Abstract
Meiotic recombination rates are potentially affected by cis- and trans-acting factors, i.e., genotype-specific modifiers that do or do not reside in the recombining interval, respectively. Effects of trans modifiers on recombination across the ∼140-kb maize a1–sh2 interval of chromosome 3L were studied in the absence of polymorphic cis factors in three genetically diverse backgrounds into which a sequence-identical a1–sh2 interval had been introgressed. Genetic distances across a1–sh2 varied twofold among genetic backgrounds. Although the existence of regions exhibiting high and low rates of recombination (hot and cold spots, respectively) was conserved across backgrounds, the absolute rates of recombination in these sequence-identical regions differed significantly among backgrounds. In addition, an intergenic hot spot had a higher rate of recombination as compared to the genome average rate of recombination in one background and not in another. Recombination rates across two genetic intervals on chromosome 1 did not exhibit the same relationships among backgrounds as was observed in a1–sh2. This suggests that at least some detected trans-acting factors do not equally affect recombination across the genome. This study establishes that trans modifier(s) polymorphic among genetic backgrounds can increase and decrease recombination in both genic and intergenic regions over relatively small genetic and physical intervals.
MEIOTIC recombination mediates the proper disjunction of chromosomes, generates novel allelic combinations across chromosomes, and provides the genetic diversity upon which selection can act. According to the double-strand break repair (DSBR) model (Szostak et al. 1983; Sun et al. 1991), meiotic recombination initiates with a double-strand break (DSB). Fungal, animal, and plant chromosomes exhibit regions of recombination hyperactivity, i.e., hot spots, and hypoactivity, i.e., cold spots (reviewed in Lichten and Goldman 1995; Schnable et al. 1998; Petes 2001; Nachman 2002), as compared to the genomewide average rate of recombination in the organism being studied. Consistent with the DSBR model, recombination hot spots in Saccharomyces cerevisiae are clearly associated with DSBs (reviewed in Lichten and Goldman 1995) and these DSB hot spots are not distributed randomly across the yeast genome (Gerton et al. 2000).
Nonrandom patterns of recombination have been identified in all organisms studied to date. For example, in most species recombination rates per megabase are suppressed in and surrounding centromeres (Lambie and Roeder 1986; Werner et al. 1992; Mahtani and Willard 1998; Puechberty et al. 1999; Kagawa et al. 2002). Similarly, in most species, with the exception of Caenorhabditis elegans, for which recombination rates are negatively correlated with gene density (Barnes et al. 1995), recombination rates are higher in gene-rich than in gene-poor regions of the genome (reviewed in Lichten and Goldman 1995; Schnable et al. 1998). In wheat and barley, for example, analyses of integrated genetic and cytogenetically based physical maps (Gill et al. 1993, 1996a,b; Hohmann et al. 1994; Delaney et al. 1995a,b; Mickelson-Young et al. 1995; Kunzel et al. 2000) have shown that most recombination occurs in relatively small gene-rich regions. Similarly, in maize recombination nodules cluster in regions of high gene density (Anderson et al. 2006). In addition, it is estimated that recombination in a gene-dense portion of the bz1 region on chromosome 9S in maize is two orders of magnitude greater than in a flanking region of high retrotransposon density (Fu et al. 2002).
Although it is well established that high frequencies of recombination are positively correlated with gene-rich regions in the grasses, recombination breakpoints within these hot spots might resolve equally in genic and intergenic sequences. A high-resolution recombination mapping study across the ∼140-kb multigenic maize a1–sh2 interval supports that genes per se are preferred recombination hot spots and intergenic regions are cold spots; even so, nongenic hot spots and genic cold spots do exist (Yao et al. 2002). These maize genic hot spots can have either nonrandom distributions of breakpoints as observed in a1 (Xu et al. 1995) and other genes (Eggleston et al. 1995; Patterson et al. 1995) or uniform distributions across the gene, as in bz1 (Dooner and Martinez-Ferez 1997).
Variations in meiotic recombination rates occur not only among various regions of the genome but also among genetic backgrounds (reviewed in Simchen and Stamberg 1969) as evidenced in both animals (Koehler et al. 2002) and plants. In Arabidopsis, recombination rates vary among accessions (Sanchez-Moran et al. 2002). In maize, heterogeneity in recombination frequencies has been well documented within corn belt (Beavis and Grant 1991; Tulsieram et al. 1992), corn belt × exotic germplasm (Tulsieram et al. 1992), synthetic (Fatmi et al. 1993), exotic, and teosinte-maize hybrid (Williams et al. 1995) mapping populations and has often confounded the generation of composite genetic maps. Recombination rates in intervals with heterogenous frequencies varied two- to threefold among diverse mapping populations (Williams et al. 1995). Within a mapping population, recombination frequencies both in adjacent (Tulsieram et al. 1992) and between genetically unlinked (Beavis and Grant 1991; Fatmi et al. 1993) regions were often not correlated, demonstrating that recombination can be differentially regulated across a chromosome. But in addition, several intervals on different chromosomes were shown to have either negatively or positively correlated recombination frequencies (Fatmi et al. 1993). Together, these studies suggest that the control of variation in recombination rates might be polygenic and that some factors might control specific genetic intervals.
The heterogeneity in recombination frequencies among genetic backgrounds can be attributed to two general classes of genetic factors: cis and trans. Cis-acting elements are genetic factors that affect recombination in the region in which the factor resides. A study of recombination across several teosinte a1–sh2 intervals, each having large insertion/deletion polymorphisms (IDPs) as compared to maize, has revealed significant differences in recombination rates and distribution of recombination breakpoints across the interval caused by the action of cis factors (Yao and Schnable 2005). Similarly, a not yet molecularly characterized cis factor increases recombination in the large A188 Sh1–Bz1 interval on maize chromosome 9S (Timmermans et al. 1997).
In contrast to cis elements, trans-acting factors are genetic modifiers that are not closely linked to the interval in which they affect recombination. Trans modifiers might include factors involved in chromatin remodeling, proteins involved in the recombination machinery, and autonomous transposons. For example, the rate of crossover (CO) increases near a Mu insertion in a1 in the presence of MuDR transposase (Yandeau-Nelson et al. 2005). Also, recombination is affected on the maize chromosome 9 c1–sh1 and bz1–wx1 intervals as well as chromosome 7 by a single unidentified trans factor (Timmermans et al. 1997). The maize mapping studies described earlier established recombination rate heterogeneity among genetic backgrounds and suggest that this variation is under polygenic control (i.e., multiple trans factors). However, these studies (Tulsieram et al. 1992; Fatmi et al. 1993) were unable to distinguish between the effects of cis- and trans-acting genetic modifiers, both of which potentially influence recombination rates. Also, available data only suggest that rates of recombination differ among genetic backgrounds, but the manner in which factors can affect the distribution of recombination breakpoints across a small physical distance containing both genic and nongenic intervals is unknown.
To elucidate the effects of trans-acting modifiers specifically, these factors must be studied in the absence of polymorphic cis-acting effects. This can be achieved by studying recombination across an interval that is sequence identical in several different genetic backgrounds. The ∼140-kb a1–sh2 interval on maize chromosome 3L is an ideal system in which to study how trans factors modulate both recombination rates and the distribution of recombination breakpoints across an interval because (1) recombination events across the interval are easily identified by nonparental kernel phenotypes, (2) high-resolution mapping of recombination breakpoints is straightforward due to a significant degree of sequence polymorphism between the A1 Sh2 haplotypes and the availability of previously developed IDP markers across much of the interval (Xu et al. 1995; Yao et al. 2002), and (3) the interval is multigenic (Yao et al. 2002), which allows for the study of how trans-acting factors differentially affect recombination across genic and nongenic regions.
A sequence-identical a1∷rdt sh2 interval that had been introgressed into three unique genetic backgrounds (maize inbreds A632, Oh43, and W64A) was used to assess the extent to which trans-acting genetic modifiers that were polymorphic among the backgrounds affect meiotic recombination in the ∼0.1-cM a1–sh2 interval. This study reveals that trans-acting factors can influence both rates of recombination and distribution of recombination breakpoints within conserved hot and cold spots and can convert an average spot to a hot spot. The modifier(s) affecting recombination across this small interval on chromosome 3L does not appear to differentially affect recombination among genetic backgrounds in the same way across an interval on chromosome 1S, suggesting that at least some trans-acting factors affect specific regions of the genome.
MATERIALS AND METHODS
Genetic stocks:
The shrunken-2 (sh2) gene is located on chromosome 3 ∼0.1 cM centromere-distal to the a1 locus (Civardi et al. 1994). Mutations at these loci condition shrunken and colorless kernel phenotypes, respectively. The a1∷rdt sh2 haplotype (GenBank accession no. AF072704) was described previously (reviewed in Xu et al. 1995). The A1-LC Sh2 haplotype (GenBank accession nos. AF434192, AF347696, AF363390, X05068, and AF363391) is derived from the inbred line C (LC), a color-converted version of W22.
The a1∷rdt sh2 (A632)7, a1∷rdt sh2 (Oh43)9, and a1∷rdt sh2 (W64A)6 genetic stocks were gifts from David Glover (Purdue University). To generate these stocks, the inbreds A632, Oh43, and W64A were crossed by a line that was homozygous for the a1∷rdt sh2 haplotype. Using the a1 and sh2 mutants as markers, the a1∷rdt sh2 haplotype was backcrossed into the inbreds for seven, nine, and six generations to generate the trans stocks a1∷rdt sh2 (A632)7, a1∷rdt sh2 (Oh43)9, and a1∷rdt sh2 (W64A)6, respectively. The trans stocks were self-pollinated to generate homozygous a1∷rdt sh2 sources in each of the three genetic backgrounds.
Isolation of genetic recombinants and estimations of genetic distances:
Recombinants across a1–sh2:
To identify recombinants that resolve between a1 and sh2, each of the three inbred trans stocks was used as a male onto the inbred line C (cross 1). Recombination was measured in the three resulting trans stock/line C F1's, i.e., A632/LC, Oh43/LC, and W64A/LC, via testcrosses (cross 2) as described by Yao et al. (2002) and Yao and Schnable (2005). To control environmental effects on recombination, these testcrosses were performed during a single season (summer, 1997) in a single field.
Cross 1: A1-LC Sh2/A1-LC Sh2 (line C) × a1∷rdt sh2/a1∷rdt sh2 (trans stock)
Cross 2: A1-LC Sh2/a1∷rdt sh2 (F1 from cross 1) × a1∷rdt sh2/a1∷rdt sh2
Progeny from cross 2 segregated for parental colored round and colorless shrunken kernels. Rare kernels with nonparental phenotypes (viz., colored shrunken and colorless round) putatively carry recombination events between a1 and sh2. Because the original A632, Oh43, and W64A inbreds are recessive for r1, a gene that encodes a transcription factor that acts upstream of a1 in anthocyanin biosynthesis, colorless round (a1′Sh2) kernels could be generated due to the absence of r1 and not because of resolution of a recombination breakpoint between a1 and sh2. For this reason, only the colored shrunken (A1′sh2) recombinant class was analyzed in this study. Putative A1′sh2 recombinant kernels from each inbred background were tested, confirmed, and made homozygous as described in Xu et al. (1995). The actual number of A1′sh2 recombinants isolated from each genetic background was estimated from the frequency of confirmed recombinants out of the total number analyzed.
Recombinants across chromosome 1S:
To identify genetic intervals unlinked to the a1–sh2 interval on chromosome 3L, ∼1000 IDP genetic markers developed by the Maize Genetic Mapping Project (http://maize-mapping.plantgenomics.iastate.edu/) and genetically mapped to chromosomes 1–2, and 4–10 were surveyed. The criteria for selecting genetic intervals were that: (1) they be defined by two IDP markers that were 1–20 cM apart, (2) each IDP marker was polymorphic between line C (the A1 Sh2 haplotype used in cross 1) and the three trans stocks, and (3) the genetic interval could be assayed in each of the three trans-stock experiments. Only two intervals that met these criteria were identified; both were located on chromosome 1S. Interval 1S.1 is defined by genetic markers IDP194 and IDP254 and 1S.2 is defined by markers IDP112 and IDP643. The IDP194 and IDP254 markers were designed from MEST54-C06 (GenBank accession no. BM072826) and MEST139-B10 (GenBank accession no. BM334583), respectively, and map to positions 115.6 and 115.0 cM, respectively, on the maize IDP_body map version 4 (http://magi.plantgenomics.iastate.edu/cgi-bin/cmap). Markers IDP112 and IDP643 were designed from MEST19-B03 (GenBank accession no. BG841229) and MEST129-G08 (GenBank accession no. BM333984) and map to positions 82.8 and 84.6 cM, respectively.
Mapping populations were generated by crossing a single F1 plant from cross 1 for each of the trans stocks to the inbred B77 (cross 3).
Cross 3: B77 × A1-LC Sh2/a1∷rdt sh2 (F1 from cross 1)
Consequently, the progeny of cross 3 potentially carry alleles from the specific trans stock, line C, and B77. B77 was selected as the female parent for cross 3 because when tested with a sample of IDP markers across the genome it showed a high frequency of polymorphisms relative to line C and each of the three trans stocks (data not shown). Recombination rates across 1S.1 and 1S.2 were assayed in populations of ∼700 seedlings derived from progeny of cross 3 in each genetic background. Recombinants across each interval were identified as those individuals having nonparental combinations of genotypes at the two linked loci in each interval. Genetic distances in each background were calculated using the total number of recombinants identified in each trans stock and the total number of seedlings analyzed.
Genotypes were determined using PCR primers IDP112F, 5′-CTGTGACATGTTTGATGCCC-3′, IDP112R, 5′-GGTGATGACCACGTACAAGC-3′, and IDP112AW, 5′-CCC TGC TGA TAG TGA TAG AC-3′; IDP643F, 5′-ACCCTCATCTTCAGCAG TCG-3′ and IDP643R, 5′-GGTGAAACGGCAGTACAAGG-3′; IDP194F, 5′-GACAGATCCCTAACACTTGGG-3′ and IDP194R, 5′-AACAAGGCAACCTGTGAAGC-3′; and IDP254F, 5′-ATGTTGGTTGAGCCTCTTGG-3′ and IDP254R, 5′-GATTCAGAGAGAGTGCATGGC-3′.
Seed sterilization, germination, and DNA isolation:
Colored shrunken kernels that were homozygous for recombinant A1′sh2 haplotypes originally isolated from cross 2 and ∼700 kernels from each genetic background derived from cross 3 were germinated in 96-well flats. PCR-ready DNA was isolated as described (Dietrich et al. 2002).
Due to fungal contamination, many shrunken kernels do not germinate in the soil. For these recombinants, a second aliquot of shrunken kernels was sterilized with pure bleach (Clorox) for 1 min and then rinsed repeatedly (10–15 times) with sterile water. Sterilized kernels were plated on moistened autoclaved germination paper (Anchor Paper, St. Paul) in sterile Petri dishes, treated with 0.5% Captan 50-W fungicide (Platte Chemical, Fremont, NE), and covered with activated charcoal. Seeds were germinated at 28° for 5–7 days. Seeds were transferred to new plates, watered, and covered with fresh charcoal every 1–2 days. Tissue was collected from coleoptiles and DNA isolation was as described above.
Physically mapping recombination breakpoints:
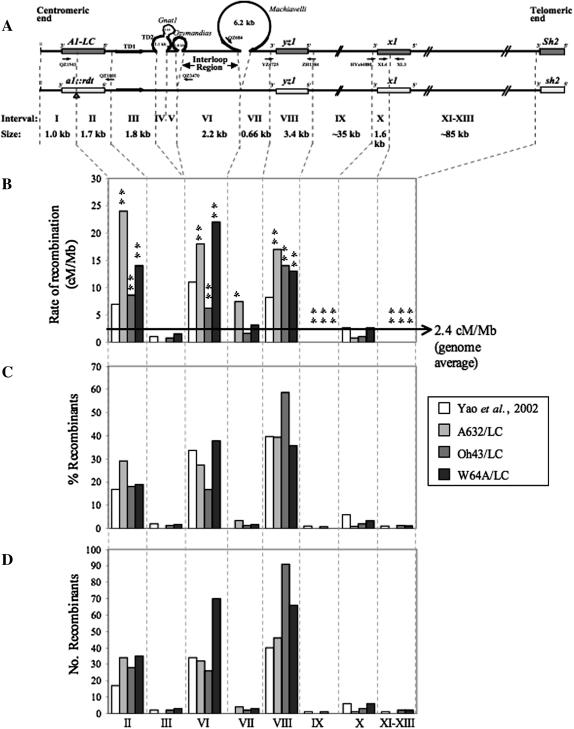
Recombination breakpoints across the a1–sh2 interval were mapped relative to eight IDP markers that had previously been identified between the A1-LC Sh2 and a1∷rdt sh2 haplotypes (YAO et al. 2002). These markers separated the a1–sh2 interval into eight informative subintervals (Figure 1A); intervals are numbered according to Yao et al. (2002). Allele-specific primers not described by Yao et al. (2002) are listed below. Primers that when paired recognize a size polymorphism between the two haplotypes are XL3, 5′-ATGAGCGGGAGCCTATG-3′, and XL4, 5′-TCAGCATCCATACCATTG-3′. Allele-specific and size polymorphism primers are shown in Figure 1A.
Figure 1.—
Distributions of recombination breakpoints across the a1–sh2 interval in the A632/LC, Oh43/LC, and W64A/LC genetic backgrounds. (A) A schematic of the A1-LC Sh2 and a1∷rdt sh2 haplotypes in which boxes represent genes. Subintervals are defined by allele-specific PCR primers designed at sequence polymorphisms that distinguish the haplotypes and are numbered according to Yao et al. (2002). Allele-specific primers are indicated by arrows above the haplotype they specifically amplify. The size of each subinterval in the line C haplotype is listed. As compared to Yao et al. (2002), subintervals XI, XII, and XIII have been combined into a single interval (XI–XIII). Because subintervals IV and V are very small and no recombinants mapped to these subintervals, they are considered not informative in this study. (B) Rates of recombination in each subinterval. The * and ** denote significant differences relative to the genomewide average recombination rate at the 0.05 and 0.01 levels, respectively. The rates of recombination in subintervals IX and XI–XIII are so low that they are not visible on this graph. Subinterval X contains the 3′ portion of x1. Even if all of the recombinants that resolved within subintervals XI–XIII resolved within x1, the rate of recombination in x1 would still be significantly lower than the genomewide average in each of the three backgrounds (P-value < 0.002). (C) The percentage of recombination breakpoints that mapped to each subinterval. (D) The number of recombination breakpoints that mapped to each subinterval. (C and D) In some cases, no breakpoints mapped to a subinterval in a given genetic background and therefore no bar is shown. (B–D) Data from Yao et al. (2002) are provided for reference.
The breakpoints associated with >85% of the confirmed recombinants from the A632 (90%; 117/130), Oh43 (87%; 155/178), and W64A (85%; 185/219) genetic backgrounds were physically mapped. It was not possible to map the remainder due to difficulties in obtaining homozygous recombinant chromosomes or in germinating shrunken kernels.
Statistical methods:
For each physical interval examined, genetic distances (in centimorgans) were compared among genetic backgrounds using χ2 homogeneity tests. For the genetic distances between a1 and sh2, the corrected number of A1′sh2 recombinants (Table 1) was doubled because the reciprocal class, a1′Sh2, was not analyzed. This analysis makes the reasonable assumption that the rates at which the two recombinant classes arise are equal.
TABLE 1.
Genetic distances across a common a1–sh2 interval in three genetic backgrounds
| Genetic background | No. isolateda | No. recoveredb | No. testedc | No. confirmedd | No. correct recombinantse | Population sizes | Genetic distances (cM)f |
|---|---|---|---|---|---|---|---|
| A632/LC | 229 | 138 | 130 | 130 | 229 | 320,718 | 0.143 ± 0.000667 |
| Oh43/LC | 323 | 188 | 188 | 178 | 306 | 758,271 | 0.081 ± 0.00033 |
| W64A/LC | 442 | 219 | 219 | 219 | 442 | 704,077 | 0.126 ± 0.000442 |
Number of putative colored shrunken recombinants isolated from cross 2 (materials and methods).
Number of putative recombinants successfully propagated.
Number of putative recombinants whose validity was tested (materials and methods).
Number of putative recombinants confirmed.
Number of correct recombinants = no. isolated × (no. confirmed/no. tested).
Genetic distance = 100 × no. correct recombinants × 2/population size. Because colorless round recombinants were not isolated, genetic distances were calculated by multiplying the number of colored shrunken recombinants by two. Standard errors were calculated according to the formula (pq/n)1/2. The genetic distances among genetic backgrounds varied significantly (P < 0.03).
Statistical methods and population size adjustments were performed as described by Yao and Schnable (2005). The actual numbers of mapped recombinants and the adjusted population sizes were used for subsequent χ2 homogeneity tests. Pearson χ2 tests were conducted as described by Yao and Schnable (2005) to compare the actual recombinant breakpoint distribution in each genetic background across a1–sh2 to (1) the expected distribution if recombination across the interval was equal to the genomewide average (2.4 cM/Mb) and (2) the expected distribution of recombinants if the rate of recombination across each subinterval was equal to the average recombination rate across the entire a1–sh2 interval. In this study, rates of recombination were compared to a genomewide average recombination rate of 2.4 cM/Mb (Table 2). This is an updated rate compared to the 2.1 cM/Mb rate (YAO et al. 2002) based on a more recent maize genetic map that consists of 5917 cM (Lee et al. 2002).
TABLE 2.
Statistical analyses of recombination across a1–sh2
| Informative subintervals | Genetic backgrounds | Comparisons to the average of a1–sh2a | Comparisons to the genome's averagea | Recombination activitiesb |
|---|---|---|---|---|
| II | A632/LC | 2.8e−156↑ (22×) | 4.2e−63↑ (10×) | Hot spot (global, local) |
| Oh43/LC | 8.8e−77↑ (14×) | 5.9e−13↑ (3.6×) | Hot spot (global, local) | |
| W64A/LC | 3.5e−100↑ (15×) | 3.0e−32↑ (5.8×) | Hot spot (global, local) | |
| III | A632/LC | 0.28 | 0.10 | Average spot |
| Oh43/LC | 0.73 | 0.094 | Average spot | |
| W64A/LC | 0.41 | 0.41 | Average spot | |
| VI | A632/LC | 6.8e−104↑ (17×) | 2.1e−40↑ (7.5×) | Hot spot (global, local) |
| Oh43/LC | 8.0e−49↑ (10×) | 6.3e−07↑ (2.6×) | Hot spot (global, local) | |
| W64A/LC | 0↑ (23×) | 3.1e−110↑ (9.2×) | Hot spot (global, local) | |
| VII | A632/LC | 7.1e−6↑ (6.9×) | 0.018↑ (3.1×) | Hot spot (global, local) |
| Oh43/LC | 0.16 | 0.55 | Average spot | |
| W64A/LC | 0.030↑ (3.3×) | 0.66 | Hot spot (local) | |
| VIII | A632/LC | 4.4e−137↑ (16×) | 3.3e−52↑ (7.1×) | Hot spot (global, local) |
| Oh43/LC | 0↑ (23×) | 9.7e−81↑ (5.8×) | Hot spot (global, local) | |
| W64A/LC | 4.9e−175↑ (14×) | 1.2e−54↑ (5.4×) | Hot spot (global, local) | |
| IX | A632/LC | 3.1e−8↓ (NA)c | 1.1e−16↓ (NA) | Cold spot (global, local) |
| Oh43/LC | 5.1e−10↓ (40×) | 1.5e−36↓ (160×) | Cold spot (global, local) | |
| W64A/LC | 3.1e−12↓ (NA) | 9.2e−29↓ (NA) | Cold spot (global, local) | |
| X | A632/LC | 0.74 | 0.23 | Average spot |
| Oh43/LC | 0.40 | 0.11 | Average spot | |
| W64A/LC | 0.011 ↑(2.7×) | 0.89 | Hot spot (local) | |
| XI–XIII | A632/LC | 4.1e−17↓ (NA) | 2.9e−38↓ (NA) | Cold spot (global, local) |
| Oh43/LC | 2.8e−21↓ (51×) | 2.1e−86↓ (200×) | Cold spot (global, local) | |
| W64A/LC | 3.9e−25↓ (59×) | 1.6e−66↓ (150×) | Cold spot (global, local) |
P-values are shown for χ2 goodness-of-fit tests used to compare the observed recombination rate in each subinterval with (a) the average rate of recombination (cM/Mb) across the a1-sh2 interval in each genetic background and (b) the average rate of recombination in the genome (2.4 cM/Mb). This genome rate of 2.4 cM/Mb is an updated estimate based on a new maize genetic map that consists of 5917 cM (Lee et al. 2002) compared to the rate previously used (Yao et al. 2002). Arrows (↑ and ↓) denote a significantly higher or lower rate of recombination (P-value < 0.05), respectively, than the rate to which it was compared. For subintervals with significant P-values, the fold difference between rates is shown in parentheses.
Hot and cold spots are intervals that have higher and lower rates of recombination, respectively, than the rates to which they are compared. Global and local spots significantly differ from the average genome and the a1–sh2 interval rates of recombination, respectively. Average spots do not significantly differ from either the global or local rates.
For genetic backgrounds that have no recombination in a given subinterval, the fold difference with the rate to which it is compared cannot be calculated. NA, not applicable.
The expected numbers of recombinants for each subinterval were calculated using the subinterval distances measured in the line C haplotype (Figure 1A). The sizes of each subinterval were previously determined via sequencing or DNA gel blot analyses (Yao et al. 2002). For the a1∷rdt sh2 haplotype, interval sizes were determined using the genomic sequence from a1 to yz1 of this haplotype (GenBank accession no. AF072704) for subintervals II (1.7 kb), III (1.34 kb), IV (0.4 kb), V (80 bp), VI (2.2 kb), VII (620 bp), and VIII (3.7 kb). To test if the use of subinterval sizes from line C affects comparisons of recombination rates to the genomewide and to the a1–sh2 interval average rates of recombination, the expected numbers of recombinants and the corresponding statistical analyses were also conducted using subinterval sizes from the a1∷rdt sh2 haplotype. Because the significance or nonsignificance for each subinterval was not altered by which haplotype was used to determine sizes of each subinterval, only statistical analyses using subinterval sizes measured in line C are presented.
To investigate which subintervals contribute to the different breakpoint distributions among the backgrounds, an exploratory statistical analysis was conducted. Freeman–Halton tests as described in Yao and Schnable (2005) were conducted in which each subinterval was successively removed (and then replaced) from the analysis. If the recombination rates in a subinterval were contributing to differences in breakpoint distributions across the entire interval, then upon removal of this subinterval (or group of subintervals) the backgrounds should no longer differ significantly.
RESULTS
The genetic distance between a1 and sh2 and the distribution of recombination breakpoints varies among genetic backgrounds:
A common a1∷rdt sh2 interval was introgressed into the inbred lines A632, Oh43, and W64A for multiple generations (materials and methods). Each of the resulting trans stocks contains a sequence-identical a1∷rdt sh2 interval on chromosome 3L, while the bulk of its genome is expected to be derived from the recurrent parent: i.e., A632, Oh43, and W64A. These stocks are ideal for studying the effects of trans-acting genetic modifiers on recombination across the a1–sh2 interval because the presence of an identical a1–sh2 interval in each of the three backgrounds ensures that the cis factors that affect recombination within the a1–sh2 interval are identical in each of the inbred backgrounds. To analyze the effects of genetic background on recombination across the a1–sh2 interval, each of the trans stocks was crossed to the inbred line C. Because each of the resulting F1's is identically heterozygous at the a1–sh2 interval (A1-LC Sh2/a1∷rdt sh2), the only modifiers of recombination that are expected to differ among these three F1's should be outside of the a1–sh2 interval. Hence, any observed differences in meiotic recombination across the a1–sh2 interval among the three genetic backgrounds can be attributed specifically to trans factors.
Meiotic recombination events that resolved between a1 and sh2 and yielded colored shrunken kernels (A1′sh2) were identified from cross 2 (materials and methods). Colorless round (a1′Sh2) kernels were not analyzed for reasons described in materials and methods. Each of the genetic backgrounds exhibited a statistically different rate of recombination across the a1–sh2 interval and these rates varied almost twofold (Table 1).
Rates of recombination across the a1–sh2 interval differ significantly due to the action of trans-acting modifier(s). To determine whether these trans-acting factors also affect the distributions of meiotic recombination breakpoints across genic and nongenic regions in this interval, recombination breakpoints isolated from each genetic background were mapped relative to sequence polymorphisms between the A1-LC Sh2 and a1∷rdt sh2 haplotypes (materials and methods). Because the a1–sh2 intervals present in each of the genetic backgrounds were identical, breakpoints could be mapped relative to the same sequence polymorphisms, thereby facilitating comparisons among genetic backgrounds.
In each background recombination breakpoints were not randomly distributed across the a1–sh2 interval (P < 4.0e−39). Indeed, >95% of the recombinants isolated from each background mapped within the a1–yz1 interval (subintervals II–VIII; Figure 1, C and D) that composes only ∼10% of the physical length of a1–sh2. The majority of the remaining recombinants resolved at the 3′ end of x1 (subinterval X; Figure 1, C and D). These observations are consistent with previously described distributions of recombination breakpoints across the a1–sh2 interval (Yao et al. 2002; Yao and Schnable 2005).
The distributions of recombination breakpoints across the eight a1–sh2 subintervals defined by sequence polymorphisms between the A1-LC Sh2 and a1∷rdt sh2 haplotypes (Figure 1) were compared among genetic backgrounds (materials and methods). The distribution of breakpoints in the Oh43/LC background differed significantly from the distributions in both the A632/LC and W64A/LC backgrounds (Table 3). This finding indicates that trans-acting factors that are polymorphic in the Oh43/LC background as compared to the other two genetic backgrounds affect not only rates of recombination across the entire a1–sh2 interval but also the distribution of recombination breakpoints within this interval. Using the exploratory statistical analysis described in materials and methods it was possible to establish that in the Oh43/LC background the interloop (subinterval VI) and the yz1 gene (subinterval VIII) were the most significant contributors to the differences in the distribution of breakpoints across the a1–sh2 interval.
TABLE 3.
Statistical analysis of rates of recombination among genetic backgrounds
| Comparisons among genetic backgroundsa
|
|||
|---|---|---|---|
| Subintervalsb | A632/LC vs. Oh43/LC | A632/LC vs. W64A/LC | Oh43/LC vs. W64A/LC |
| II | A > O (1.8e−5) | A > W (0.019) | O ≃ W (0.052) |
| III | A ≃ O (0.36) | A ≃ W (0.20) | O ≃ W (0.45) |
| VI | A > O (2.6e−5) | A ≃ W (0.36) | W > O (5.4e−9) |
| VII | A > O (0.049) | A ≃ W (0.24) | O ≃ W (0.45) |
| VIII | A ≃ O (0.35) | A ≃ W (0.24) | O ≃ W (0.73) |
| IX | A ≃ O (0.51) | A ≃ W (ND)c | O ≃ W (0.38) |
| X | A ≃ O (0.83) | A ≃ W (0.24) | O ≃ W (0.16) |
| XI–XIII | A ≃ O (0.36) | A ≃ W (0.29) | O ≃ W (0.79) |
For each subinterval, statistically significant relationships among rates of recombination in the A632/LC (A), Oh43/LC (O), and W64A/LC (W) genetic backgrounds are shown. The P values associated with χ2 homogeneity tests are provided in parenthesis. In a given subinterval, rates of recombination are indicated as being not distinguishable (≃) if P > 0.05.
The distribution of breakpoints across the entire a1–sh2 interval in the Oh43/LC background differed significantly from the distributions in both the A632/LC (P = 0.013) and W64A/LC (P = 0.0008) backgrounds.
Because no recombinant breakpoints mapped to this interval in either background, rates of recombination could not be compared. ND, not determined.
Recombination hot and cold spots are mostly conserved among genetic backgrounds:
As defined by Yao and Schnable (2005) “global” recombination hot and cold spots are regions that exhibit rates of recombination that are either higher or lower, respectively, than the genomewide average rate of recombination (2.4 cM/Mb). “Local” recombination hot and cold spots are regions that have rates of recombination that are higher and lower, respectively, than the average rate of recombination across the entire a1–sh2 interval in the genetic background being examined. In each of the genetic backgrounds examined in this study the average recombination rate across the a1–sh2 interval (i.e., the local average) is lower than the genomewide average rate of recombination. “Average” spots exhibit rates of recombination that do not differ significantly from the genomewide average.
All three recombination hot spots previously identified by Yao et al. (2002) in the same A1-LC Sh2/a1∷rdt sh2 heterozygote used in this study but in another genetic background [the a1 gene (subinterval II), the yz1 gene (subinterval VIII), and the apparently nongenic interloop (subinterval VI)] are both global and local recombination hot spots in all three genetic backgrounds (Figure 1B; Table 2). Similarly, two intergenic regions and one gene (x1) identified by Yao et al. (2002) as recombination cold spots (subintervals IX, X, and XI–XIII, Figure 1B; Table 2), are also cold spots in all three genetic backgrounds analyzed in this study.
Trans-acting factors affect rates of recombination within hot spots:
As discussed above, the a1 (subinterval II) and yz1 (subinterval VIII) genes and the nongenic interloop (subinterval VI) are conserved global recombination hot spots in each of the three genetic backgrounds (Table 2). If trans-acting modifiers affect each subinterval equally, the rates of recombination in each subinterval should be highest and lowest in the genetic backgrounds that have the largest (i.e., A632/LC) and smallest genetic distances (i.e., Oh43/LC), respectively, across the entire a1–sh2 interval (Table 1). Because this is often not true (Figure 1; Table 3), we conclude that trans-acting factor(s) that are polymorphic among the genetic backgrounds are differentially affecting rates of recombination within the conserved hot spots.
The most notable example of differential control of recombination among genetic backgrounds occurs in yz1 (subinterval VIII; Figure 1A). Among the three backgrounds, the rate of recombination across a1–sh2 is lowest in Oh43/LC (0.607 cM/Mb) and consistent with this low rate most subintervals show the lowest rate of recombination in this background (Figure 1B). However, more than half the recombination events isolated in the Oh43/LC background resolve in yz1 (Figure 1, C and D) and the corresponding rate of recombination is 23-fold higher than the rate across a1–sh2 (P-value ∼0; Table 2) in this background, making yz1 the most recombinationally active region in Oh43/LC. In contrast, only ∼40% of the breakpoints associated with recombinants isolated in the A632/LC and W64A/LC genetic backgrounds resolve within yz1 (Figure 1C). The recombination rate in yz1 is approximately seven-, approximately six-, and approximately fivefold hotter than the genomewide rate (Table 2) in the A632/LC, Oh43/LC, and W64A/LC genetic backgrounds, respectively, with no significant differences in recombination rates among the backgrounds (Figure 1B; Table 3). In each of the backgrounds, the rate of recombination in each of the subintervals of yz1 did not differ significantly from the rate of recombination for the entire gene (P-value > 0.05) and, further, the recombination rates in each of the subintervals of yz1 did not differ significantly among the backgrounds (data not shown).
Trans-acting modifiers can convert a region with an average recombination rate into a hot spot:
Although their levels of recombination activity vary, recombination hot and cold spots are generally conserved among the three genetic backgrounds. The exception, however, is subinterval VII, a nongenic region proximal to yz1. While no recombination was observed in this interval in another genetic background (Yao et al. 2002), in the A632/LC genetic background subinterval VII is approximately threefold more recombinationally active than the genomewide average (7.4 cM/Mb; P-value = 0.018; Table 2; Figure 1B) and approximately sevenfold hotter than the average rate across a1–sh2 in the A632/LC background (P-value = 7.1e−06; Table 2). In the W64A/LC background, however, subinterval VII is an average spot when compared to the genomewide average, but locally it is approximately three times more recombinationally active than the average rate across a1–sh2 (P-value = 0.03; Table 2). In Oh43/LC, subinterval VII is an average spot both globally and locally (Figure 1B; Table 2). In summary, this interval is more recombinationally active in each of the genetic backgrounds in this study compared to the sequence-identical interval in the line C background (Yao et al. 2002).
Genetic distances of chromosomal intervals on chromosome 1:
The genetic distances between a1 and sh2 differ significantly among the three genetic backgrounds tested in this study. To determine whether trans-acting modifiers that affect recombination across the a1–sh2 interval of chromosome 3 are global or region-specific modifiers of recombination, recombination was assayed in ∼700 progeny of cross 3 in two intervals on chromosome 1 (intervals 1S.1 and 1S.2) in the three genetic backgrounds. Intervals 1S.1 and 1S.2 are separated by 30.4 cM on chromosome 1S in the maize IBM mapping population (IBM_IDP+MMP_body map version 4.0; http://magi.plantgenomics.iastate.edu/cgi-bin/cmap). In both intervals, the genetic distances were highest in the A632/LC background consistent with results in a1–sh2 and, unlike in the a1–sh2 interval, lowest in W64A/LC (Table 4). Therefore, the relationships between genetic distances among the genetic backgrounds differ between the intervals examined on chromosome 1S and the a1–sh2 interval on 3L.
TABLE 4.
Genetic distances across two genetic intervals unlinked to a1–sh2 in three genetic backgrounds
| 1S.1 interval
|
1S.2 interval
|
|||||
|---|---|---|---|---|---|---|
| Genetic background | No. recombinantsa | Population sizes | Genetic distances (cM) | No. recombinantsa | Population sizes | Genetic distances (cM) |
| A632/LC | 15 | 704 | 2.13 ± 0.06b | 5 | 729 | 0.69 ± 0.03c |
| Oh43/LC | 15 | 717 | 2.09 ± 0.05 | 1 | 705 | 0.14 ± 0.01 |
| W64A/LC | 6 | 678 | 0.89 ± 0.04b | 0 | 678 | —c |
Total number from the two expected classes of recombinants.
The 2.4-fold larger genetic distance in A632 as compared to W64A is weakly supported (P-value = 0.058).
The genetic distance in A632 is higher than in W64A (P-value = 0.03).
DISCUSSION
Trans-acting genetic modifiers affect rates of recombination in the a1–sh2 interval:
The heterogeneity in rates of recombination observed in previous studies among maize genomes (Beavis and Grant 1991; Tulsieram et al. 1992; Fatmi et al. 1993; Williams et al. 1995) could be due to cis- and/or trans-acting genetic modifiers. In contrast, the genetic stocks used in this study ensured that elements that affect recombination in cis are the same in each background. This is, therefore, the first molecular analysis in plants, animals, or yeast for which differences in recombination across a multigenic interval can be directly attributed to trans-acting modifiers that differ among genetic backgrounds.
To enhance our ability to identify inbred backgrounds that exhibit polymorphic trans-acting factors, recombination was assayed in three genetically distinct maize inbreds. A632 is a Stiff Stalk (SS) Reid Yellow Dent inbred, while the Non-Stiff Stalk (NSS) inbreds Oh43 and W64A (http://www.maizegenetics.net/index.php?page=germplasm/lines.html) are members of the Lancaster Sure Crop and the Hy:T8:Wf9 subgrouping (Liu et al. 2003), respectively. A632 and Oh43 are among the six inbreds that contribute to ∼70% of all hybrids in the United States (Nass and Paterniani 2000).
Rates of recombination between a1 and sh2 varied up to approximately twofold among these three genetic backgrounds (Table 1). This establishes that trans-acting modifiers can affect rates of recombination and, not surprisingly, the effects of these factors vary among the inbreds within different heterotic groups.
Trans effects on a multigenic interval:
Unlike previous studies of trans-acting effects that used large genetic (and physical) intervals with uncharacterized molecular structures (Timmermans et al. 1997) or studies that compare genetic sizes among different genetic maps (Beavis and Grant 1991; Tulsieram et al. 1992; Fatmi et al. 1993; Williams et al. 1995), analysis of the well-characterized multigenic a1–sh2 interval (Figure 1A) allowed for observation of the effects of trans-acting modifiers in defined genic and intergenic regions.
Although recombination hot and cold spots are largely conserved among the genetic backgrounds (Figure 1B; Table 2), the rates within recombination hot spots are affected by trans-acting factors that are polymorphic among the backgrounds (Table 3). For example, in the Oh43/LC genetic background trans-acting factor(s) have seemingly redirected recombination events that might normally resolve within the nongenic interloop (subinterval VI) to yz1 (subinterval VIII) such that the resulting rate of recombination in yz1 does not differ significantly from the rates in the A632/LC and W64A/LC genetic backgrounds. In each of the conserved recombination hot spots, the relationships between recombination rates among the backgrounds vary (Table 3). This demonstrates that the trans-acting modifier(s) act by increasing or decreasing the rates of recombination within particular hot spots.
Trans-acting modifier(s) affecting recombination in this study are affecting not only the overall rate of recombination between a1 and sh2 but also the distributions of recombinants across this interval. This observation is in direct contrast to the trans-acting autonomous transposon MuDR that increased the rate of recombination at an a1 allele containing a Mu1 insertion but did not change the distribution of recombinant breakpoints compared to patterns observed in other a1 alleles (Yandeau-Nelson et al. 2005). This suggests, not surprisingly, that different types of trans-acting modifiers will affect recombination in different ways.
Trans-acting modifier(s) can act as hot and cold switches in a region of lower sequence similarity:
Although there are exceptions (Yao et al. 2002), in general plant genes are recombination hot spots and nongenic regions are cold spots (reviewed in Puchta and Hohn 1996; Schnable et al. 1998). Indeed, nongenic subintervals IX and XI–XIII are both local and global cold spots (Figure 1B; Table 2) in each of the three genetic backgrounds. In striking contrast, the nongenic subinterval VII is both a global and local hot spot in A632/LC, providing a second example of an apparently nongenic recombination hot spot. Because subinterval VII is only a local hot spot in the W64A/LC and an average spot in the Oh43/LC genetic backgrounds (Table 2), trans-acting modifiers not only are able to alter rates of recombination within hot spots but also can switch hot spots on and off.
Recombination hot spots have been shown to be “dialed down” by sequence heterology (cis-acting elements) in many organisms, including within the maize a1–sh2 interval (Yao and Schnable 2005), by a mechanism involving mismatch repair proteins (reviewed in Modrich and Lahue 1996; Borts et al. 2000; Evans and Alani 2000; Schofield and Hsieh 2003). At the maize bz1 locus, as little as 1.5% sequence divergence reduces rates of recombination twofold (Dooner 2002). In subinterval VII, the level of sequence heterology between the line C haplotype and the a1∷rdt-sh2 haplotype present in each genetic background is 3.1%. Even with this relatively high degree of heterology, the rate of recombination in A632 is approximately threefold higher than the average rate of recombination in the genome (7.4 vs. 2.4 cM/Mb; Table 2). This demonstrates that trans-acting modifiers in the A632/LC and W64A/LC (where recombination in this subinterval is also increased but to a lesser extent) backgrounds can overcome the suppression of recombination that often occurs between heterologous sequences. Further, this establishes that high sequence identity, a cis-acting factor, is not the only criterion that dictates whether a region is a recombination hot or cold spot.
Comparison of cis- and trans-acting effects on recombination:
Both cis- and trans-acting factors can potentially affect the rate of recombination in a given interval. Only by studying each type of factor in the absence of the other (e.g., trans in the absence of polymorphic cis effects) is it possible to assess how these two categories similarly and differently affect recombination. To that end, the a1–sh2 interval in maize is the first region for which the effects of trans-acting modifiers on recombination (this study) can be compared to the effects of cis-acting elements on recombination that were characterized among different a1–sh2 teosinte intervals introgressed into maize (Yao and Schnable 2005).
Both the cis- and trans-acting modifiers in the two studies affect recombination similarly in that in both studies most of the recombination breakpoints resolve in the proximal ∼10% of the a1–sh2 interval. Together these studies suggest that although cis and trans effects on recombination in the a1–sh2 interval are, in general, similar, the cis elements represented in the teosinte a1–sh2 intervals affect recombination to a greater extent than do the trans-acting modifiers that are polymorphic in the A632/LC, Oh43/LC, and W64A/LC genetic backgrounds. For example, in a subinterval containing the interloop (subinterval VI in this study), which is a recombination hot spot in each of the trans stocks, cis elements act to make this region an average, cold, and hot spot in the three different intervals studied. Such comparisons are limited, however, because the trans-acting modifiers in the cis studies most likely differ from those in the inbred backgrounds in this study.
Do the trans-acting factors act globally or on specific genetic intervals?
Trans-acting modifiers of meiotic recombination (reviewed in Wahls 1998) have been divided into two categories: those that affect recombination across the entire genome (i.e., global modifiers) and those that act only on specific (i.e., region-specific modifiers) genetic intervals (Catcheside 1977). Global trans-acting modifiers are most likely proteins intimately involved in the mechanisms of recombination (reviewed in van den Bosch et al. 2002). For example, in yeast rates of meiotic CO are globally reduced in mutants of DNA polymerase δ (Maloisel et al. 2004). In maize, desynaptic (Ji et al. 1999), which is involved in crossover control, affects recombination across the genome (Bass et al. 2003).
To test whether or not the trans-acting factors that affect the a1–sh2 interval are global modifiers, in each genetic background recombination was assayed in two intervals genetically unlinked to a1–sh2. In both intervals on chromosome 1, the relationships among genetic distances were the same. However, these relationships differed from that seen in the a1–sh2 interval (compare Tables 1 and 4). This could be due to the same trans-acting modifier(s) within a genetic background differentially affecting recombination in chromosomes 1S and 3L. Alternatively, different region-specific modifiers could be acting on at least portions of each of the two chromosomes. In addition, because the 1S.1 and 1S.2 intervals are most likely not sequence-identical among the backgrounds we cannot rule out the possibility that a combination of both trans- and cis-acting elements is affecting recombination in these intervals. Hence, these results suggest, but do not prove, that the trans-acting modifiers detected in this study do not have global effects and are instead region specific.
Region-specific trans-acting modifiers have been characterized in Schizosaccharomyces pombe (De Veaux et al. 1992; De Veaux and Smith 1994; Li et al. 1997; Krawchuk et al. 1999; Pryce et al. 2005) and Saccharomyces cerevisiae (Rockmill and Roeder 1990). The effects of chromatin organization on DSB formation and subsequent meiotic recombination repair have been extensively demonstrated in many organisms (reviewed in Lichten 2001; Petes 2001), including plant species (reviewed in Schuermann et al. 2005). In S. pombe, rec10, a protein involved in the formation of lateral elements, is required for the activation of some but not all M26-containing recombination hot spots (Pryce et al. 2005). In this case, higher-order chromatin structure affects recombination at a known hot spot. Trans-acting modifiers involved in chromatin organization or remodeling might be responsible for the transformation of the nongenic subinterval VII from an average spot in Oh43/LC to a hot spot in the A632/LC genetic background by opening the chromatin and allowing access by the recombination machinery. If so, trans-acting modifiers of chromatin most likely act in context of local structures (e.g., sequence motifs). Alternatively, genotype-specific patterns of crossover interference (reviewed in van Veen and Hawley 2003; Hillers 2004; Copenhaver 2005) could differently affect the distributions of recombination hot spots across a chromosome among genetic backgrounds.
Region-specific trans-acting modifiers could also include proteins (e.g., transcription factors) for which binding to specific cis-acting elements [e.g., unique sequence motifs (Wahls and Smith 1994; Kon et al. 1997; Pryce et al. 2005) or promoters] is necessary for activation of recombination hot spots as seen in α-hot spots in yeast (reviewed in Lichten and Goldman 1995; Petes 2001). Such interactions between cis-acting elements and trans-acting modifiers, if they occur in a1–sh2, must be due to differences among the trans-acting modifiers (e.g., binding specificity or affinity for the cis element) because the a1–sh2 intervals are sequence-identical among the genetic backgrounds.
Acknowledgments
We thank Dan Nettleton (Iowa State University) for advice regarding statistical analyses, Heather Smith for the isolation and purification of recombinant alleles, undergraduate students Brian Reinertson, Timothy Dunham, John Tenhunfeld, and Lakeysha Toomer for technical assistance, and David Glover (Purdue University) for the gift of the trans stocks. This research was supported in part by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education and Extension Service, grant numbers 9701407 and 9901579 to P.S.S. and B.J.N. and 0101869 and 0300940 to P.S.S., and supported by Hatch Act and State of Iowa funds.
References
- Anderson, L., A. Lai, S. M. Stack, C. Rizzon and B. S. Grant, 2006. Uneven distribution of expressed sequence tag loci on maize pachytene chromosomes. Genome Res. 16: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H. W., S. J. Bordoli and E. M. Foss, 2003. The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L) cause distinct telomere-misplacement phenotypes during meiotic prophase. J. Exp. Bot. 54: 39–46. [DOI] [PubMed] [Google Scholar]
- Beavis, W. D., and D. Grant, 1991. A linkage map based on information from four F2 populations of maize (Zea mays L.). Theor. Appl. Genet. 82: 636–644. [DOI] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Catcheside, D. G., 1977. The Genetics of Recombination. Edward Arnold Limited, London.
- Civardi, L., Y. Xia, K. J. Edwards, P. S. Schnable and B. J. Nikolau, 1994. The relationship between genetic and physical distances in the cloned a1-sh2 interval of the Zea mays L. genome. Proc. Natl. Acad. Sci. USA 91: 8268–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., 2005. Plant genetics: when not to interfere. Curr. Biol. 15: R290–R291. [DOI] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. a Cytogenetically based physical maps of the group-2 chromosomes of wheat. Theor. Appl. Genet. 91: 568–573. [DOI] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. b Cytogenetically based physical maps of the group-3 chromosomes of wheat. Theor. Appl. Genet. 91: 780–782. [DOI] [PubMed] [Google Scholar]
- DeVeaux, L. C., and G. R. Smith, 1994. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 8: 203–210. [DOI] [PubMed] [Google Scholar]
- DeVeaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. R., F. Cui, M. L. Packila, J. Li, D. A. Ashlock et al., 2002. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., 2002. Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martinez-Ferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston, W. B., M. Alleman and J. L. Kermicle, 1995. Molecular organization and germinal instability of R-stippled maize. Genetics 141: 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E., and E. Alani, 2000. Roles for mismatch repair factors in regulating genetic recombination. Mol. Cell. Biol. 20: 7839–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatmi, A., C. G. Poneleit and T. W. Pfeiffer, 1993. Variability of recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L.). Theor. Appl. Genet. 86: 859–866. [DOI] [PubMed] [Google Scholar]
- Fu, H., Z. Zheng and H. K. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill and T. R. Endo, 1993. A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102: 374–381. [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and E. V. Boyko, 1996. a Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143: 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and T. Taylor, 1996. b Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., 2004. Crossover interference. Curr. Biol. 14: R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., T. R. Endo, K. S. Gill and B. S. Gill, 1994. Comparison of genetic and physical maps of group 7 chromosomes from Triticum aestivum L. Mol. Gen. Genet. 245: 644–653. [DOI] [PubMed] [Google Scholar]
- Ji, Y., D. M. Stelly, M. De Donato, M. M. Goodman and C. G. Williams, 1999. A candidate recombination modifier gene for Zea mays L. Genetics 151: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa, N., K. Nagaki and H. Tsujimoto, 2002. Tetrad-FISH analysis reveals recombination suppression by interstitial heterochromatin sequences in rye (Secale cereale). Mol. Genet. Genomics 267: 10–15. [DOI] [PubMed] [Google Scholar]
- Koehler, K. E., J. P. Cherry, A. Lynn, P. A. Hunt and T. J. Hassold, 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith and W. P. Wahls, 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94: 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk, M. D., L. C. DeVeaux and W. P. Wahls, 1999. Meiotic chromosome dynamics dependent upon the rec8+, rec10+ and rec11+ genes of the fission yeast Schizosaccharomyces pombe. Genetics 153: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie, E. J., and G. S. Roeder, 1986. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114: 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Li, Y. F., M. Numata, W. P. Wahls and G. R. Smith, 1997. Region-specific meiotic recombination in Schizosaccharomyces pombe: the rec11 gene. Mol. Microbiol. 23: 869–878. [DOI] [PubMed] [Google Scholar]
- Lichten, M., 2001. Meiotic recombination: breaking the genome to save it. Curr Biol 11: R253–R256. [DOI] [PubMed] [Google Scholar]
- Lichten, M., and A. S. Goldman, 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Liu, K., M. Goodman, S. Muse, J. S. Smith, E. Buckler et al., 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani, M. M., and H. F. Willard, 1998. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 8: 100–110. [DOI] [PubMed] [Google Scholar]
- Maloisel, L., J. Bhargava and G. S. Roeder, 2004. A role for DNA polymerase δ in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167: 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelson-Young, L., T. R. Endo and B. S. Gill, 1995. A cytogenetic ladder-map of wheat homoeologous group-4 chromosomes. Theor. Appl. Genet. 90: 1007–1011. [DOI] [PubMed] [Google Scholar]
- Modrich, P., and R. Lahue, 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., 2002. Variation in recombination rate across the genome: evidence and implications. Curr. Opin. Genet. Dev. 12: 657–663. [DOI] [PubMed] [Google Scholar]
- Nass, L. L., and E. Paterniani, 2000. Pre-breeding: A link between genetic resources and maize breeding. Scientia Agricola 57: 581–587. [Google Scholar]
- Patterson, G. I., K. M. Kubo, T. Shroyer and V. L. Chandler, 1995. Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics 140: 1389–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Pryce, D. W., A. Lorenz, J. B. Smirnova, J. Loidl and R. J. McFarlane, 2005. Differential activation of M26-containing meiotic recombination hot spots in Schizosaccharomyces pombe. Genetics 170: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H., and B. Hohn, 1996. From centiMorgans to base pairs: homologous recombination in plants. Trends Genet. 1: 340–348. [Google Scholar]
- Puechberty, J., A. M. Laurent, S. Gimenez, A. Billault, M. E. Brun-Laurent et al., 1999. Genetic and physical analyses of the centromeric and pericentromeric regions of human chromosome 5: recombination across 5cen. Genomics 56: 274–287. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran, E., S. J. Armstrong, J. L. Santos, F. C. Franklin and G. H. Jones, 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P. S., A. P. Hsia and B. J. Nikolau, 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1: 123–129. [DOI] [PubMed] [Google Scholar]
- Schofield, M. J., and P. Hsieh, 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57: 579–608. [DOI] [PubMed] [Google Scholar]
- Schuermann, D., J. Molinier, O. Fritsch and B. Hohn, 2005. The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181. [DOI] [PubMed] [Google Scholar]
- Simchen, G., and J. Stamberg, 1969. Fine and coarse controls of genetic recombination. Nature 222: 329–332. [DOI] [PubMed] [Google Scholar]
- Sun, H., D. Treco and J. W. Szostak, 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Timmermans, M. C., O. P. Das, J. M. Bradeen and J. Messing, 1997. Region-specific cis- and trans-acting factors contribute to genetic variability in meiotic recombination in maize. Genetics 146: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulsieram, L., W. A. Compton, R. Morris, M. Thomas-Compton and K. Eskridge, 1992. Analysis of genetic recombination in maize populations using molecular markers. Theor Appl Genet 84: 65–72. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., P. H. Lohman and A. Pastink, 2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383: 873–892. [DOI] [PubMed] [Google Scholar]
- van Veen, J.E., and R.S. Hawley, 2003. Meiosis: when even two is a crowd. Curr. Biol. 13: R831–R833. [DOI] [PubMed] [Google Scholar]
- Wahls, W. P., 1998. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37: 37–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls, W. P., and G. R. Smith, 1994. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 8: 1693–1702. [DOI] [PubMed] [Google Scholar]
- Werner, J. E., T. R. Endo and B. S. Gill, 1992. Toward a cytogenetically based physical map of the wheat genome. Proc. Natl. Acad. Sci. USA 89: 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C. G., M. M. Goodman and C. W. Stuber, 1995. Comparative recombination distances among Zea mays L. inbreds, wide crosses and interspecific hybrids. Genetics 141: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., A. P. Hsia, L. Zhang, B. J. Nikolau and P. S. Schnable, 1995. Meiotic recombination break points resolve at high rates at the 5′ end of a maize coding sequence. Plant Cell 7: 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson, M. D., Q. Zhou, H. Yao, X. Xu, B. J. Nikolau et al., 2005. MuDR transposase increases the frequency of meiotic crossovers in the vicinity of a Mu insertion in the maize a1 gene. Genetics 169: 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., and P. S. Schnable, 2005. Cis-effects on meiotic recombination across distinct a1-sh2 intervals in a common Zea genetic background. Genetics 170: 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]