Abstract
In goat milk the most abundant proteins are the casein genes, CSN1S1, CSN2, CSN1S2, and CSN3. Mutations have been identified within these genes affecting the level of gene expression, and effects on milk production traits have been reported. The aim of this study was to detect polymorphisms (SNPs) in the casein genes of Norwegian goats, resolve haplotype structures within the loci, and assess the effect of these haplotypes on milk production traits. Four hundred thirty-six Norwegian bucks were genotyped for 39 polymorphic sites across the four loci. The numbers of unique haplotypes present in each locus were 10, 6, 4, and 8 for CSN1S1, CSN2, CSN1S2, and CSN3, respectively. The effects of the CSN1S1 haplotypes on protein percentage and fat kilograms were significant, as were the effects of CSN3 haplotypes on fat percentage and protein percentage. A deletion in exon 12 of CSN1S1, unique to the Norwegian goat population, explained the effects of CSN1S1 haplotypes on fat kilograms, but not protein percentage. Investigation of linkage disequilibrium between all possible pairs of SNPs revealed higher levels of linkage disequilbrium for SNP pairs within casein loci than for SNP pairs between casein loci, likely reflecting low levels of intragenic recombination. Further, there was evidence for a site of preferential recombination between CSN2 and CSN1S2. The value of the haplotypes for haplotype-assisted selection (HAS) is discussed.
GOAT milk is a valuable source of protein in many countries, including a large number of African and Asian countries and European countries such as Norway, France, and Italy. The most abundant proteins in goat milk, as in other milks, are the caseins. The four caseins expressed in goat milk, αS1-, β-, αS2-, and κ-casein, are coded by the loci CSN1S1, CSN2, CSN1S2, and CSN3, respectively, located within a 250-kb segment of caprine chromosome 6 (Martin et al. 2002a).
A number of genetic variants of the casein genes that affect milk production traits have been described. CSN1S1 is the most complex and is highly polymorphic (Martin et al. 2002b). As many as 18 different alleles have been identified for this gene (Leroux et al. 2003; Devold 2004), including alleles with “strong” effects, “medium” effects, and “weak” effects and “null” alleles associated with no synthesis of the protein (Leroux et al. 1990; Neveu et al. 2002). Goats carrying the strong variants have been reported to produce milk with a significantly higher casein, total protein, and fat content than goats carrying the weak variants (Manfredi et al. 1993; Remeuf 1993; Barbieri et al. 1995). However, reports of the effects of these alleles on milk yield are somewhat contradictory; for example, Mahe et al. (1994) reported that genetic variants of CSN1S1 had no effects on milk yield. A unique deletion in CSN1S1 (here termed the 04 allele) has been reported in Norwegian goats and is present in the Norwegian population at high frequency [0.86 (Ådnøy et al. 2003)]. In most other European breeds the null alleles are at low frequency.
At least five variants of CSN2 are described, including a null allele associated with no protein expression (Mahe and Grosclaude 1993; Galliano et al. 2004). Likewise at least five different alleles have been described for CSN1S2 (Recio et al. 1997; Martin et al. 1999; Erhardt et al. 2002). Prinzenberg et al. (2005) recently described alleles of CSN3, including two new alleles and a new nomenclature for the 16 previously described alleles. The influence of CSN3 on milk production traits still remains to be evaluated.
While the mutations described above can dramatically affect the levels of expression of the genes they are found in, effects on total protein production are in some cases less pronounced. One hypothesis is that when expression of one casein gene is downregulated, the others can be upregulated to compensate (Leroux et al. 2003). Bovenhuis et al. (1992) suggested that the conflicting results of the effects of mutations in casein genes for cattle at least might be due to linkage between mutations in different caseins, as well as the different statistical models used in the analyses. They proposed a multigene model as an alternative to single-gene models. As mutations with effects on quantitative traits such as milk production can occur in exons, introns (e.g., Andersson and Georges 2004), promoters, and other regulatory sequences (e.g., Hoogendoorn et al. 2003), it is possible that the functional mutation(s) will not be within the set of mutations (such as single-nucleotide polymorphisms, SNPs) genotyped in the data set. However, the functional mutation(s) will have occurred in an ancestral chromosome segment, copies of which persist in the current generation of animals. These identical-by-descent (IBD) chromosome segments can be identified in the current population by unique haplotypes of SNP or marker alleles. This suggests investigating the effects of haplotypes of the mutation alleles across the casein loci on protein production as an alternative to considering the genes in isolation.
In this article, we investigate effects of haplotypes of polymorphisms in the casein genes on production traits in the Norwegian dairy goat population. We first sequenced fragments of all four of the casein loci in seven Norwegian dairy bucks to detect additional polymorphisms in the casein loci. We also sequenced fragments of the promoters for these genes. We then genotyped the 436 bucks representative of the commercial population, for these new SNPs as well as previously reported mutations, and constructed haplotypes within each casein locus. The pattern of linkage disequilibrium between the haplotypes suggested preferential recombination between CSN2 and CSN1S2; this was supported by analysis of linkage disequilibrium between individual markers. The haplotypes were found to have large effects on the milk production traits. The possibility of using these haplotypes in haplotype-assisted selection (HAS) is discussed.
METHODS
SNP detection and genotyping:
Among polymorphisms (six SNPs and two deletions) selected from the literature six turned out to be nonpolymorphic in the Norwegian goat population (see supplemental information at http://www.genetics.org/supplemental/). Primers for resequencing of casein loci were designed for the promoters, selected exons, and introns of CSN1S1, CSN2, and CSN3, including exon 16 of CSN1S2 and exon 7 of CSN2. Seven Norwegian goats, known to be variable in CSN1S1 from isoelectric focusing (IEF) of milk samples (Vegarud et al. 1989), were targeted for the resequencing. In addition, coding parts of CSN1S1 were resequenced in 15 goats using primers CSNS1mRNA-F and CSNS1mRNA-R. For this part biopsies were taken from the mammary gland and reverse transcribed into cDNA using primer T720 and a Superscript II RT kit (Invitrogen, Carlsbad, CA).
Primers for amplification and resequencing are given in supplemental material at http://www.genetics.org/supplemental/. Samples were sequenced using Dye terminator chemistry and an ABI3730 sequencer (Applied Biosystems, Foster City, CA). For the identification of SNPs a pipeline based on the phred, phrap, and polyphred programs was used as described by Olsen et al. (2005). Contig assembly and putative SNPs were visually inspected using consed (Gordon et al. 1998) before assays were constructed and SNPs were genotyped using matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS) (Sequenom, San Diego). Assays for the genotyping are provided as supplemental information (http://www.genetics.org/supplemental/). For simplicity, polymorphisms are labeled SNP1–SNP39, in order along the chromosome segment containing the caseins. Three polymorphisms in exon 12 of CSN1S1 were genotyped in the same assay and coded as alleles 1, 3, and 6. Sequences for the three polymorphisms in exon 12 are as follows:
Allele 1: GAACAGCTTCTCAGACTGAAAAATACAACGTGCCCCAGCTG
Allele 3: GAACAGCTTCTCAGACTGAAGAAATACAACGTGCCCCAGCTG
Allele 6: GAACAGCTTCTCAGACTGAAAAAATACAACGTGCCCCAGCTG.
Allele 1 is characterized by a single-point deletion coding for very low levels of mRNA (our unpublished data) and a truncated protein undetectable by IEF of milk samples (Vegarud et al. 1989). So far this deletion is reported only in Norwegian goats with a surprisingly high frequency of 0.86 (Ådnøy et al. 2003).
Haplotype construction:
To construct haplotypes from SNP genotypes of 436 bucks, two different programs were used in sequence. SimWalk (Sobel and Lange 1996) uses pedigree information to reconstruct haplotypes, while PHASE (Stephens et al. 2001) uses linkage disequilibrium and allele frequencies. Sufficient pedigree for successful haplotype construction with SimWalk was available only for a subset of the data, including 240 bucks belonging to half-sib families (common sire) of at least six individuals. The identified haplotypes of these 240 bucks were then assumed to be phase-known genotypes in the PHASE program, along with the phase-unknown genotypes of the rest of the 196 Norwegian bucks. Haplotypes were predicted within each casein locus. Haplotypes with a frequency of <1% were omitted from the data set.
Level of linkage disequilibrium:
Once the haplotypes were constructed, we estimated the level of linkage disequilibrium between all pairs of loci using the r2-statistic (Hudson 1985). The result was visualized using the Haploview program (Barret et al. 2005).
To determine if there were differences in intragenic and intergenic levels of linkage disequilibrium (LD) and to determine if levels of LD were significantly lower between pairs of markers flanking a potential site of preferential recombination suggested from the visualization of results (between CSN2 and CSN1S2), we fitted the following model to the r2-values between all pairs of markers,
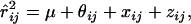 |
where 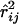 is the estimate of r2 between markers i and j; xij is an indicator variable that takes the value of 0 if both markers are in the same gene and 1 if the markers are in a different gene; zij is an indicator variable that takes the value of 1 if both markers are in either CSN1 or CSN2, 2 if both markers are in CSN1S2 or CSN3, and 3 otherwise; and
is the estimate of r2 between markers i and j; xij is an indicator variable that takes the value of 0 if both markers are in the same gene and 1 if the markers are in a different gene; zij is an indicator variable that takes the value of 1 if both markers are in either CSN1 or CSN2, 2 if both markers are in CSN1S2 or CSN3, and 3 otherwise; and 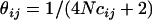 , where cij is the distance between markers i and j in megabases, and N is a parameter reflecting the effective population size (e.g., Sved 1971). We ran the above model in ASREML (Gilmour et al. 1999) with values of N from 1 to 25,000. The parameter estimates were taken from the model with the value of N that maximized the log likelihood.
, where cij is the distance between markers i and j in megabases, and N is a parameter reflecting the effective population size (e.g., Sved 1971). We ran the above model in ASREML (Gilmour et al. 1999) with values of N from 1 to 25,000. The parameter estimates were taken from the model with the value of N that maximized the log likelihood.
Estimation of haplotype and SNP effects:
The effects of the haplotypes on the buck's daughter-yield deviations (DYDs) were calculated for kilograms and percentage of fat, protein, and lactose, in addition to kilograms of milk. DYDs were calculated using data from the Norwegian Dairy Goat Control. Milk production records for each goat were first corrected for the effects of days in milk (DIM), lactation number, herd-year-test day (hy-td), and permanent environment (p-env), calculated from all goat control records, using the model
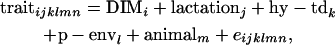 |
where traitijklmn is record ijklmn of milk, fat, protein, or lactose. DIMi is a fixed effect of stage of lactation i when the record was registered (the lactation period was split into 97 3-day intervals, starting with DIM = 16 if the record was measured at day 15, 16, or 17). Lactationj is a fixed effect of which lactation the goat was in when the record was measured (j = 1 or 2); hy-tdk is a random effect of herd, the year when the record was registered and the test-day k (k = 1, … , 30,321); p-envl is a random effect of animal within lactation l (l = 1, … , 240,176); animalm is a random effect of animal m (m = 1, … , 173,179); and eijklmn is the random residual effect of record ijklmn. The following (co)variance structure was assumed for the model,
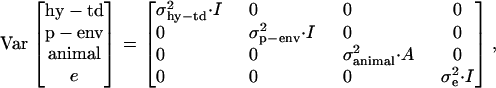 |
where 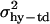 ,
, 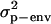 ,
, 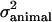 , and
, and 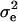 are variance components estimated simultaneously with the effects, I is the identity matrix, and A is the additive genetic relationship matrix, including 173,179 animals.
are variance components estimated simultaneously with the effects, I is the identity matrix, and A is the additive genetic relationship matrix, including 173,179 animals.
Of the bucks with reliable haplotypes, only 207 had daughters in the goat control. These daughters had 29,032 test-day records for milk, 18,465 test-day records for protein, 18,246 test-day records for fat, and 18,600 records for lactose. The DYDs for the 207 bucks were calculated by averaging the daughters' corrected milk records. Next we estimated the effect of the haplotypes on the DYDs for the seven traits. Weighted analyses were performed in ASREML (Gilmour et al. 1999) by the model
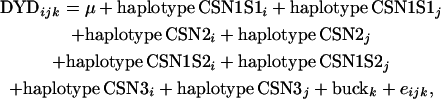 |
where DYDijk is the daughter-yield deviation for buck k, μ is a fixed effect of the mean, haplotypei and haplotypej are random effects of the paternal and maternal haplotypes carried by buck k (for each casein locus), and eijk is the random residual effect for observation ijk. DYDs were weighted by standardized reliabilities ranging between 0 and 1. The reliability is inversely proportional to the variance of the DYDs, which is defined as
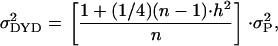 |
where n is the number of daughters contributing to the DYD, h2 is the heritability, and 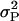 is the phenotypic variance (Bovenhuis and Meuwissen 1996). However, for protein percentage the weight statement in the ASREML analyses had to be removed, due to the fact that there was no error variance left after fitting the haplotype and the buck. The (co)variance matrix was
is the phenotypic variance (Bovenhuis and Meuwissen 1996). However, for protein percentage the weight statement in the ASREML analyses had to be removed, due to the fact that there was no error variance left after fitting the haplotype and the buck. The (co)variance matrix was
 |
The additive genetic relationship matrix A included 2270 animals from six generations. A likelihood-ratio test was performed to evaluate if the haplotypes had significant effects on the milk production traits. Let L0 be the likelihood value for the model under H0, where the haplotype for a particular casein is omitted from the model. Then L1 is the likelihood value for the alternative model, that is, when all haplotypes are included in the model. The test statistic was defined as 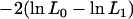 . The haplotype effects for a particular casein locus were taken as significant if the test statistic was >2.71 (Almasy and Blangero 1998). Interactions between casein locus haplotypes were also fitted if the haplotypes for more than one casein locus were significant.
. The haplotype effects for a particular casein locus were taken as significant if the test statistic was >2.71 (Almasy and Blangero 1998). Interactions between casein locus haplotypes were also fitted if the haplotypes for more than one casein locus were significant.
In addition to testing effects of haplotypes, ASREML was used to test if the individual SNPs had significant effect on the milk production traits, using the following model for each of the 39 SNPs,
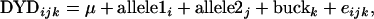 |
where DYDijk is the daughter-yield deviation for buck k with allele1 i and allele2 j (note that for SNP14 there are three levels as described above), μ is a fixed effect of the mean, allele1i and allele2j are random effects of alleles i and j (i, j = A, C, G, T, or deletion for every SNP except for SNP14, where i, j = 1, 3, or 6), buckk is a random effect of buck k (k = 1, … , 207) and eijk is the random residual effect for observation ijk. The (co)variance matrix was similar to the matrix for the haplotype analyses. The weights were also the same as for the haplotype analyses, although for protein percentage the weight statement again had to be removed, as there was no error variance left after fitting the allele and the buck. Permutation testing was used to determine an appropriate significance threshold when testing the effect of multiple individual SNPs on each trait. In a single permutation, the phenotypic records were randomly shuffled across SNP genotypes. The above variance component model was run across the 39 SNPs, and the highest log likelihood was stored. Five hundred permutations were conducted, and the 450th largest log-likelihood value was taken as the 10% chromosome segmentwide threshold.
RESULTS
SNP detection results:
We detected 39 SNPs in the exons, the introns, and the promoter regions of the four casein genes. Table 1 gives an overview of each of the 39 SNPs, the genes and region they were found in, alleles present at the SNP, and frequencies. A number of the SNPs were only tens of bases apart, for example, SNP2 and SNP3. The largest number of SNPs was in CSN1S1. The lowest frequency of the rare allele of any SNP, when genotyped in the 436 bucks, was found in SNP19 and was 0.01. The frequency of the deletion in exon 12 of CSN1S1 in the Norwegian population was similar to that found in earlier studies by Ådnøy et al. (2003).
TABLE 1.
SNP positions and relative allele frequencies for each SNP
| SNP | Gene | Position | Alleles (rare in parentheses) | Frequency of rare allele |
|---|---|---|---|---|
| 1 | CSN1S1 | Promoter | A (G) | 0.018 |
| 2 | CSN1S1 | Promoter | C (T) | 0.078 |
| 3 | CSN1S1 | Promoter | C (T) | 0.199 |
| 4 | CSN1S1 | Promoter | (A) G | 0.172 |
| 5 | CSN1S1 | Promoter | (A) G | 0.159 |
| 6 | CSN1S1 | Promoter | (A) G | 0.2 |
| 7 | CSN1S1 | Promoter | C (T) | 0.195 |
| 8 | CSN1S1 | Promoter | (A) G | 0.203 |
| 9 | CSN1S1 | Exon 4 | (C) T | 0.166 |
| 10 | CSN1S1 | Exon 4 | C (G) | 0.083 |
| 11 | CSN1S1 | Exon 9 | C (D) | 0.082 |
| 12 | CSN1S1 | Intron 9 | A (G) | 0.198 |
| 13 | CSN1S1 | Exon 10 | C (G) | 0.199 |
| 14 | CSN1S1 | Exon 12 | 1, 3, 6 | a |
| 15 | CSN1S1 | Exon 17 | C (T) | 0.111 |
| 16 | CSN2 | Exon 7 | (C) T | 0.090 |
| 17 | CSN2 | Promoter | A (G) | 0.082 |
| 18 | CSN2 | Promoter | (A) G | 0.010 |
| 19 | CSN2 | Promoter | A (G) | 0.105 |
| 20 | CSN2 | Promoter | (A) T | 0.101 |
| 21 | CSN2 | Promoter | C (T) | 0.101 |
| 22 | CSN1S2 | Exon 3 | (A) G | 0.017 |
| 23 | CSN1S2 | Exon 15 | C (T) | 0.167 |
| 24 | CSN1S2 | Intron 15 | C (G) | 0.001 |
| 25 | CSN1S2 | Intron 15 | C (T) | 0.258 |
| 26 | CSN1S2 | Exon 16 | A (T) | 0.271 |
| 27 | CSN3 | Promoter | A (G) | 0.500 |
| 28 | CSN3 | Promoter | (A) G | 0.480 |
| 29 | CSN3 | Promoter | (A) G | 0.017 |
| 30 | CSN3 | Promoter | (A) T | 0.467 |
| 31 | CSN3 | Promoter | (A) T | 0.488 |
| 32 | CSN3 | Promoter | C (G) | 0.478 |
| 33 | CSN3 | Promoter | (G) T | 0.425 |
| 34 | CSN3 | Promoter | G (T) | 0.491 |
| 35 | CSN3 | Promoter | A (G) | 0.070 |
| 36 | CSN3 | Promoter | (C) T | 0.226 |
| 37 | CSN3 | Promoter | G (T) | 0.227 |
| 38 | CSN3 | Promoter | A (G) | 0.157 |
| 39 | CSN3 | Promoter | (A) G | 0.067 |
Relative frequencies are 0.745, 0.111, and 0.144 for alleles 1, 3, and 6, respectively.
Haplotype reconstruction and extent of linkage disequilibrium:
There were 10 unique haplotypes for CSN1S1, 6 for CSN2, 4 for CSN1S2, and 8 for CSN3 (Table 2). For each casein locus, the two most frequent haplotypes accounted for the majority of haplotypes.
TABLE 2.
Haplotypes and their frequencies in 436 Norwegian dairy bucks
| Gene | Haplotype | Haplotype alleles | Frequency |
|---|---|---|---|
| CSN1S1 | 1 | ACCGGGCGTCCAC1C | 0.72 |
| 2 | ATTAAATACCCGG3T | 0.07 | |
| 3 | ACTAAATACGDGG6C | 0.06 | |
| 4 | ACCGGGCGTCCAC6C | 0.06 | |
| 5 | GCTAAATACGCGG3T | 0.02 | |
| 6 | ACTAAATACCDGG6C | 0.01 | |
| 7 | ATTAAATACGCGG3T | 0.01 | |
| 8 | ACTAAATACCCGG6C | 0.01 | |
| 9 | ACCGAATACCCAC3T | 0.01 | |
| 10 | ACTAAATACGCGG3T | 0.01 | |
| CSN2 | 1 | TAGATC | 0.8 |
| 2 | TAGGAT | 0.09 | |
| 3 | CGGATC | 0.09 | |
| 4 | TAAGAT | 0.01 | |
| CSN1S2 | 1 | GCCCA | 0.64 |
| 2 | GCCTT | 0.25 | |
| 3 | GTCCA | 0.07 | |
| 4 | GTCTT | 0.01 | |
| 5 | ACCCA | 0.01 | |
| CSN3 | 1 | AAGTACGGATGAG | 0.48 |
| 2 | GGGATGTTACTAG | 0.23 | |
| 3 | GGGATGTTATGGG | 0.16 | |
| 4 | GGGATGTTGTGGA | 0.07 | |
| 5 | GAGTTCTGATGAG | 0.02 | |
| 6 | GGAATGTTATGGG | 0.02 | |
| 7 | GAGTTCTTATGAG | 0.01 | |
| 8 | GGGATGTTATGAG | 0.01 |
Only haplotypes with a frequency >4 are included. The D in SNP12 is a deletion, and the alleles of SNP14 are described in the text.
The low number of haplotypes in the population (of 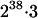 possible haplotypes) indicates considerable LD in the segment of chromosome containing the SNPs. LD between pairs of loci varied from complete disequilibrium to almost no disequilibrium (Figure 1). As distances between loci increased, both the variability and the level of LD declined. Regions of high LD were not equally spread across the chromosome segment. LD was much higher between SNPs in CSN1S1 and CSN2 and SNPs in CSN1S2 and CSN3 than between SNPs in CSN2 and CSN1S2.
possible haplotypes) indicates considerable LD in the segment of chromosome containing the SNPs. LD between pairs of loci varied from complete disequilibrium to almost no disequilibrium (Figure 1). As distances between loci increased, both the variability and the level of LD declined. Regions of high LD were not equally spread across the chromosome segment. LD was much higher between SNPs in CSN1S1 and CSN2 and SNPs in CSN1S2 and CSN3 than between SNPs in CSN2 and CSN1S2.
Figure 1.—
LD across the chromosome segment visualized using the Haploview program (Barrett et al. 2005). Each diamond contains the level of LD measured by r2 between the markers specified. Darker tones correspond to increasing levels of r2.
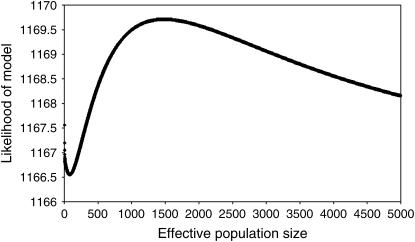
The log likelihood of the model fitted to the estimates of r2-parameters including the effect of distance, intra- or intergenic location, and flanking the region between CSN2 and CSN1S2 was maximized when the effective population size was 1484 (Figure 2). While this value maximizes the likelihood of the model and therefore is the value where the estimates of the above parameters are most accurate, it is unlikely to be a good estimate of the effective population size per se for a number of reasons: the likelihood surface was comparatively flat, indicating lower power to accurately predict N; the estimate is likely to be historical rather than recent, as the distance between markers is comparatively small (e.g., Hayes et al. 2003); and N here is estimated from linkage disequilibrium in only a very small section of the genome.
Figure 2.—
Log likelihood of model fitted to pairwise r2-values with different values for N, a parameter that reflects effective population size.
Whether a pair of SNPs spanned an intra- or intergenic region had a highly significant effect on r2 (Table 3). SNPs spanning intergenic regions had considerably lower r2-values. SNPs that spanned the region between CSN2 and CSN1S2 also had significantly lower r2 than SNPs that did not span this region.
TABLE 3.
Effects of parameters and on r2-values between all possible pairs of SNPs
| Parameter | F-value | Effect | Estimate |
|---|---|---|---|
| μ | 243.30 | 0.12 | |
| θ | 110.27 | 0.17 | |
| Intragenic vs. intergenic SNP pairs | 38.51 | Intragenic | 0 |
| Intergenic | −0.059 | ||
| Between-locus SNP pairs | 10.03 | SNP pairs within CSN1S1 and CSN2 | 0 |
| SNP pairs between CSN2 and CSN1S2 | −0.039 | ||
| SNP pairs within CSN1S2 and CSN3 | 0.021 |
All parameters were significant at P < 0.001.
Effects of haplotypes on milk production traits:
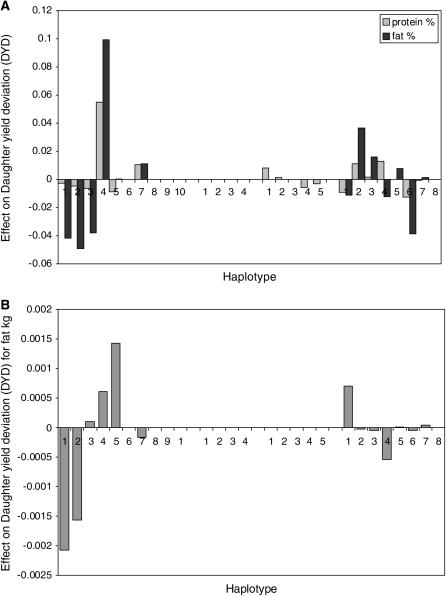
Haplotypes at both the CSN1S1 and the CSN3 loci had significant effects on the production traits (Table 4). CSN1S1 haplotypes had a significant effect on protein percentage, fat percentage, and fat kilograms, while CSN3 haplotypes significantly affected protein percentage and fat percentage. We also tested the effect of the interaction of haplotypes at CSN1S1 and CSN3 on protein percentage; this was not significant with a test statistic of 1.2. Haplotype 1 of the CSN1S1 SNPs had a large negative effect both on fat kilograms and on protein percentage and fat percentage (Figure 3). This is somewhat surprising, given that this haplotype is at very high frequency in the Norwegian goat population (Table 2). Haplotype 4 of the CSN1S1 SNPs had the largest positive effect for protein percentage and fat percentage.
TABLE 4.
Significance of effect of haplotypes on production traits
| Gene
|
||||
|---|---|---|---|---|
| Trait | CSN1S1 | CSN2 | CSN1S2 | CSN3 |
| Milk kg | 0 | 0 | 0.42 | 1.79 |
| Prot% | 10.86* | 0 | 0.62 | 3.73* |
| Prot kg | 0.86 | 0 | 0 | 1.9 |
| Fat% | 5.13* | 0 | 0 | 1.96 |
| Fat kg | 6.49* | 0 | 0 | 1.69 |
| Lact kg | 0 | 0 | 0 | 0 |
| Lact% | 0 | 0 | 0.9 | 2.69* |
Milk kg, kilograms of milk; Prot%, protein percentage; Prot kg, kilograms of protein; Fat%, fat percentage; Fat kg, kilograms of fat; Lact kg, kilograms of lactose; Lact%, lactose percentage.
Significant at P < 0.05.
Figure 3.—
(A) Effect of each haplotype on DYDs for protein and fat percentage. (B) Effect of haplotype on kilograms of fat.
Haplotype 4 of the CSN3 SNPs had an interesting pattern of effects, increasing protein percentage while decreasing fat percentage. Haplotype 6 had large negative effects on both traits.
Effects of individual SNPs:
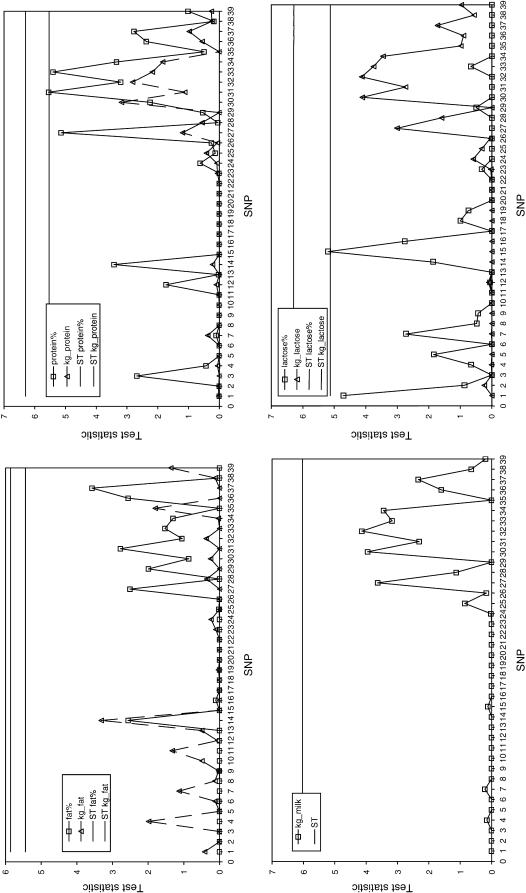
The significance of effects of the SNPs on four milk production traits is shown in Figure 4. None of the SNPs were significant at the 5% chromosome segmentwise threshold. At the 10% threshold, only two SNPs were significant—SNP31 had a significant effect on protein percentage and SNP15 had a significant effect on lactose percentage. In general two areas of the chromosome segment appeared to have some effect on the traits: a cluster of SNPs in CSN3 and SNP14 in CSN1S1.
Figure 4.—
For every trait and SNP the test statistic (two times the difference between the full and the reduced model) was plotted. The straight line indicates the 0.10 chromosome segmentwise threshold of significance. Every SNP with a test statistic value above or on this line is considered significant for the specific trait.
We also investigated whether the Norwegian-specific deletion (SNP14) accounted for the effects of CSN1S1 haplotypes. By fitting a model with both SNP14 and the haplotype effects fitted, we were able to determine that the deletion does explain the haplotype effects on fat kilograms but not on protein percentage (for protein percentage, there was an improvement in the log likelihood from fitting the haplotypes to a model with SNP14 fitted of 10.02, while for fat kilograms the improvement from fitting the haplotypes was only 0.516).
Haplotype tagging:
Only 11 of the 39 SNPs were required to capture all the information contained in the haplotypes according to the SNPtagger software (Ke and Cardon 2003). This reflects the extensive LD in this chromosome segment. The SNPs required are given in supplemental information (http://www.genetics.org/supplemental/).
DISCUSSION
Until now the impact of the goat casein genes on milk production traits has been evaluated by single- or multigene analyses, reviewed by Martin et al. (2002). As the caseins are extensively coregulated, downregulation of protein expression as a result of a mutation in one casein gene may result in upregulation of the other caseins (Leroux et al. 2003). In this study, we have taken a haplotype approach, which considers the 39 mutations from the literature and our SNP discovery in all caseins simultaneously. The effects of CSN1S1 haplotypes on protein percentage and fat kilograms were significant, as were the effects of CSN3 haplotypes on protein percentage and fat percentage.
Both the haplotype analysis and the analysis of effects of individual SNPs were consistent in indicating two casein loci with effects on milk production traits in Norwegian goats: CSN1S1 and CSN3. The analyses of the effects of the individual SNPs indicated that two sites on the chromosome segment containing the caseins were having suggestive effects on the milk production traits: SNP14 in CSN1S1 with effects on protein and fat percentage and kilograms of fat, SN15 on lactose percentage, and a cluster of SNPs in the promoter of CSN3 with effects on protein and fat percentage and on kilograms of milk and lactose. The SNP14 deletion mutation in CSN1S1 leads to very low gene expression (our unpublished data) and is found at a high frequency, 0.86, in the Norwegian dairy goat population (Ådnøy et al. 2003). The high frequency of this mutation, which decreases dry matter yield, is difficult to explain in light of the fact that the breeding goal for this goat population is an increased dry matter content in milk. One explanation could be that the founders of the Norwegian goat population, the number of which was likely to be small, carried the deletion at high frequency. Other deletions resulting in null or low levels of expression of CSN1 have been reported, as reviewed in Neveu et al. (2005). Neveu et al. (2005) and others proposed that lack of CSN1S1 disrupts the intracellular transport of caseins, leading to accumulation of caseins in the cisternae, which in turn disturbs the whole secretion process, including lipids. Our observation of reduced fat kilograms in the presence of the SNP14 deletion adds further weight to this hypothesis. Haplotype 1 of CSN1S1 carried the SNP14 deletion. However, while the SNP14 effect was sufficient to explain the effect of the CSN1S1 haplotypes on fat kilograms, it was not sufficient to explain the effect of these haplotypes on protein percentage. One explanation would be that this haplotype is in linkage disequilibrium with an as yet undetected mutation that is causing the other portion of the effect on protein percentage.
The effect of mutations in CSN3 on production traits in goats has not been previously reported. In our single SNP analysis, a cluster of SNPs in the promoter region of CSN3 had suggestive effects on protein percentage and fat percentage. However, the effect of the haplotypes of these SNPs had a higher test statistic. This suggests that none of the SNPs we have detected in this gene are the causative mutation, rather the SNPs, and even more so the haplotypes, are in linkage disequilibrium with the true causative mutation. The SNPs in CSN3 are in very strong linkage disequilibrium.
The variability in LD between the SNPs (r2 ranging between 1 and almost 0), particularly between those only tenss of bases apart, was striking. Mechanisms such as gene conversion have been proposed to explain the high variability between very closely spaced SNPs (e.g., (Frisse et al. 2001). We found that the level of linkage disequilibrium for pairs of markers within each casein locus was higher than for pairs of markers in different loci, even though a correction was made for declining linkage disequilibrium with increasing distance between a pair of markers. This finding concurs with observations of reduced recombination in genic regions compared with that in nongenic regions (e.g., Myers et al. 2005).
LD was not evenly spread across the chromosome segment containing the caseins—high levels of LD were observed at either end of the segment, with low levels of LD in the middle of the segment. Levels of linkage disequilibrium for marker pairs spanning CSN2–CSN1S2 were significantly lower than those for marker pairs located within the two segments, even when a correction was made for declining LD with distance. Preferential recombination in the region of the chromosome segment containing the caseins would ensure continuous generation of new combinations of casein gene alleles. There has been a previous report of recombination generating new alleles in caprine caseins (Bevilacqua et al. 2002), although the proposed site of recombination was within the CSNS1 locus.
Milk from Norwegian dairy goats is used almost entirely for cheese production. Farmers are paid for kilograms of milk, but with a bonus for increased dry matter content. However, many farmers exceed their quota, so they would receive extra returns only with increased dry matter percentage. As we have identified haplotypes that increase protein percentage and fat percentage and decrease milk volume, for example, haplotype 4 in CSN1S1 and haplotype 2 in CSN3, HAS would seem to have potential in Norwegian dairy goats, particularly as such haplotypes appear to be at only moderate frequency in the population. The cost of HAS would be greatly reduced by the use of the 11 tagging SNPs, rather than the entire set of 39 SNPs.
Acknowledgments
We thank Silje Karoliussen, Kristil Sundaasen, and Arne Roseth for their technical assistance. We thank TINE Norwegian Dairies for covering lab costs and a portion of the salaries for the experiments in this study. Data from the Norwegian Goat Control were made available free of cost.
References
- Ådnøy, T., G. Vegarud, T. G. Devold, R. Nordbø, I. Colbjørnsen et al., 2003. Effects of the 0- and F-alleles of alpha S1 casein in two farms of northern Norway. Proceedings of the International Workshop on Major Genes and QTL in Sheep and Goat, Toulouse, France.
- Almasy, L., and J. Blangero, 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, L., and M. Georges, 2004. Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5: 202–212. [DOI] [PubMed] [Google Scholar]
- Barbieri, M. E., E. Manfredi, J. M. Elsen, G. Ricordeau, J. Bouillon et al., 1995. Effects of the alpha(S1)-casein locus on dairy performances and genetic-parameters of alpine goats. Genet. Sel. Evol. 27: 437–450. [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, C., P. Ferranti, G. Garro, C. Veltri, R. Lagonigro et al., 2002. Interallelic recombination is probably responsible for the occurrence of a new alpha(s1)-casein variant found in the goat species. Eur. J. Biochem. 269: 1293–1303. [DOI] [PubMed] [Google Scholar]
- Bovenhuis, H., and T. H. E. Meuwissen, 1996. Detection and mapping of quantitative trait loci. Workshop, Animal Genetics and Breeding, University of New England, Armidale, New South Wales, Australia.
- Bovenhuis, H., J. A. M. Vanarendonk and S. Korver, 1992. Associations between milk protein polymorphisms and milk-production traits. J. Dairy Sci. 75: 2549–2559. [DOI] [PubMed] [Google Scholar]
- Devold, T. G., 2004. Effect of milk polymorphism on protein composition in milk from Norwegian breed of dairy goat and Norwegian dairy cattle. D.Sc. Thesis, Norwegian University of Life Sciences, Ås, Norway.
- Erhardt, G., S. Jager, E. Budelli and A. Caroli, 2002. Genetic polymorphism of goat alpha(S2)-casein (CSN1S2) and evidence for a further allele. Milchwissenschaft 57: 137–140. [Google Scholar]
- Frisse, L., R. R. Hudson, A. Bartoszewicz, J. D. Wall, J. Donfack et al., 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliano, F., R. Saletti, V. Cunsolo, S. Foti, D. Marletta et al., 2004. Identification and characterization of a new beta-casein variant in goat milk by high-performance liquid chromatography with electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 18: 1972–1982. [DOI] [PubMed] [Google Scholar]
- Gilmour, A. R., B. R. Cullis, S. J. Welham and R. Thompson, 1999. ASREML reference manual. NSW Agric. Biometric Bull. 3: 210. [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Hayes, B. J., P. E. Visscher, H. McPartlan and M. E. Goddard, 2003. A novel multi-locus measure of linkage disequilibrium and its use to estimate past effective population size. Genome Res. 13: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn, B., S. L. Coleman, C. A. Guy, K. Smith, T. Bowen et al., 2003. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 12: 2249–2254. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 1985. The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics 109: 611–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, X. Y., and L. R. Cardon, 2003. Efficient selective screening of haplotype tag SNPs. Bioinformatics 19: 287–288. [DOI] [PubMed] [Google Scholar]
- Leroux, C., P. Martin, M. F. Mahe, H. Leveziel and J. C. Mercier, 1990. Restriction-fragment-length-polymorphism identification of goat alpha-S1-casein alleles—a potential tool in selection of individuals carrying alleles associated with a high-level protein-synthesis. Anim. Genet. 21: 341–351. [DOI] [PubMed] [Google Scholar]
- Leroux, C., F. Le Provost, E. Petit, L. Bernard, Y. Chilliard et al., 2003. Real-time RT-PCR and cDNA macroarray to study the impact of the genetic polymorphism at the alpha(s1)-casein locus on the expression of genes in the goat mammary gland during lactation. Reprod. Nutr. Dev. 43: 459–469. [DOI] [PubMed] [Google Scholar]
- Mahe, M. F., and F. Grosclaude, 1993. Polymorphism of beta-casein in the Creole goat of Guadeloupe—evidence for a null allele. Genet. Sel. Evol. 25: 403–408. [Google Scholar]
- Mahe, M. F., E. Manfredi, G. Ricordeau, A. Piacere and F. Grosclaude, 1994. Effects of the alpha-S1-casein polymorphism on goat dairy performances—a within-sire analysis of alpine bucks. Genet. Sel. Evol. 26: 151–157. [Google Scholar]
- Manfredi, E., M. E. Barbieri, J. Bouillon, A. Piacere, M. F. Mahe et al., 1993. Effects of alpha(S1) casein variants on dairy performance in goats. Lait 73: 567–572. [Google Scholar]
- Martin, P., M. Ollivier-Bousquet and F. Grosclaude, 1999. Genetic polymorphism of caseins: a tool to investigate casein micelle organization. Int. Dairy J. 9: 163–171. [Google Scholar]
- Martin, P., M. Szymanowska, L. Zwierzchowski and C. Leroux, 2002. The impact of genetic polymorphism on the protein composition of ruminant milks. Reprod. Nutr. Dev. 42: 433–459. [DOI] [PubMed] [Google Scholar]
- Myers, S., L. Bottolo, C. Freeman, G. McVean and P. Donnelly, 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 14(310): 321–324. [DOI] [PubMed] [Google Scholar]
- Neveu, C., A. Riaublanc, G. Miranda, J. F. Chich and P. Martin, 2002. Is the apocrine milk secretion process observed in the goat species rooted in the perturbation of the intracellular transport mechanism induced by defective alleles at the alpha(s1)-Cn locus? Reprod. Nutr. Dev. 42: 163–172. [DOI] [PubMed] [Google Scholar]
- Olsen, H. G., S. Lien, M. Gautier, H. Nilsen, A. Roseth et al., 2005. Mapping of a milk production quantitative trait locus to a 420-kb region on bovine chromosome 6. Genetics 169: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzenberg, E. M., K. Gutscher, S. Chessa, A. Caroli and G. Erhardt, 2005. Caprine kappa-casein (CSN3) polymorphism: new developments in molecular knowledge. J. Dairy Sci. 88: 1490–1498. [DOI] [PubMed] [Google Scholar]
- Recio, I., M. L. PerezRodriguez, L. Amigo and M. Ramos, 1997. Study of the polymorphism of caprine milk caseins by capillary electrophoresis. J. Dairy Res. 64: 515–523. [DOI] [PubMed] [Google Scholar]
- Remeuf, F., 1993. Influence of genetic-polymorphism of caprine alpha(S1)-casein on physicochemical and technological properties of goats milk. Lait 73: 549–557. [Google Scholar]
- Sobel, E., and K. Lange, 1996. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58: 1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved, J. A., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2: 125–141. [DOI] [PubMed] [Google Scholar]
- Vegarud, G. E., T. S. Molland, M. J. Brovold, T. G. Devold, P. Alestrom et al., 1989. Rapid separation of genetic-variants of caseins and whey proteins using urea-modified gels and fast electrophoresis. Milchwissenschaft 44: 689–691. [Google Scholar]