Abstract
Background
More than 85% of Rett syndrome (RTT) patients have heterozygous mutations in the X-linked MECP2 gene which encodes methyl-CpG-binding protein 2, a transcriptional repressor that binds methylated CpG sites. Because MECP2 is subject to X chromosome inactivation (XCI), girls with RTT express either the wild type or mutant MECP2 in each of their cells. To test the hypothesis that MECP2 mutations result in genome-wide transcriptional deregulation and identify its target genes in a system that circumvents the functional mosaicism resulting from XCI, we performed gene expression profiling of pure populations of untransformed T-lymphocytes that express either a mutant or a wild-type allele.
Methods
Single T lymphocytes from a patient with a c.473C>T (p.T158M) mutation and one with a c.1308-1309delTC mutation were subcloned and subjected to short term culture. Gene expression profiles of wild-type and mutant clones were compared by oligonucleotide expression microarray analysis.
Results
Expression profiling yielded 44 upregulated genes and 77 downregulated genes. We compared this gene list with expression profiles of independent microarray experiments in cells and tissues of RTT patients and mouse models with Mecp2 mutations. These comparisons identified a candidate MeCP2 target gene, SPOCK1, downregulated in two independent microarray experiments, but its expression was not altered by quantitative RT-PCR analysis on brain tissues from a RTT mouse model.
Conclusion
Initial expression profiling from T-cell clones of RTT patients identified a list of potential MeCP2 target genes. Further detailed analysis and comparison to independent microarray experiments did not confirm significantly altered expression of most candidate genes. These results are consistent with other reported data.
Background
Rett syndrome (RTT, OMIM 312750) is an X-linked neurodevelopmental disorder that affects 1 in 10,000 to 15,000 females [1,2]. Girls with RTT have an apparently normal early development, followed by deceleration of head growth, loss of language skills, loss of purposeful hand movements and impaired social contact. As the disease progresses they develop respiratory abnormalities, autistic features, stereotypic hand movements, scoliosis, general growth delay, seizures and ataxia [3,4]. RTT is caused by heterozygous mutations in the methyl-CpG-binding protein 2 gene (MECP2), an X-linked gene subject to X chromosome inactivation (XCI) [5]. Mutations in the coding region of this gene are detected in 85% of patients with classic RTT [6-9]. An additional 10% have large deletions affecting several exons of MECP2 [10-12]. Alternative splice variants of MECP2 have been identified [9,13,14] that result in two protein isoforms. MeCP2-e1 (MeCP2α/B) is encoded by exons 1, 3 and 4 and is more abundant in brain than the previously identified MeCP2-e2 (MeCP2β/A) isoform, which is encoded by exons 2, 3 and 4. Interestingly, mutations in exon 1 are only rarely found in RTT patients [9,15,16]. Both isoforms of MeCP2 are identical beyond exon 2 and contain an 84-amino acid methyl-CpG-binding domain [17] and a 104-amino acid transcriptional repression domain (TRD) [18] as well as a C-terminal protein interaction domain. MeCP2 has been shown to bind DNA, preferentially at methylated CpG dinucleotides with resulting transcriptional repression of nearby genes through the recruitment of a histone deacetylase (HDAC1 and 2) and a Sin3A-containing corepressor complex [19,20]. MeCP2 also associates with histone methyltransferase activity and the DNA methyltransferase DNMT1 [21,22]. Brahma (Brm), the catalytic component of the SWI/SNF ATPase-dependent remodelling complex, was found to interact with MeCP2 [23], extending the mechanistic link between DNA methylation, chromatin remodelling and transcriptional repression. Recently, MecP2 has also been demonstrated to regulate alternative splicing and interact with an RNA-binding protein (Y box-binding protein 1) [24].
Despite active research since the discovery of MECP2 mutations in RTT, it has proven difficult to identify other direct target genes for the proposed functions of MeCP2. Candidate gene-based approaches using vertebrate models with disrupted MeCP2 have resulted in the identification of brain-derived neurotrophic factor (Bdnf) [25-27] and Hairy2a [28] as MeCP2 targets. MeCP2 binds to methylated CpG sites near promoter III of BDNF in resting neurons [25,26], and disease progression in a RTT mouse model correlates inversely with Bdnf expression [27]. Hairy2a is upregulated in the absence of MeCP2 in Xenopus embryos [28]. Following the hypothesis that MeCP2 functions primarily as a transcriptional repressor, several groups have attempted to screen for its targets by transcriptional profiling using RNA from postmortem brain tissues or cell lines derived from RTT patients, or from tissues of mice with engineered mutations in Mecp2. In one study, 70 transcripts were found to have altered gene expression in mutant versus wild-type fibroblast clones and lymphoblastoid cells lines [29]. The authors concluded that MeCP2 deficiency did not lead to global deregulation of gene expression and suggested that clonal fibroblast lines may show substantial variation, making them an unstable resource for expression profiling studies. In addition, lymphoblastoid cell lines are immortalized by Epstein-Barr virus (EBV) transformation, which can alter their transcriptional profile and methylation status. Expression profiling of brain from male mice with a deletion of Mecp2 also yielded only few genes with altered expression between wild-type and mutant mice [30]. Upon further analysis, those identified fell well within the range of the high false-positive rate [30]. In a third transcriptional profiling microarray study on postmortem RTT brains, significant changes in expression for 135 genes on three different cDNA microarrays were found [31]. Yet, the small sample size analyzed and the validation of the array data with the same samples instead of independent samples may have led to false-positive results [30]. It has been hypothesized that transcriptional profiling of MECP2-deficient brains has failed to identify MeCP2-regulated genes because of the high complexity in regional organization and the admixture of neuronal and non-neuronal cell types that is inherent to brain tissue [32]. Perhaps MeCP2 targets are also difficult to identify by microarray analysis because the absolute changes in expression level for individual genes are very small [33]. A more direct approach that circumvents this problem is the use of a chromatin immunoprecipitation (ChIP) strategy to directly search for the sites of MeCP2-binding to DNA. This led to the recent identification of the DLX5 gene as a MeCP2 target and other genes such as CNTN2, FOXA3, and SIAT4A as candidate genes regulated by MeCP2 [34].
In the present study we sought to overcome some of the limitations of transcriptional profiling on complex tissues, such as brain, as well as those of transformed lymphoblastoid cell lines by studying clonal cultures of non-transformed lymphocytes from individual RTT patients. These cells can be easily obtained through blood sampling, readily cultured and subjected to single-cell subcloning. Because MECP2 undergoes XCI, this last feature allows the separation of cells that express the wild-type MECP2 from the active X from those cells that express the mutant MECP2 from the active X. To identify downstream targets of MeCP2, we compared global gene expression patterns in matched pairs of clonally derived mutant or wild-type MECP2-expressing lymphocyte cultures from two girls with classic Rett syndrome. Our microarray analysis revealed 121 genes with changes in expression between cells expressing the wild-type MECP2 and those expressing the mutant MECP2. We compared this gene list to those of other transcriptional profiling experiments and focused further analysis on a downregulated putative MeCP2 target, SPOCK1. However, quantitative RT-PCR analysis on RNA from the clonal T-lymphocytes, differentiating mouse embryoid bodies (EB) and various mouse brain regions did not reveal significantly different expression between mutant and wild-type RNAs.
Methods
T-lymphocyte single cell cloning
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque from fresh blood samples of Rett syndrome patients, collected under a research protocol approved by the Baylor College of Medicine Institutional Review Board for Human Subjects Research. Isolation of T-lymphocyte clones has been described previously [35-37]. Briefly, PBMCs from the patients were plated at limiting dilution in 96-well plates in the presence of irradiated allogeneic PBMCs in RPMI media containing 20% pooled human sera (Sigma-Aldrich, St. Louis, MO), 2 μg/mL PHA.P (Murex diagnostics, Inc. Dartford Kent, UK) and 5% Human T-stim (Collaborative Biomedical Products, Bedford, MA). Nine to ten days following original culture, wells showing clonal positive growth were restimulated and cultured for an additional 7–14 days. Additional restimulations were performed every 10–12 days until >107 cells were obtained from each clone.
RNA isolation and reverse transcription
Total RNA was isolated from T-cell clones using Trizol reagent (Invitrogen Corp., Carlsbad, CA). RNA was treated with 1 U of RNase-free DNase I (Ambion Inc., Austin, TX). RNA samples were stored at -80°C until later use. Analysis of allelic expression of MECP2 by RT-PCR and restriction digestion to verify clonality of expanded lymphocyte clones from patients RT208 (c.1308-1309delTC mutation) and RT211 (p.T158M mutation) has been previously described [37].
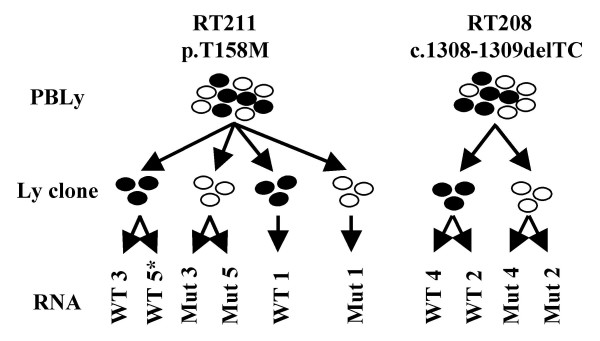
Animal research was performed under a protocol approved by the Baylor College of Medicine animal research review board. After euthanasia, brain regions (cortex, cerebellum, olfactory bulb) were quickly removed from 5 week-old mice and immediately frozen in liquid nitrogen. E16.5 embryos were collected and heads and bodies immediately frozen in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Mini kit (Qiagen Inc., Valencia, CA). Prior to array hybridization, the RNA quality and degradation was verified using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). In addition, quality control parameters were assessed throughout the experimental process to assure the efficiency of transcription, integrity of hybridization, and consistency of qualitative calls. Assessment during the synthesis of the hybridization transcript was accomplished by spectrophotometric analysis of the starting RNA and cRNA, and by gel electrophoresis following synthesis of the cDNA, cRNA and fragmentation of the cRNA. A 3'/5' ratio of GAPDH less than 2 was considered acceptable for efficiency of transcription. Spiked control transcripts were also monitored to verify hybridization integrity. Scaling factors for each sample ranged between 1.72 and 5.39. All quality control measures were consistent with the manufacturers recommended procedures and conformed to the recommended cut-off values except for one of the RT211 (p.T158M) wild-type clonal lymphocyte samples (Wt5) which was not included in subsequent analysis because the RNA was found to be partially degraded.
Oligonucleotide expression array hybridization
Total RNA was converted to double-stranded cDNA (ds-cDNA) in the presence of an oligo-dT primer containing a T7 RNA polymerase promoter. In vitro transcription was performed to generate biotin-labeled cRNA from the ds-cDNA template in the presence of a mixture of biotin-labeled ribonucleotides. Biotin-labeled cRNA (15 μg) was fragmented to a size range between 50–200 bases in length and used to hybridize to Affymetrix human HG-U133A chips. After hybridization, the arrays were washed and stained with Streptavidin-phycoerythrin. Arrays were read at a resolution of 3 microns using a HP Gene Array Scanner (Hewlett Packard, Inc., Palo Alto CA) The average background for each chip was determined to be below the common threshold of 100 for all analyzed chips.
Microarray data analysis
For the array hybridizations with RNA from clonal lymphocyte cultures, raw data was obtained from an HP Gene Array Scanner and normalized by the GC Robust Multichip Average or GC-RMA analysis. Briefly, in RMA analysis [38,39], described by Irazarry et al. [40,41], the background correction is based on the distribution of probe mismatch (MM) values amongst all probes on the array and compensates for non-specific binding using "perfect match" (PM) distributions rather than PM-MM values. The RMA method uses probe-level multichip quantile normalization to unify PM distributions across all chips and robust probe-set summary of the log-normalized probe-level data by median polishing. GC-RMA is similar to RMA, except that it reduces the bias of not substracting MM values in the background correction by an algorithm that takes into account that the non-specific affinity of a probe is related to its sequence [39,42]. GC-RMA normalizations were performed using GeneSpring GX 7.3.1 software (Agilent Technologies, Palo Alto, CA) with default parameters as follows: values below 0.01 were set to 0.01; each measurement was divided by the 50.0th percentile of all measurements in that sample; each gene was divided by the median of its measurements in all samples; if the median of the raw values was below 10 then each measurement for that gene was divided by 10 if the numerator was above 10, otherwise the measurement was thrown out. Log2-transformed data files from all GC-RMA normalizations were analyzed for significance using the GeneSpring GX 7.3.1 software with the most recent annotations of the HG-U133A human genome chip. Samples from 9 chips were divided in 4 groups according to genotype (wild-type versus mutant) and specific patient (RT211 with the p.T158M mutation versus RT208 with the c.1308-1309delTC). As outlined above, we excluded one wild-type chip (Wt5) from analysis because of poor RNA quality (Figure 1). Parametric testing (one-way ANOVA with a p-value cut-off of <0.05) was used; variances were calculated with the cross-gene error model. When multiple-testing correction was performed, the Benjamini-Hochberg false discovery rate (FDR) analysis was used. The lists were also filtered for two-fold and 1.5-fold up or down-regulation.
Figure 1.
Experimental design of T-lymphocyte clone isolation. Two technical replicate hybridizations for the wild type MECP2 and mutant MECP2-expressing clonal cell-cultures (RT211 with p.T158M mutation and RT208 with c. 1308-1309delTC mutation) were performed, as well as a biological replicate hybridization with an independent clone of patient RT211. (PBLy = peripheral blood lymphocytes; Ly clone = T-lymphocyte clone; * indicates unused in array hybridization analysis because of insufficient RNA quality)
Gene lists of all up and downregulated genes at the p < 0.05 level were then compared to lists of genes from analysis of another transcriptional profiling experiment in progress in our laboratory that compares gene expression profiles of Mecp2R308/Y-mutant [43] and wild-type embryoid bodies (EB) at day 5 after retinoic acid-induced in vitro differentiation of mouse embryonic stem cells (unpublished data). We also compared the gene lists from the clonal lymphocyte cultures to those from previously published microarray experiments on RNA samples of Rett syndrome patients or of Mecp2-mutant mouse models [29-31,44], and genes found to be MeCP2-targets by chromatin immunoprecipitation experiments [34]. To allow direct comparison of all different datasets using consistent nomenclature, we replaced the gene annotations in all these lists with the official symbol for the human genes (or human orthologue of mouse genes) from the NCBI Entrez Gene browser [45] (See additional file 1: supplementary table 1).
Quantitative real-time PCR
For quantitative real-time PCR on mouse tissues, an equal amount of total RNA isolated from brain regions (cortex, cerebellum and olfactory bulb) of 5 week-old mice or E16.5 embryos was converted to cDNA with SuperScript II reverse transcriptase (Invitrogen Corp., Carlsbad, CA). For quantitative real-time PCR on human samples, total RNA from the clonal lymphocyte cultures used for microarray hybridization was similarly converted to cDNA. We used the ABI Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA) to perform quantitative real time PCR with SYBR Green as the detection agent. The primers used were as follows. Murine Spock1 (NM_009262): Spock1 forward primer: TGCACGGACAAGGAGCTGCG, Spock1 reverse primer: GAACCAGTCCTTCAGCCGG; Murine Gapdh (NM_001001303): Gapdh forward primer: CATGGCCTTCCGTGTTCCTA, Gapdh reverse primer: GCGGCACGTCAGATCCA; human SPOCK1 (NM_004598): SPOCK1 forward primer: TGCACAGACAAGGAGTTGCG, SPOCK1 reverse primer: AAACCAATCCTTCAGCCGG; human GAPDH (NM_002046): GAPDH forward primer: TGGGCTACACTGAGCACCAG, and GAPDH reverse primer: GGGTGTCGCTGTTGAAGTCA. After PCR amplification, a dissociation protocol was performed to determine the melting curve of the PCR product. Only reactions with melting curves indicative of a single amplification product were analyzed further. The identity and expected size of the single PCR product was also confirmed by agarose gel electrophoresis. Relative quantification of the abundance of each gene at every time point was performed by the comparative ΔΔCT method as described in the Applied Biosystem user bulletin #2 [46]. The values obtained by this method are a measure of the fold-change in expression of the gene of interest compared to the calibrator sample, all normalized to Gapdh (mouse samples) or GAPDH (human samples). The Student's t test was used to determine statistical significance of expression differences between average CT values of wild-type and mutant samples. A p-value < 0.05 was considered significant.
Results
Isolation of MECP2-mutant and MECP2-wild type T-lymphocyte clones
Because MECP2 is an X-linked gene that undergoes XCI in females, each cell expresses MECP2 exclusively from only one of the two X chromosomes (the active X), while the copy of the gene on the inactive X chromosome is silenced. In order to separate cells that express the wild type MECP2 from those that express the mutant MECP2 allele, we performed single-cell cloning of T-lymphocytes isolated from peripheral blood mononuclear cells of RTT patients [37]. To allow for comparison between two types of mutations, samples from two girls with classic RTT, one (RT211) with a p.T158M missense mutation and one (RT208) with a c.1308-1309delTC frameshift mutation, were used for this experiment. Following two rounds of mitogenic stimulation, all clonal cultures with greater than 107 cells were harvested for RNA isolation. As previously reported for these cell cultures, RT-PCR followed by digestion of the amplified products to completion with restriction enzymes specific for the mutant allele, demonstrated that we had obtained pure populations of clonal cells, expressing either a mutant or a wild-type MECP2 [37].
Microarray analysis of matched lymphocyte clones
Two technical replicate hybridizations were performed for one of each clone type, as well as an additional biological replicate hybridization for the wild-type MECP2 and mutant MECP2 clones of RT211 (p.T158M) on HG-U133A human genome chips containing 22,283 genes (Affymetrix, Santa Clara, CA) (Figure 1). Signals of all hybridized chips were normalized using the GC-RMA method [42]. We then performed one-way ANOVA analysis to find genes with significantly altered expression in Mut2 and Mut4 from patient RT208 (c.1308-1309delTC), and Mut1, Mut3 and Mut5 from patient RT211 (p.T158M) compared to WT2 and WT4 from patient RT208, and WT1 and WT3 from patient RT211.
One-way ANOVA analysis, with a p < 0.05 yielded a total of 121 genes with significantly different expression between wild-type and mutant samples. Of these, 44 had a reduced level of expression (Table 1) and 77 had an increased level of expression (Table 2) in mutant compared to wild-type clones. When these lists were filtered for absolute fold-expression change, there were 28 genes with 1.5 fold higher expression in mutant compared to wild-type (13 genes with 2-fold higher expression) and 21 genes with 1.5 fold lower expression in mutant compared to wild-type (12 genes with 2-fold lower expression). It is however important to note that, although these expression changes are statistically significant at a cut-off of p < 0.05, 1,134 (5%) of the 22,283 genes present in the HG-U133A chip, would randomly be expected to show altered expression. In addition, FDR analysis using the Benjamini-Hochberg algorithm did not retain any genes with significantly altered expression. Because FDR analysis may be too stringent for genes with low-level expression, we next determined if any of the 121 genes found to have altered expression without the FDR correction might be more relevant to Rett syndrome. We compared our data to those of other published expression microarray experiments [29-31,44], to a published list of putative direct MeCP2 targets found by chromatin immunoprecipitation [34], and to results from another ongoing microarray experiments in our laboratory that compares murine EBs with a truncating Mecp2 mutation (Mecp2R308/Y) to wild-type EBs at day 5 after retinoic-acid induced in vitro differentiation of mouse ES cells (unpublished data). Very limited overlap was found between each of these lists (See additional file 2: Supplementary table 2). One downregulated gene, BEXL1, encoding brain expressed X-linked-like 1, a protein of unknown function of the BEX family of proteins [47], was also found downregulated in the expression profiling performed by Traynor et al. [29]. However, its expression was not altered in our microarray experiments on Mecp2R308/Y mutant samples compared to wild-type differentiating mouse EBs, and it was not prioritized for further analysis. In contrast, two genes, LPIN1 and SPOCK1 with significantly reduced expression in this microarray analysis also showed lower expression in Mecp2R308/Y mutant samples compared to wild-type in our microarray experiments on differentiating mouse EBs. LPIN1 encodes lipin, a protein important for adipocyte differentiation and function that plays a role in glucose and lipid homeostasis and has been associated with human lipodystrophy [48,49]. Based on this known function, we considered it a less likely candidate for an important role in the neurological phenotype of RTT. We focused on SPOCK1, the only gene found downregulated in our own two microarray experiments that has a function that is compatible with a possible role in the RTT phenotype. SPOCK1 encodes the 439-amino acid SPOCK1 protein (Sparc/osteonectin-like domains, CWCV and Kazal-like domains proteoglycan 1 precursor), also known as Testican [50]. It is a predominantly extracellular matrix chondroitin sulphate-linked proteoglycan that is related to protein families that have protease inhibitor function and are involved in cell adhesion, migration, and proliferation. Human SPOCK1 and mouse Spock1 share 95% identity. SPOCK1 is highly enriched in neurons and endothelial cells of the central nervous system, absent in quiescent astroglia, but upregulated in activated astroglia [51]. Mouse Spock1 is highly expressed at the start of neurogenesis during neuron migration and axonal outgrowth and subsequently in developing synaptic fields [52]. Spock1 has also been found to inhibit adhesion and neurite outgrowth of cultured Neuro2A cells [53]. In the adult, it localizes predominantly to the postsynaptic region of a subpopulation of pyramidal neurons in the CA3 region of the hippocampus [54]. These features suggested a possible role in motor or behavioural aspects of the RTT phenotype. Therefore, we focused on this gene for further analysis.
Table 1.
Genes with lower expression in mutant samples on microarray analysis. This table contains a list of genes with lower expression in mutant T-lymphocyte clones by one-way ANOVA analysis (p < 0.05). * indicates >1.5-fold change in expression level and ** indicates >2-fold change in expression level.
| No. | Gene Name | P-value | Common name | Genbank No. | Description |
| 1** | 201160_s_at | 0.000593 | CSDA | AL556190 | cold shock domain protein A |
| 2* | 201417_at | 0.00108 | SOX4 | AL136179 | SRY (sex determining region-Y) box 4 |
| 3 | 202565_s_at | 0.00385 | SVIL | NM_003174 | supervillin |
| 4 | 204860_s_at | 0.00453 | BIRC1 | AI817801 | Transcribed seq with strong similarity BIR1 |
| 5 | 215333_x_at | 0.00459 | GSTM1 | X08020 | glutathione S-transferase M1 |
| 6* | 210763_x_at | 0.00546 | NCR3 | AF031137 | natural cytotoxicity triggering receptor 3 |
| 7** | 217979_at | 0.00821 | TM4SF13 | NM_014399 | transmembrane 4 superfamily member 13 |
| 8 | 208524_at | 0.0117 | GPR15 | NM_005290 | G protein-coupled receptor 15 |
| 9** | 215440_s_at | 0.012 | BEXL1 | AL523320 | hypothetical protein FLJ10097 |
| 10 | 213193_x_at | 0.0157 | TRBC1 | AL559122 | T cell receptor beta chain BV20S1 BJ1-5 BC1 mRNA |
| 11 | 220131_at | 0.0174 | FXYD7 | NM_022006 | FXYD domain containing ion transport regulator 7 |
| 12 | 204550_x_at | 0.0175 | GSTM1 | NM_000561 | glutathione S-transferase M1 |
| 13 | 212739_s_at | 0.0186 | NME4 | AL523860 | non-metastatic cells 4, protein expressed in |
| 14* | 220684_at | 0.0187 | TBX21 | NM_013351 | T-box 21 |
| 15 | 203030_s_at | 0.0195 | PTPRN2 | AF007555 | protein tyrosine phosphatase, receptor type N polypeptide 2 |
| 16 | 219693_at | 0.0232 | AGPAT4 | NM_020133 | 1-acylglycerol-3-phosphate O-acyltransferase 4 |
| 17* | 219654_at | 0.0245 | PTPLA | NM_014241 | protein tyrosine phosphatase-like, member a |
| 18 | 217764_s_at | 0.0245 | RAB31 | AF183421 | RAB31, member RAS oncogene family |
| 19* | 217671_at | 0.0247 | RFX3 | BE466926 | regulatory factor X, 3 (Influences HLA class II expression) |
| 20** | 204232_at | 0.025 | FCER1G | NM_004106 | Fc fragment of IgE, high affinity receptor for γ-polypeptide |
| 21* | 1405_i_at | 0.0298 | CCL5 | M21121 | chemokine (C-C motif) ligand 5 |
| 22** | 206170_at | 0.0308 | ADRB2 | NM_000024 | adrenergic, beta-2-, receptor, surface |
| 23** | 211583_x_at | 0.0317 | NCR3 | AF031136 | natural cytotoxicity triggering receptor 3 |
| 24 | 202279_at | 0.0328 | C14orf2 | NM_004894 | chromosome 14 open reading frame 2 |
| 25 | 214012_at | 0.0333 | ARTS-1 | BE551138 | type 1 TNF receptor shedding aminopeptidase regulator |
| 26 | 204396_s_at | 0.0346 | GRK5 | NM_005308 | G protein-coupled receptor kinase 5 |
| 27** | 211010_s_at | 0.0349 | NCR3 | AF031138 | natural cytotoxicity triggering receptor 3 |
| 28 | 208791_at | 0.0351 | CLU | M25915 | clusterin |
| 29** | 219529_at | 0.0383 | CLIC3 | NM_004669 | chloride intracellular channel 3 |
| 30 | 217104_at | 0.0385 | LOC283687 | AL109714 | hypothetical protein LOC283687 |
| 31 | 213280_at | 0.0397 | GARNL4 | AK000478 | GTPase activating RANGAP domain-like 4 |
| 32 | 202234_s_at | 0.04 | SLC16A1 | BF511091 | solute carrier family 16, member 1 |
| 33** | 215006_at | 0.0404 | AK023816 | CDNA FLJ13754 fis, clone PLACE3000362 | |
| 34 | 204883_s_at | 0.0409 | HUS1 | AI968626 | HUS1 checkpoint homolog (S. pombe) |
| 35** | 217963_s_at | 0.0409 | NGFRAP1 | NM_014380 | nerve growth factor receptor (TNFRSF16) ass. protein 1 |
| 36** | 212070_at | 0.0419 | GPR56 | AL554008 | G protein-coupled receptor 56 |
| 37** | 202363_at | 0.042 | SPOCK1 | AF231124 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) |
| 38* | 206974_at | 0.0427 | CXCR6 | NM_006564 | chemokine (C-X-C motif) receptor 6 |
| 39 | 200965_s_at | 0.0433 | ABLIM1 | NM_006720 | actin binding LIM protein 1 |
| 40 | 219457_s_at | 0.045 | RIN3 | NM_024832 | Ras and Rab interactor 3 |
| 41** | 213915_at | 0.0452 | NKG7 | NM_005601 | natural killer cell group 7 sequence |
| 42 | 215411_s_at | 0.0463 | C6orf4 | AL008730 | |
| 43* | 212274_at | 0.0484 | LPIN1 | AV705559 | AV705559ADB Homo sapiens cDNA clone ADBAPE04 5' |
| 44 | 219155_at | 0.049 | PITPNC1 | NM_012417 | phosphatidylinositol transfer protein, cytoplasmic 1 |
Table 2.
Genes with higher expression in mutant samples on microarray analysis. This table contains a list of genes with higher expression in mutant T-lymphocyte clones by one-way ANOVA analysis (p < 0.05). * indicates >1.5-fold change in expression level and ** indicates >2-fold change in expression level.
| No. | Gene Name | P-value | Common name | Genbank No. | Description |
| 1 | 219759_at | 6.66E-11 | LRAP | NM_022350 | leukocyte-derived arginine aminopeptidase |
| 2 | 206148_at | 0.000152 | IL3RA | NM_002183 | interleukin 3 receptor, alpha (low affinity) |
| 3* | 213655_at | 0.000209 | YWHAE | AA502643 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein |
| 4* | 212952_at | 0.000278 | MNT | AA910371 | calreticulin |
| 5 | 218340_s_at | 0.000346 | FLJ10808 | NM_018227 | hypothetical protein FLJ10808 |
| 6* | 200765_x_at | 0.000507 | CTNNA1 | NM_001903 | catenin (cadherin-associated protein), alpha 1, 102 kDa |
| 7* | 210732_s_at | 0.000704 | LGALS8 | AF342816 | lectin, galactoside-binding, soluble, 8 (galectin 8) |
| 8 | 219974_x_at | 0.00131 | ECHDC1 | NM_018479 | enoyl Coenzyme A hydratase domain containing 1 |
| 9 | 202732_at | 0.00142 | PKIG | NM_007066 | protein kinase (cAMP-dependent, catalytic) inhibitor γ |
| 10 | 204418_x_at | 0.00183 | GSTM4 | NM_000848 | glutathione S-transferase M2 (muscle) |
| 11 | 221754_s_at | 0.00187 | CORO1B | AI341234 | coronin, actin binding protein, 1B |
| 12* | 219648_at | 0.00266 | FLJ10116 | NM_018000 | hypothetical protein FLJ10116 |
| 13* | 212099_at | 0.00528 | RHOB | AI263909 | ras homolog gene family, member B |
| 14 | 213135_at | 0.00579 | TIAM1 | U90902 | T-cell lymphoma invasion and metastasis 1 |
| 15** | 210844_x_at | 0.00603 | CTNNA1 | D14705 | catenin (cadherin-associated protein), alpha 1, 102 kDa |
| 16 | 208743_s_at | 0.00617 | YWHAB | BC001359 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein |
| 17* | 204201_s_at | 0.00621 | PTPN13 | NM_006264 | protein tyrosine phosphatase, non-receptor type 13 |
| 18* | 222146_s_at | 0.00628 | TCF4 | AK026674 | transcription factor 4 |
| 19* | 214130_s_at | 0.00717 | PDE4DIP | AI821791 | phosphodiesterase 4D interacting protein (myomegalin) |
| 20** | 211893_x_at | 0.00833 | CD6 | U66145 | CD6 antigen |
| 21** | 210136_at | 0.00975 | MBP | AW070431 | myelin basic protein |
| 22** | 209012_at | 0.0106 | TRIO | AV718192 | triple functional domain (PTPRF interacting) |
| 23 | 222173_s_at | 0.0109 | TBC1D2 | AK026105 | TBC1 domain family, member 2 |
| 24 | 218435_at | 0.0112 | DNAJD1 | NM_013238 | DnaJ (Hsp40) homolog, subfamily D, member 1 |
| 25 | 210354_at | 0.0122 | IFNG | M29383 | Human mRNA for HuIFN-gamma interferon. |
| 26 | 217744_s_at | 0.0132 | PERP | NM_022121 | PERP, TP53 apoptosis effector |
| 27* | 213822_s_at | 0.0133 | UBE3B | BE856776 | ubiquitin protein ligase E3B |
| 28 | 204412_s_at | 0.015 | NEFH | NM_021076 | neurofilament, heavy polypeptide 200 kDa |
| 29 | 200862_at | 0.0151 | DHCR24 | NM_014762 | 24-dehydrocholesterol reductase |
| 30 | 210949_s_at | 0.0153 | EIF3S8 | BC000533 | eukaryotic translation initiation factor 3, subunit 8, 110 kDa |
| 31 | 218393_s_at | 0.0172 | SMU1 | NM_018225 | homolog of C. elegans smu-1 |
| 32 | 221550_at | 0.018 | COX15 | BC002382 | COX15 homolog, cytochrome c oxidase assembly protein |
| 33* | 203413_at | 0.0192 | NELL2 | NM_006159 | NEL-like 2 (chicken) |
| 34 | 221036_s_at | 0.0192 | PSFL | NM_031301 | anterior pharynx defective 1B-like |
| 35* | 210915_x_at | 0.0208 | PRSS1 | M15564 | T-cell receptor precursor |
| 36 | 218943_s_at | 0.0212 | DDX58 | NM_014314 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide |
| 37 | 207813_s_at | 0.0215 | FDXR | NM_004110 | ferredoxin reductase |
| 38 | 221021_s_at | 0.0216 | CTNNBL1 | NM_030877 | catenin, beta like 1 |
| 39 | 214316_x_at | 0.0216 | MNT | AI378706 | Calreticulin |
| 40** | 209728_at | 0.022 | HLA-DRB4 | BC005312 | major histocompatibility complex, class II, DR beta 3 |
| 41 | 215230_x_at | 0.0229 | EIF3S8 | AA679705 | eukaryotic translation initiation factor 3, subunit 8, 110 kDa |
| 42 | 219269_at | 0.024 | FLJ21616 | NM_024567 | hypothetical protein FLJ21616 |
| 43** | 207533_at | 0.0246 | CCL1 | NM_002981 | chemokine (C-C motif) ligand 1 |
| 44 | 207275_s_at | 0.0255 | ACSL1 | NM_001995 | acyl-CoA synthetase long-chain family member 1 |
| 45 | 217856_at | 0.0274 | RBM8A | AF182415 | RNA binding motif protein 8A |
| 46 | 200647_x_at | 0.0277 | EIF3S8 | NM_003752 | eukaryotic translation initiation factor 3, subunit 8, 110 kDa |
| 47* | 214395_x_at | 0.028 | PP3856 | AI335509 | eukaryotic translation elongation factor 1 delta |
| 48** | 211900_x_at | 0.0297 | CD6 | U66146 | CD6 antigen |
| 49 | 215606_s_at | 0.0307 | ELKS | AB029004 | Rab6-interacting protein 2 |
| 50 | 218945_at | 0.0311 | MGC2654 | NM_024109 | hypothetical protein MGC2654 |
| 51 | 209790_s_at | 0.0314 | CASP6 | BC000305 | caspase 6, apoptosis-related cysteine protease |
| 52 | 201143_s_at | 0.0325 | EIF2S1 | BC002513 | eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa |
| 53 | 211965_at | 0.0332 | ZFP36L1 | BE620915 | zinc finger protein 36, C3H type-like 1 |
| 54** | 201242_s_at | 0.0339 | ATP1B1 | BC000006 | ATPase, Na+/K+ transporting, beta 1 polypeptide |
| 55 | 213388_at | 0.0346 | PDE4DIP | H15535 | ym27c01.s1 Soares infant brain 1NIB cDNA clone IMAGE:49385 3' |
| 56 | 214182_at | 0.036 | ARF6 | AA243143 | ADP-ribosylation factor 6 |
| 57 | 212190_at | 0.0362 | SERPINE2 | AL541302 | AL541302 Homo sapiens PLACENTA cDNA clone CS0DE006YI10 5' |
| 58* | 204015_s_at | 0.0365 | DUSP4 | BC002671 | dual specificity phosphatase 4 |
| 59** | 206366_x_at | 0.0372 | XCL1 | U23772 | chemokine (C motif) ligand 1 |
| 60* | 216609_at | 0.0374 | TXN | AF065241 | Thioredoxin |
| 61 | 219836_at | 0.038 | ZBED2 | NM_024508 | zinc finger, BED domain containing 2 |
| 62** | 208602_x_at | 0.0383 | CD6 | NM_006725 | CD6 antigen |
| 63** | 208302_at | 0.0385 | HB-1 | NM_021182 | minor histocompatibility antigen HB-1 |
| 64 | 209433_s_at | 0.0393 | PPAT | AI457120 | phosphoribosyl pyrophosphate amidotransferase |
| 65** | 221618_s_at | 0.0404 | TAF9L | AF220509 | TAF9-like RNA polymerase II |
| 66 | 202807_s_at | 0.0427 | TOM1 | NM_005488 | target of myb1 (chicken) |
| 67 | 222145_at | 0.043 | AK027225 | Similar to PI-3-kinase-related kinase SMG-1 isoform 1; (LOC390682) | |
| 68** | 212599_at | 0.0435 | AUTS2 | AK025298 | autism susceptibility candidate 2 |
| 69 | 209392_at | 0.0447 | ENPP2 | L35594 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) |
| 70 | 204137_at | 0.0453 | TM7SF1 | NM_003272 | transmembrane 7 superfamily member 1 (upregulated in kidney) |
| 71 | 209015_s_at | 0.0455 | DNAJB6 | BC002446 | DnaJ (Hsp40) homolog, subfamily B, member 6 |
| 72 | 209013_x_at | 0.0482 | TRIO | AF091395 | triple functional domain (PTPRF interacting) |
| 73 | 215920_s_at | 0.0488 | LOC283970 | AC002544 | |
| 74 | 221069_s_at | 0.0492 | LOC51204 | NM_016360 | clone HQ0477 PRO0477p |
| 75 | 214741_at | 0.0494 | ZNF131 | AW968301 | zinc finger protein 131 (clone pHZ-10) |
| 76 | 207485_x_at | 0.0495 | BTN3A1 | NM_007048 | butyrophilin, subfamily 3, member A1 |
| 77 | 220937_s_at | 0.0496 | SIAT7D | NM_014403 | sialyltransferase 7D |
Expression of the mouse orthologue of SPOCK1 is not altered in a RTT mouse model
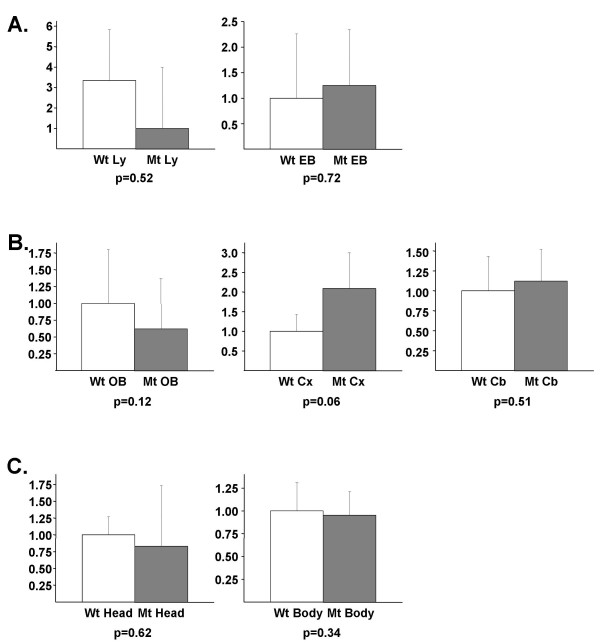
Although not statistically significant, a trend for lower expression was found by qPCR analysis using the RNAs from the clonal lymphocyte cultures that were used to perform the microarray hybridizations, but no difference was seen in RNAs from differentiating wild-type and mutant mouse EBs (Figure 2A). To test if there is altered expression in the brain, the organ most affected by RTT-causing mutations, we performed qPCR reactions on RNA samples from mutant Mecp2308/Y and wild-type cerebral cortex, cerebellum and olfactory bulb from 5 week old mice (Figure 2B), as well as on heads and bodies from wild-type and mutant E16.5 embryos (Figure 2C). No statistically significant differences in expression of Spock1 between any of these mutant and wild type samples were detected.
Figure 2.
Quantitative RT-PCR (qPCR) analysis of Spock1 expression. Quantitative RT-PCR analysis of the expression of (A.) human SPOCK1 in the clonal lymphocyte RNAs (Mt Ly, Wt Ly) used for the microarray experiments and of mouse Spock1 in differentiating wild-type (Wt EB) and Mecp2R308/Y mutant (Mt EB) male mouse embryoid bodies; (B.) cerebral cortex (Wt Cx, Mt Cx), cerebellum (Wt Cb, Mt Cb) and olfactory bulb (Wt OB, Mt OB) of adult (5 week-old) wild-type and Mecp2R308/Y mutant male mice; and (C.) E16.5 embryo head (Wt head, Mt head) and body (Wt body, Mt body) (4 independent samples were tested for each tissue and genotype). The Y-axis shows the average fold change in expression +/- standard deviation) of mutant compared to wild-type, which is set as one-fold baseline, except for the Ly samples, where the one-fold baseline was set in the mutant for figure clarity (light bars = wild-type; dark bars = mutant), p-values are given under each respective graph.
Discussion
Heterozygous mutations in X-linked MECP2 are found in most girls with RTT. The MECP2 gene is subject to XCI and classic RTT patients typically have random XCI patterns [55,56]. Hence, patients with RTT have a mosaic distribution of cells expressing the wild-type MECP2 and cells expressing the mutant MECP2 allele in all of their tissues. Analysis of gene expression profiles on tissues from deceased girls with RTT will therefore be compromised by unpredictable patterns of XCI. In addition, the presence of functional "wild-type" cells will also dampen small effects on the expression levels of MeCP2 target genes. Postmortem tissues are mostly from girls at later stages of the disorder and their gene expression profiles may reflect the presence of concurrent disease.
Because central nervous system neurons are the primary cell types where the RTT syndrome pathology occurs, a pure population of neurons with the mutant MECP2 on the active X chromosome, compared to one with the wild-type MECP2 on the active X chromosome from the same individual would constitute an ideal sample set for analysis of gene expression profiles in the presence of MECP2 mutations, but such samples are unavailable from human patients and alternative strategies are needed.
MECP2 is expressed, albeit at lower levels, in nearly all other tissues, including T-lymphocytes. Although T-lymphocytes are not the primary cell type where the RTT phenotype manifests, previous experiments have shown that lymphocytes with the mutant MECP2 on the active X chromosome are at a growth disadvantage compared to those with the wild-type MECP2 on the active X chromosome [37]. This suggested to us that at least a subset of the MeCP2 target genes are dysregulated in this cell type and pure T-lymphocyte clonal cell lines may be a valuable source of RNA for gene expression profiling in Rett syndrome.
We isolated single T-lymphocytes from blood samples of RTT patients and clonally expanded them in order to compare MECP2 mutant and wild type expression in the same patient. Since only one form of MECP2 would be expressed in each isolated clone, we expected that effects on gene expression profiles would be more pronounced. Furthermore, by comparing these clonal T-lymphocytes from the same patient, we could eliminate changes resulting from interindividual genetic differences. A similar approach was taken by Traynor et al., 2002, but instead of lymphocytes they used lymphoblastoid cell lines and fibroblast clones [29]. Lymphoblastoid cells have the disadvantage that they are immortalized which can affect gene expression patterns, while fibroblast cultures can gain epigenetic differences as they divide in culture. Although the T-lymphocytes used in our experiment were also cultured, the number of cell divisions was small and the cells were not transformed.
Because we did not study the primary cell type that is responsible for the RTT pathology, we strove to increase the likelihood that any observed differences in gene expression levels between mutant and wild-type MECP2-expressing clones were relevant to Rett syndrome. We used samples from patients with two different types of mutations (p.T158M and c.1308-1309delTC), combined the results from these samples to focus only on genes with a consistent pattern of increased or decreased levels of expression in all mutant clones compared to wild type. We next compared these results to those of our other independent microarray experiment on Mecp2-mutant mouse EBs and to those published in the literature from expression profiling [29-31,44] or large-scale ChIP experiments [34]. The limited overlap in identified genes with up or downregulated expression between all these different lists cannot be easily explained, but could have a number of different reasons. The tissues, or cell lines studied and specific mutations varied between the studies. Human as well as mouse samples were analyzed and they originated from different stages of the disease process. More likely reasons may be that overall expression differences between mutant and wild-type MeCP2 RNA samples are subtle but affect a large number of genes. An alternate hypothesis is that MeCP2 primarily regulates expression of a limited number of genes in very specific cells or in a transient manner. Such changes would be difficult to detect with the strategies used in any of these expression profiling studies.
One of the genes identified by our comparative analysis was SPOCK1. Its mouse homologue encodes Spock1/testican, a Ca(2+)-binding proteoglycan predominantly located in the extracellular matrix. Although its function is incompletely understood, Spock1/testican contains six functional domains, several of which have a protease-inhibitor function [57]. There is an N-terminal hydrophobic signal sequence for secretion of the protein in the extracellular matrix, a cysteine-rich region, a Kazal-like homology domain with a putative seine-protease inhibitor role, comprised in several regions of homology to SPARC/osteonectin, two calcium-binding EF hand motifs, and a C-terminal domain with homology to thyrotropin cysteine protease inhibitors (CVCW or thyorglobulin-like domain) that has been shown to inhibit the lysosomal cysteine protease cathepsin-L [58]. Spock1 is highly enriched in brain and is associated with the postsynaptic region of a subpopulation of pyramidal neurons in the CA3 area of the hippocampus [54]. It is also found in the choroid plexus and is highly enriched in the central nervous system (CNS) during neuronal proliferation and migration [52]. It affects neuronal attachment and neurite outgrowth likely through a modulation of the extracellular matrix [53]. Quantitative RT-PCR experiments on the clonal lymphocyte cultures showed only a trend for downregulation. Quantitative RT-PCR experiments on brain regions of Mecp2 mutant mice also did not show consistent changes in expression. This suggests that SPOCK1 is not a direct target of MeCP2 and its downregulation in the microarray experiment might have been a false positive finding.
Conclusion
To find new putative MeCP2 targets, we performed expression profiling on clonal lymphocyte cultures from RTT patients with two different MECP2 mutations and compared the gene expression profiles to those of other screens for MeCP2 targets. No statistically significant differences in expression were found upon stringent analysis of the data. A putative downregulated MeCP2-target gene, SPOCK1, did not have altered expression in brain regions in a mouse model of Rett syndrome.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
KNT and JML prepared the lymphocyte cultures, verified their clonality and prepared RNA. DSK prepared samples for microarray analysis. IJD did the microarray data analysis, RT-PCR experiments and wrote the initial draft of the manuscript. JML and IBV were responsible for the concept, supervision and coordination of the experiments. IBV participated in data analysis and finalized the writing of the manuscript. All authors have contributed comments to the contents of the manuscript and have seen and approved the final submitted version.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Transformation of gene annotations to official human gene symbols for across-microarray comparisons.
Comparison of gene lists from different microarray experiments.
Acknowledgments
Acknowledgements
This work was supported by a grant from the National Institutes of Health (NIH P01-HD40301) and the Microarray Expression Core of the Baylor Mental Retardation and Developmental Disabilities Research Center (NIH HD24064). The authors thank Huda Zoghbi for sharing the Mecp2R308 mutant mice and Miguel Landers for valuable input during preparation of this manuscript.
Contributor Information
Ivan J Delgado, Email: idelgado@identigene.com.
Dong Sun Kim, Email: doskim@knu.ac.kr.
Karen N Thatcher, Email: knthatcher@ucdavis.edu.
Janine M LaSalle, Email: jmlasalle@ucdavis.edu.
Ignatia B Van den Veyver, Email: iveyver@bcm.tmc.edu.
References
- Akesson HO, Hagberg B, Wahlstrom J. Rett syndrome, classical and atypical: genealogical support for common origin. J Med Genet. 1996;33:764–766. doi: 10.1136/jmg.33.9.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B, Hagberg G. Rett syndrome: epidemiology and geographical variability. Eur Child Adolesc Psychiatry. 1997;6:5–7. [PubMed] [Google Scholar]
- Armstrong DD. Review of Rett syndrome. J Neuropathol Exp Neurol. 1997;56:843–849. doi: 10.1097/00005072-199708000-00001. [DOI] [PubMed] [Google Scholar]
- Weaving LS, Ellaway CJ, Gecz J, Christodoulou J. Rett syndrome: clinical review and genetic update. J Med Genet. 2005;42:1–7. doi: 10.1136/jmg.2004.027730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl- CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Miltenberger-Miltenyi G, Laccone F. Mutations and polymorphisms in the human methyl CpG-binding protein MECP2. Hum Mutat. 2003;22:107–115. doi: 10.1002/humu.10243. [DOI] [PubMed] [Google Scholar]
- Van den Veyver IB, Zoghbi HY. Methyl-CpG-binding protein 2 mutations in Rett syndrome. Curr Opin Genet Dev. 2000;10:275–279. doi: 10.1016/S0959-437X(00)00083-6. [DOI] [PubMed] [Google Scholar]
- Van den Veyver IB, Zoghbi HY. Genetic basis of Rett syndrome. Ment Retard Dev Disabil Res Rev. 2002;8:82–86. doi: 10.1002/mrdd.10025. [DOI] [PubMed] [Google Scholar]
- Amir RE, Fang P, Yu Z, Glaze DG, Percy AK, Zoghbi HY, Roa BB, Van den Veyver IB. Mutations in exon 1 of MECP2 are a rare cause of Rett Syndrome. J Med Genet. 2005;42:e15. doi: 10.1136/jmg.2004.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laccone F, Junemann I, Whatley S, Morgan R, Butler R, Huppke P, Ravine D. Large deletions of the MECP2 gene detected by gene dosage analysis in patients with Rett syndrome. Hum Mutat. 2004;23:234–244. doi: 10.1002/humu.20004. [DOI] [PubMed] [Google Scholar]
- Archer HL, Whatley SD, Evans JC, Ravine D, Huppke P, Kerr A, Bunyan D, Kerr B, Sweeney E, Davies SJ, Reardon W, Horn J, Macdermot KD, Smith RA, Magee A, Donaldson A, Crow Y, Hermon G, Miedzybrodzka Z, Cooper DN, Lazarou L, Butler R, Sampson J, Pilz DT, Laccone F, Clarke AJ. Gross rearrangements of the MECP2 gene are found in both classical and atypical Rett syndrome patients. J Med Genet. 2006;43:451–456. doi: 10.1136/jmg.2005.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Shibayama A, Liu Q, Nguyen VQ, Feng J, Santos M, Temudo T, Maciel P, Sommer SS. Detection of heterozygous deletions and duplications in the MECP2 gene in Rett syndrome by Robust Dosage PCR (RD-PCR) Hum Mutat. 2005;25:505. doi: 10.1002/humu.9338. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32:1818–1823. doi: 10.1093/nar/gkh349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, Jones JR, Scherer SW, Schanen NC, Friez MJ, Vincent JB, Minassian BA. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet. 2004;36:339–341. doi: 10.1038/ng1327. [DOI] [PubMed] [Google Scholar]
- Philippe C, Villard L, De Roux N, Raynaud M, Bonnefond JP, Pasquier L, Lesca G, Mancini J, Jonveaux P, Moncla A, Chelly J, Bienvenu T. Spectrum and distribution of MECP2 mutations in 424 Rett syndrome patients: a molecular update. Eur J Med Genet. 2006;49:9–18. doi: 10.1016/j.ejmg.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Evans JC, Archer HL, Whatley SD, Kerr A, Clarke A, Butler R. Variation in exon 1 coding region and promoter of MECP2 in Rett syndrome and controls. Eur J Hum Genet. 2005;13:124–126. doi: 10.1038/sj.ejhg.5201270. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol Cell. 2003;12:425–435. doi: 10.1016/S1097-2765(03)00276-4. [DOI] [PubMed] [Google Scholar]
- Traynor J, Agarwal P, Lazzeroni L, Francke U. Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med Genet. 2002;3:12. doi: 10.1186/1471-2350-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Jeon OH, Hyder K, Chenchik A, Khimani AH, Narayanan V, Hoffman EP, Kaufmann WE, Naidu S, Pevsner J. Gene expression profiling in postmortem Rett Syndrome brain: differential gene expression and patient classification. Neurobiol Dis. 2001;8:847–865. doi: 10.1006/nbdi.2001.0428. [DOI] [PubMed] [Google Scholar]
- Pescucci C, Meloni I, Renieri A. Is Rett syndrome a loss-of-imprinting disorder? Nat Genet. 2005;37:10–11. doi: 10.1038/ng0105-10. [DOI] [PubMed] [Google Scholar]
- Klose R, Bird A. Molecular biology. MeCP2 repression goes nonglobal. Science. 2003;302:793–795. doi: 10.1126/science.1091762. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Balmer D, LaSalle JM. Clonal maintenance of imprinted expression of SNRPN and IPW in normal lymphocytes: correlation with allele-specific methylation of SNRPN intron 1 but not intron 7. Hum Genet. 2001;108:116–122. doi: 10.1007/s004390000455. [DOI] [PubMed] [Google Scholar]
- LaSalle JM, Ritchie RJ, Glatt H, Lalande M. Clonal heterogeneity at allelic methylation sites diagnostic for Prader-Willi and Angelman syndromes. Proc Natl Acad Sci USA. 1998;95:1675–1680. doi: 10.1073/pnas.95.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer D, Arredondo J, Samaco RC, LaSalle JM. MECP2 mutations in Rett syndrome adversely affect lymphocyte growth, but do not affect imprinted gene expression in blood or brain. Hum Genet. 2002;110:545–552. doi: 10.1007/s00439-002-0724-4. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioconductor http://www.bioconductor.org
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F. A model based background adjustment for oligonucleotide expression arrays. Johns Hopkins Dept of Biostatistics Working Papers. 2004. (paper 1).
- Shahbazian MD, Young YI, Yuva-Paylor LA, Spencer CM, Antalffy BA, Noebels JL, Armstrong DL, Paylor R, Zoghbi HY. Mice with Truncated MeCP2 Recapitulate Many Rett Syndrome Features and Display Hyperacetylation of Histone H3. Neuron. 2002;35:243–254. doi: 10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, Monros E, Esteller M. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005;116:91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnoology Information Entrez Gene http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
- Applied Biosystems user bulletin #2: ABI PRISM 7700 Sequence Detection System http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf
- Alvarez E, Zhou W, Witta SE, Freed CR. Characterization of the Bex gene family in humans, mice, and rats. Gene. 2005;357:18–28. doi: 10.1016/j.gene.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Suviolahti E, Reue K, Cantor RM, Phan J, Gentile M, Naukkarinen J, Soro-Paavonen A, Oksanen L, Kaprio J, Rissanen A, Salomaa V, Kontula K, Taskinen MR, Pajukanta P, Peltonen L. Cross-species analyses implicate Lipin 1 involvement in human glucose metabolism. Hum Mol Genet. 2006;15:377–386. doi: 10.1093/hmg/ddi448. [DOI] [PubMed] [Google Scholar]
- Alliel PM, Perin JP, Jolles P, Bonnet FJ. Testican, a multidomain testicular proteoglycan resembling modulators of cell social behaviour. Eur J Biochem. 1993;214:347–350. doi: 10.1111/j.1432-1033.1993.tb17930.x. [DOI] [PubMed] [Google Scholar]
- Marr HS, Basalamah MA, Bouldin TW, Duncan AW, Edgell CJ. Distribution of testican expression in human brain. Cell Tissue Res. 2000;302:139–144. doi: 10.1007/s004410000277. [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Chanoine C, Cifuentes-Diaz C, Gallien CL, Rieger F, Alliel PM, Perin JP. Expression of the proteoglycan SPOCK during mouse embryo development. Mech Dev. 2000;90:317–321. doi: 10.1016/S0925-4773(99)00255-5. [DOI] [PubMed] [Google Scholar]
- Marr HS, Edgell CS. Testican-1 inhibits attachment of Neuro-2a cells. Matrix Biology. 2003;22:259–266. doi: 10.1016/S0945-053X(03)00036-2. [DOI] [PubMed] [Google Scholar]
- Bonnet F, Perin JP, Charbonnier F, Camuzat A, Roussel G, Nussbaum JL, Alliel PM. Structure and cellular distribution of mouse brain testican. Association with the postsynaptic area of hippocampus pyramidal cells. J Biol Chem. 1996;271:4373–4380. doi: 10.1074/jbc.271.8.4373. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Sun Y, Zoghbi HY. Balanced X chromosome inactivation patterns in the Rett syndrome brain. Am J Med Genet. 2002;111:164–168. doi: 10.1002/ajmg.10557. [DOI] [PubMed] [Google Scholar]
- Amir RE, Zoghbi HY. Rett syndrome: methyl-CpG-binding protein 2 mutations and phenotype-genotype correlations. Am J Med Genet. 2000;97:147–152. doi: 10.1002/1096-8628(200022)97:2<147::AID-AJMG6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- BaSalamah MA, Marr HS, Duncan AW, Edgell CJ. Testican in human blood. Biochem Biophys Res Commun. 2001;283:1083–1090. doi: 10.1006/bbrc.2001.4900. [DOI] [PubMed] [Google Scholar]
- Bocock JP, Edgell CJ, Marr HS, Erickson AH. Human proteoglycan testican-1 inhibits the lysosomal cysteine protease cathepsin L. Eur J Biochem. 2003;270:4008–4015. doi: 10.1046/j.1432-1033.2003.03789.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transformation of gene annotations to official human gene symbols for across-microarray comparisons.
Comparison of gene lists from different microarray experiments.