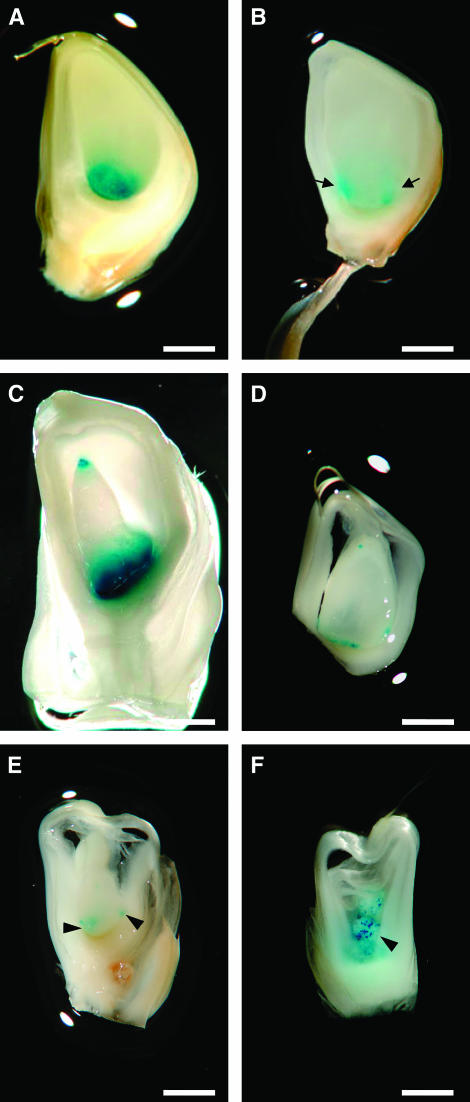
Figure 6.—
Effects of bsl1 on BETL-specific transgenic reporter expression in triploid and tetraploid endosperms. (A and B) 10-dap sibling kernels taken from a bsl1/+, ProMeg1:GUS/ProMeg1:GUS plant pollinated by a wild-type diploid plant. (A) Wild-type triploid endosperm with uniform ProMeg1:GUS expression (blue) distributed along the basal endosperm. (B) Typical mirror-image GUS expression confined to discrete basal areas of a typical bsl1/bsl1/+ endosperm (indicated by arrows). (C) 10-dap tetraploid endosperm expressing ProMeg1:GUS in the basal and apical endosperm (denoted “top–bottom”). Tetraploid endosperms were generated from crosses between wild-type ProMeg1:GUS/ProMeg1:GUS diploid plants and wild-type tetraploid plants. (D–F) Three distinct classes of 10-dap sibling kernels segregating in an ear from a bsl1/+, ProMeg1:GUS/ProMeg1:GUS plant pollinated by a wild-type tetraploid plant. (D) Typical kernel of the first class showing top–bottom ProMeg1:GUS expression patterns similar to those found in tetraploid endosperms shown in C. (E) Abnormal kernel of the second class exhibiting irregular mirror-image GUS staining patterns in the basal endosperm. (F) Kernel typical of the third class showing highly scattered ProMeg1:GUS staining throughout the endosperm (arrowheads). Bar, 200 μm.