closely related species of the obscura species group (Haine et al. 2005). A previous study that used diagnostic PCR primers to distinguish the A and B supergroups of Wolbachia reported infection by B supergroup Wolbachia in D. tropicalis, although this was not corroborated by DNA sequencing. This species may associate with Wolbachia from both supergroups, as observed in D. simulans (reviewed by Mercot and Charlat 2004).
Diversity of Spiroplasma:
Our study also revealed new strains of Spiroplasma. Haplotype 1 (from D. hydei) was closely related to the type strain of S. poulsonii from D. willistoni (Figure 3) as well as to the strains from D. nebulosa and D. melanogaster (results not shown; based on a different portion of the 16S rRNA sequence). D. hydei (subgenus Drosophila) is very distantly related to D. willistoni, D. nebulosa, and D. melanogaster (subgenus Sophophora), suggesting that horizontal transfer may have occurred in the recent past between these divergent groups. Indeed, the high similarity between the sequence from D. nebulosa (willistoni group) and a Brazilian strain from D. melanogaster (melanogaster group) has been attributed to a recent horizontal transfer from the New World native D. nebulosa to the Old World native D. melanogaster (Montenegro et al. 2005). The clade formed by S. poulsonii and haplotype 1 is most closely related to spiroplasmas found in ticks (unknown transmission mode) and in flowers and insects (horizontally transmitted). The other three haplotypes of Spiroplasma, all found in members of the repleta group (subgenus Drosophila) fall into a separate monophyletic group, whose closest relatives are S. citri, S. phoeniceum, and S. melliferum, horizontally transmitted pathogens of plants (the first two) and honeybees (the latter). The lack of monophyly of our haplotypes indicates that Spiroplasma invaded Drosophila at least twice. Despite belonging to two separate clades, all the Drosophila-derived haplotypes fell into the Citri–Melliferum–Insolitum–277F clade defined by Gasparich et al. (2004).
With few exceptions (Yamada et al. 1982; Ebbert 1991), strains of S. poulsonii cause son killing in D. willistoni, D. nebulosa, D. melanogaster, D. neocardini, D. paraguayensis, and D. ornatifrons and in species to which they have been artificially transferred (Williamson and Poulson 1979; Ebbert 1991, 1995; Williamson et al. 1999; Montenegro et al. 2005, 2006). Our preliminary results suggest that its close relative, haplotype 1, does not cause son killing in D. hydei; D. hydei was previously reported (Ota et al. 1979) to harbor a non-male killing strain of unknown relationship to the Spiroplasma of our study. The other three haplotypes (2–4) also show no evidence of son killing, suggesting that none of the Spiroplasma strains associated with repleta group flies cause son killing. Whether these Spiroplasma strains are incapable of killing males or the repleta group flies are resistant to male killing is unknown.
We observed no cases of co-infection of one individual fly or strain by more than one Wolbachia or Spiroplasma strain and no cases of co-infection by Wolbachia and Spiroplasma. Co-infection by more than one Wolbachia strain has been reported in several organisms (for example, Werren et al. 1995b; Perrot-Minnot et al. 1996; Vavre et al. 1999; Miller and Riegler 2006), and co-infection by Wolbachia and Spiroplasma has been reported only in D. melanogaster (Montenegro et al. 2005).
Conclusion:
Our study triples the number of Drosophila species screened for Wolbachia, vastly increases the screening for Spiroplasma, and is the first broad screening aimed at discovery of heritable symbionts from any bacterial phylum. Our finding of low symbiont diversity in the sampled Drosophila species suggests significant differences among insect groups in their basic proclivities for symbioses, with Drosophila possibly presenting more obstacles to the establishment of intimate associations. Some insight into reasons for this difference may be found in comparisons of gene inventories of Drosophila species with those of other arthropods, made possible by ongoing genome sequencing efforts. Our findings also raise the question of the nature of the phenotypic effects of Wolbachia and Spiroplasma in the newly discovered host species. These symbionts could play a major evolutionary role, as certain kinds of phenotypes can result in infections sweeping through populations with major consequences for levels of polymorphism and fixation of alleles (e.g., Dean et al. 2003; Riegler et al. 2005).
Acknowledgments
We thank the following for providing fly samples and positive control extracts: Tucson Stock Center, David Bruck, Bryant McAllister, Thomas Merritt, Artyom Kopp, Carolyn McBride, Jose Mojica, Laura Reed, Steve Perlman, and Scott Santos. Adam Falck, Camilo Hurtado, Frances McQueen, and Thomas Watts conducted lab work for this project. Luciano Maztkin kindly provided the xdh primers. Luis A. Hurtado and two anonymous reviewers provided helpful suggestions for the manuscript. This work was funded by National Science Foundation grant DEB-0315815 to N.A.M. and T.A.M.
References
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbutsu, H., and T. Fukatsu, 2003. Population dynamics of male-killing and non-male-killing Spiroplasmas in Drosophila melanogaster. Appl. Environ. Microbiol. 69: 1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed, F., R. Ley, J. Sonnenburg, D. Peterson and J. Gordon, 2005. Host-bacterial mutualism in the human intestine. Science 307: 1915–1920. [DOI] [PubMed] [Google Scholar]
- Baldo, L., N. Lo and J. H. Werren, 2005. Mosaic nature of the Wolbachia surface protein. J. Bacteriol. 187: 5406–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo, L., S. Bordenstein, J. J. Wernegreen and J. H. Werren, 2006. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23: 437–449. [DOI] [PubMed] [Google Scholar]
- Baumann, P., 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59: 155–189. [DOI] [PubMed] [Google Scholar]
- Bordenstein, S., and R. Rosengaus, 2005. Discovery of a novel Wolbachia supergroup in isoptera. Curr. Microbiol. 51: 393–398. [DOI] [PubMed] [Google Scholar]
- Bourtzis, K., A. Nirgianaki, G. Markakis and C. Savakis, 1996. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics 144: 1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis, K., M. M. Pettigrew and S. L. O'Neill, 2000. Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol. Biol. 9: 635–639. [DOI] [PubMed] [Google Scholar]
- Brummel, T., A. Ching, L. Seroude, A. F. Simon and S. Benzer, 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl. Acad. Sci. USA 101: 12974–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, P., 1965. Endosymbiosis of Animals With Plant Microorganisms. Interscience, New York.
- Casiraghi, M., S. R. Bordenstein, L. Baldo, N. Lo, T. Beninati et al., 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151: 4015–4022. [DOI] [PubMed] [Google Scholar]
- Cavalcanti, A. G. L., D. N. Falcão and L. E. Castro, 1957. “Sex-ratio” in Drosophila prosaltans—a character due to interaction between nuclear genes and cytoplasmic factors. Am. Nat. 91: 321–325. [Google Scholar]
- Charlat, S., J. W. Ballard and H. Mercot, 2004. What maintains noncytoplasmic incompatibility inducing Wolbachia in their hosts: a case study from a natural Drosophila yakuba population. J. Evol. Biol. 17: 322–330. [DOI] [PubMed] [Google Scholar]
- Clark, M. E., C. L. Anderson, J. Cande and T. L. Karr, 2005. Widespread prevalence of Wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170: 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, J. R., B. Chai, R. Farris, Q. Wang, S. Kulam et al., 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33: D294–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, M. D., K. J. Ballard, A. Glass and J. W. O. Ballard, 2003. Influence of two Wolbachia strains on population structure of east African Drosophila simulans. Genetics 165: 1959–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, K., and J. Jaenike, 2005. Evolutionary dynamics of a spatially structured host-parasite association: Drosphila innubila and male-killing Wolbachia. Evolution 59: 1518–1528. [PubMed] [Google Scholar]
- Ebbert, M. A., 1991. The interaction phenotype in the Drosophila willistoni spiroplasma symbiosis. Evolution 45: 971–988. [DOI] [PubMed] [Google Scholar]
- Ebbert, M. A., 1995. Variable effects of crowding on Drosophila hosts of male-lethal and non-male-lethal spiroplasmas in laboratory populations. Heredity 74: 227–240. [DOI] [PubMed] [Google Scholar]
- Folmer, O., M. Black, W. Hoeh, R. Lutz and R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294–299. [PubMed] [Google Scholar]
- Foster, J., M. Ganatra, I. Kamal, J. Ware, K. Makarova et al., 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T., and N. Nikoh, 2000. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). Appl. Environ. Microbiol. 66: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparich, G. E., 2002. Spiroplasmas: evolution, adaptation and diversity. Front. Biosci. 7: D619–D640. [DOI] [PubMed] [Google Scholar]
- Gasparich, G. E., R. F. Whitcomb, D. Dodge, F. E. French, J. Glass et al., 2004. The genus Spiroplasma and its non-helical descendants: phylogenetic classification, correlation with phenotype and roots of the Mycoplasma mycoides clade. Int. J. Syst. Evol. Microbiol. 54: 893–918. [DOI] [PubMed] [Google Scholar]
- Giordano, R., S. L. O'Neill and H. M. Robertson, 1995. Wolbachia infections and the expression of cytoplasmic incompatibility in Drosophila sechellia and D. mauritiana. Genetics 140: 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine, E. R., N. J. Pickup and J. M. Cook, 2005. Horizontal transmission of Wolbachia in a Drosophila community. Ecol. Entomol. 30: 464–472. [Google Scholar]
- Heath, B. D., R. D. J. Butcher, W. G. F. Whitfield and S. F. Hubbard, 1999. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9: 313–316. [DOI] [PubMed] [Google Scholar]
- Hoffmann, J. A., 2003. The immune response of Drosophila. Nature 426: 33–38. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and R. Ronquist, 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hurst, G. D. D., H. Anbutsu, M. Kutsukake and T. Fukatsu, 2003. Hidden from the host: Spiroplasma bacteria infecting Drosophila do not cause an immune response, but are suppressed by ectopic immune activation. Insect Mol. Biol. 12: 93–97. [DOI] [PubMed] [Google Scholar]
- Ikeda, H., 1970. The cytoplasmically-inherited ‘sex-ratio’ condition in natural and experimental populations of Drosophila bifasciata. Genetics 65: 311–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J., K. A. Dyer and L. K. Reed, 2003. Within-population structure of competition and the dynamics of male-killing Wolbachia. Evol. Ecol. Res. 5: 1023–1036. [Google Scholar]
- Jeyaprakash, A., and M. A. Hoy, 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 9: 393–405. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kyei-Poku, G. K., D. D. Colwell, P. Coghlin, B. Benkel and K. D. Floate, 2005. On the ubiquity and phylogeny of Wolbachia in lice. Mol. Ecol. 14: 285–294. [DOI] [PubMed] [Google Scholar]
- Lane, D. J., 1991. 16S/23S rRNA sequencing, pp. 115–175 in Nucleic Acid Techniques in Bacterial Systematics, edited by E. Stackebrandt and M. Goodfellow. John Wiley & Sons, New York.
- Liu, H., and A. T. Beckenbach, 1992. Evolution of the mitochondrial cytochrome oxidase II gene among 10 orders of insects. Mol. Phylogenet. Evol. 1: 41–52. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Mackauer, M., 1986. Growth and developmental interactions in some aphids and their hymenopterous parasites. J. Insect Physiol. 32: 275–280. [Google Scholar]
- Maddison, D. R., and W. Maddison, 2003. MacClade 4: Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA. [DOI] [PubMed]
- Magni, G. E., 1953. Sex-ratio; a non-Mendelian character in Drosophila bifasciata. Nature 172: 81. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. M. O'Grady, 2006. Drosophila: A Guide to Species Identification and Use. Academic Press Elsevier, London.
- McFall-Ngai, M., 2002. Unseen forces: the influence of bacteria on animal development. Dev. Biol. 242: 1–14. [DOI] [PubMed] [Google Scholar]
- Mercot, H., and S. Charlat, 2004. Wolbachia infections in Drosophila melanogaster and D. simulans: polymorphism and levels of cytoplasmic incompatibility. Genetica 120: 51–59. [DOI] [PubMed] [Google Scholar]
- Miller, S. G., B. C. Campbell, J. Becnel and L. Ehrman, 1995. Bacterial entomopathogens from the Drosophila paulistorum semispecies complex. J. Invertebr. Pathol. 65: 125–131. [DOI] [PubMed] [Google Scholar]
- Miller, W., and M. Riegler, 2006. Evolutionary dynamics of wAu-like Wolbachia variants in Neotropical Drosophila spp. Appl. Environ. Microbiol. 72: 826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro, H., V. N. Solferini, L. B. Klaczko and G. D. D. Hurst, 2005. Male-killing Spiroplasma naturally infecting Drosophila melanogaster. Insect Mol. Biol. 14: 281–288. [DOI] [PubMed] [Google Scholar]
- Montenegro, H., L. Hatadani, H. Medeiros and L. Klaczko, 2006. Male killing in three species of the tripunctata radiation of Drosophila (Diptera: Drosophilidae). J. Zool. Syst. Evol. Res. 44: 130–135. [Google Scholar]
- Moran, N. A., C. Dale, H. Dunbar, W. A. Smith and H. Ochman, 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5: 116–126. [DOI] [PubMed] [Google Scholar]
- Moran, N. A., J. Russell, T. Fukatsu and R. Koga, 2005. a Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71: 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N. A., P. Tran and N. M. Gerardo, 2005. b Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71: 8802–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson, M. A., P. Baumann, M. A. Clark, N. A. Moran, D. J. Voegtlin et al., 1991. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J. Bacteriol. 173: 6321–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis, E., and A. Aballay, 2005. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect. Immun. 73: 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C., and C. R. Fisher, 1995. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps, pp. 125–167 in Microbiology of Deep-Sea Hydrothermal Vent Habitats, edited by D. M. Karl. CRC Press, Boca Raton, FL.
- Ota, T., M. Kawabe, K. Oishi and D. F. Poulson, 1979. Non-male-killing spiroplasmas in Drosophila hydei. J. Hered. 70: 211–213. [Google Scholar]
- Perrot-Minnot, M. J., L. R. Guo and J. H. Werren, 1996. Single and double Infections with Wolbachia in the parasitic wasp Nasonia vitripennis: effects on compatibility. Genetics 143: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Poulson, D., 1966. Further cases of maternal SR in Drosophila species. Dros. Inf. Serv. 41: 77. [Google Scholar]
- Powell, J., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, New York.
- Reuter, M., and L. Keller, 2003. High levels of multiple Wolbachia infection and recombination in the ant Formica exsecta. Mol. Biol. Evol. 20: 748–753. [DOI] [PubMed] [Google Scholar]
- Riegler, M., M. Sidhu, W. Miller and S. O'Neill, 2005. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15: 1428–1433. [DOI] [PubMed] [Google Scholar]
- Russell, J. A., A. Latorre, B. Sabater-Munoz, A. Moya and N. A. Moran, 2003. Side-stepping symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Salzberg, S., J. Dunning Hotopp, A. Delcher, M. Pop, D. Smith et al., 2005. a Correction: Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 6: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg, S., J. Dunning Hotopp, A. Delcher, M. Pop, D. Smith et al., 2005. b Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 6: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, L., R. Giordano and R. F. Fialho, 2001. Male-killing, nematode infections, bacteriophage infection, and virulence of cytoplasmic bacteria in the genus Wolbachia. Annu. Rev. Ecol. Syst. 32: 519–545. [Google Scholar]
- Swofford, D. L., 1998. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson et al., 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296: 2376–2379. [DOI] [PubMed] [Google Scholar]
- Taylor, M. J., C. Bandi and A. Hoerauf, 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60: 245–284. [DOI] [PubMed] [Google Scholar]
- Tzou, P., E. De Gregorio and B. Lemaitre, 2002. How Drosophila combats microbial infection: a model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 5: 102–110. [DOI] [PubMed] [Google Scholar]
- Vavre, F., F. Fleury, D. Lepetit, P. Fouillet and M. Bouletreau, 1999. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16: 1711–1723. [DOI] [PubMed] [Google Scholar]
- Veneti, Z., M. Toda and G. Hurst, 2004. Host resistance does not explain variation in incidence of male-killing bacteria in Drosophila bifasciata. BMC Evol. Biol. 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti, Z., J. K. Bentley, T. Koana, H. R. Braig and G. D. Hurst, 2005. A functional dosage compensation complex required for male killing in Drosophila. Science 307: 1461–1463. [DOI] [PubMed] [Google Scholar]
- Weeks, A. R., R. Velten and R. Stouthamer, 2003. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proc. R. Soc. Lond. B. Biol. Sci. 270: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H., and J. D. Bartos, 2001. Recombination in Wolbachia. Curr. Biol. 11: 431–435. [DOI] [PubMed] [Google Scholar]
- Werren, J., and J. Jaenike, 1995. Wolbachia and cytoplasmic incompatibility in mycophagous Drosophila and their relatives. Heredity 75: 320–326. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., and D. M. Windsor, 2000. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. R. Soc. Lond. B. Biol. Sci. 267: 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H., D. Windsor and L. Gao, 1995. a Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B. Biol. Sci. 262: 197–204. [Google Scholar]
- Werren, J. H., W. Zhang and L. R. Guo, 1995. b Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. Lond. B. Biol. Sci. 261: 55–63. [DOI] [PubMed] [Google Scholar]
- Williamson, D. L., and D. F. Poulson, 1979. Sex ratio organisms (Spiroplasmas) of Drosophila, pp. 175–208 in The Mycoplasmas, edited by R. F. Whitcomb and J. G. Tully. Academic Press, New York.
- Williamson, D. L., B. Sakaguchi, K. J. Hackett, R. F. Whitcomb, J. G. Tully et al., 1999. Spiroplasma poulsonii sp. nov., a new species associated with male-lethality in Drosophila willistoni, a neotropical species of fruit fly. Int. J. Syst. Bacteriol. 49: 611–618. [DOI] [PubMed] [Google Scholar]
- Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy et al., 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M., S. Nawa and T. K. Watanabe, 1982. A mutant of SR organism (SRO) in Drosophila that does not kill the host males. Jpn. J. Genet. 57: 301–305. [Google Scholar]
- Zchori-Fein, E., and S. J. Perlman, 2004. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 13: 2009–2016. [DOI] [PubMed] [Google Scholar]
- Zhou, W. G., F. Rousset and S. O'Neill, 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. B. Biol. Sci. 265: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]