Abstract
Potassium channels are necessary for a number of essential biological tasks such as the generation of action potentials and setting the resting membrane potential in cells, both of which require that these channels selectively permit the passage of potassium ions while suppressing the flow of other ions. Generally, this selectivity is attributed to a narrow stretch of the channel known as the selectivity filter. Over this stretch ions are dehydrated, and the backbone oxygen atoms of the protein mimic the ion's loss of coordination by water. However, channels are long pores with spatially distinct ion-binding sites that all must be traversed during ion permeation. We have shown that selectivity of mutant Kir3.2 (GIRK2) channels can be substantially amplified by introducing acidic residues into the cavity, a binding site below the selectivity filter. Here, we carry out electrostatic calculations on homology models to quantify the degree of stabilization that these mutations have on ions in the cavity. We then construct a multiion model of ion permeation to calculate the channel's permeability to potassium relative to sodium. This kinetic model uses rates derived from the electrostatic calculations and demonstrates that nonselective electrostatic stabilization of cations in the cavity can amplify channel selectivity independently of the selectivity filter. This nonintuitive result highlights the dependence of channel properties on the entire channel architecture and suggests that selectivity may not be fully understood by focusing solely on thermodynamic considerations of ion dehydration and the energetics of the selectivity filter.
Keywords: electrostatics, model, potassium channel, simulation, ion permeation
Potassium (K+) channels permit millions of potassium ions to cross the membrane per second, while suppressing by 1,000-fold the concomitant flux of the smaller sodium ions (1). How might such remarkable selectivity be achieved given that the potassium ion diameter is only 0.4 Å larger than the sodium ion diameter, yet thermal fluctuations in the protein are on the order of angstroms? Potassium channels have a characteristic GYG amino acid sequence, and the structure of the KcsA K+ channel reveals that the backbone carbonyl oxygen atoms of these residues coordinate K+ ions in the filter (2). It has been suggested that a better approximation of the hydration shell of K+ compared with other ions underlies the basis of selectivity (2–4). Indeed, simulations predict that the filter sites are several kilocalories per mole more selective for K+ (5–7). Nonetheless, x-ray structures show that the selectivity filter binding sites are just one of several distinct positions that must be occupied as ions transit the channel. There is a water-filled cavity below the filter that ions occupy (2, 4) and charged rings in the cytoplasmic domains of inwardly rectifying potassium (Kir) channels that are likely to be additional ion-binding sites (8–10). The presence of these sequential binding sites is important for physiological properties such as the steep voltage-dependent block of outward K+ current, known as inward rectification (11–13), but is there a role for these additional sites that fall outside of the selectivity filter in determining the selectivity of the channel?
It is well understood that residues at sites above and below the selectivity filter can have a powerful effect on the permeation properties of channels. Negatively charged residues at the extracellular mouth (14), water-filled cavity (15, 16), or the cytoplasmic pore (17) increase the rate of K+ conduction in potassium channels. On the other hand, positively charged residues at sites distant from the selectivity filter in the voltage-gated CLC-0 chloride channels increase chloride conductance (18). These observations can be understood in terms of the favorable electrostatic interaction of the permeant ion and the charged residues. Long-range electrostatic interactions dramatically increase the in rates to key sites in the channel, correspondingly increasing the steady-state occupancy of these sites; however, these charged interactions are generally nonselective, affecting ions with the same charge equally. Therefore, with regard to potassium channels, one might expect that charge mutations at nonselective sites would equally increase the channel's permeability to all cations.
The presence of multiple sites in a channel provides an alternate possibility. The rate of entry into one part of the channel may be very different for two species of ions, but this difference may be masked by a rate-limiting step elsewhere in the channel, which is comparable for both ions. Removing the rate-limiting step by judiciously mutating the channel will unmask intrinsic differences in the channel and bring about unexpected results, such as increased selectivity. This scenario is particularly plausible if there is a large range in the kinetic rates for movement between different sites. Support for this hypothesis comes from kinetic models of ion permeation for K+ channels, which predict that the rate constants connecting different ion configurations span three to four orders of magnitude (19, 20). However, dissecting the relative contributions from different parts of the channel to the total conductance is a difficult task, yet important if we are to better understand how ion channels function, how they can be regulated, and how they evolved.
Together with a companion paper (21), we use a combination of experimental and computational methods to understand the importance of the cavity site in controlling ion selectivity. Starting from a mutation, S177W, behind the selectivity filter that renders the Kir3.2 channel nonselective, we discovered that introducing charged mutations at pore-facing positions in the cavity dramatically restores K+ selectivity. Here, we use molecular and kinetic modeling to explore the mechanism by which K+ selectivity is reinstated. We show that electrostatic stabilization of cations in the channel cavity increases K+ selectivity by changing rates into and out of the cavity and increasing the occupancy of the cavity-bound states, but not by changing the inherent selectivity of the selectivity filter. Thus, all of our analysis supports the notion that tuning a nonselective ion-binding site outside of the selectivity filter, from a weak cation binder to a strong cation binder, shifts the rate-limiting step for ion movement from a state that is relatively nonselective for K+ over Na+ to one that is more selective for K+. This change brings about a dramatic increase in selectivity and highlights the importance of the entire channel in controlling conduction properties.
Results
Electrostatic Interactions in the Cavity Enhance K+ Selectivity.
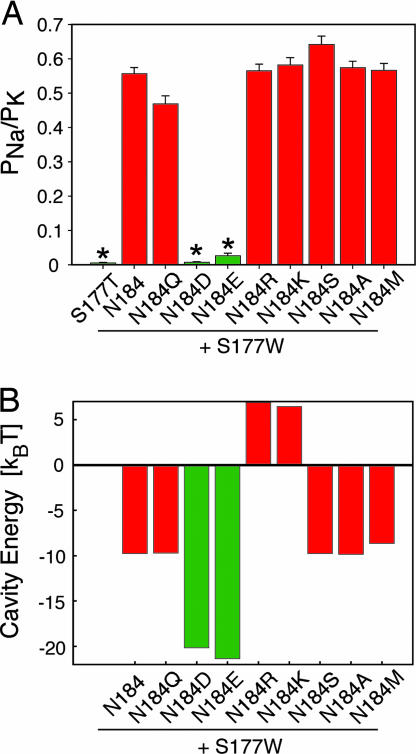
In our companion study (21), we show that substitution of S177 by tryptophan on the inner helix of Kir3.2 channels nearly abolishes the channel's ability to discriminate between Na+ and K+ ions, even though these channels still bear the intact K+ channel signature sequence. Starting from this nonselective channel, we identified the cavity facing position N184 on the inner helix (M2) as a key regulator of K+ selectivity. We replaced N184 with a wide range of amino acids, all in the background of the nonselective S177W channel, and measured changes to ion selectivity, as indicated by the relative permeability of Na+ to K+ (PNa/PK) (Fig. 1A). Interestingly, acidic substitutions restored K+ selectivity by 20- to 70-fold, whereas all other substitutions resulted in nonselective channels. We probed the mechanism underlying the enhancement of K+ selectivity by the negatively charged Asp (D) and Glu (E) on the M2 helix by quantifying the degree of electrostatic stabilization of the cavity ion with continuum-electrostatic calculations by using Kir3.2 homology models (Fig. 1B). Whereas the free energy of transfer for a monovalent cation from bulk water to the cavity is −10 kBT for WT (N) and neutral substitutions (Q, S, A and M), similar to values reported for KcsA (22), N184D/E increases the binding by −10 kBT, whereas N184R/K destabilizes the ion by ≈15 kBT (Fig. 1B). Notably, these large changes in binding energy will contribute exponentially to a bias in the distribution of ions in the cavity. However, continuum-electrostatic calculations predict no change in selectivity of K+ over Na+ at this cavity site. Looking for higher-order effects, we performed free energy perturbation calculations on fully hydrated, membrane-embedded Kir3.2 channels with or without the N184D mutation. These results revealed that there was no free energy difference between Na+ and K+ in the cavity (data not shown) leaving us with the question: how might electrostatic interactions between the channel and cavity ion affect selectivity without favoring K+ over Na+ in the cavity?
Fig. 1.
Negatively charged amino acids restore K+ selectivity and increase cation binding in the cavity. (A) Dependence of permeability ratio (PNa/PK) on amino acid substitution at position N184. Permeability ratios (PNa/PK) were deduced from the difference in reversal potential measured in oocytes bathed in 90 mM Na+ or 90 mM K+. Only acidic substitutions at N184 (N184D/E) restore K+ selectivity to S177W-containing mutant channels (values are mean ± SEM; values for selective channels are tertiapin-corrected; ∗, P < 0.001). (B) Dependence of the electrostatic free energy of the cavity ion on mutations at position N184. Negative charges (N184D/E) stabilize monovalent cations by −10 kBT, whereas basic substitutions (N184R/K) destabilize the cavity ion by 15 kBT, relative to WT channels. In agreement with experimental evidence, pKa calculations reveal that only two of the four aspartate residues facing the cavity ion are charged (36); therefore, in all calculations, we assume that only two of the four mutant residues are charged when they are either basic or acidic residues (see Materials and Methods).
Models of Ion Permeation.
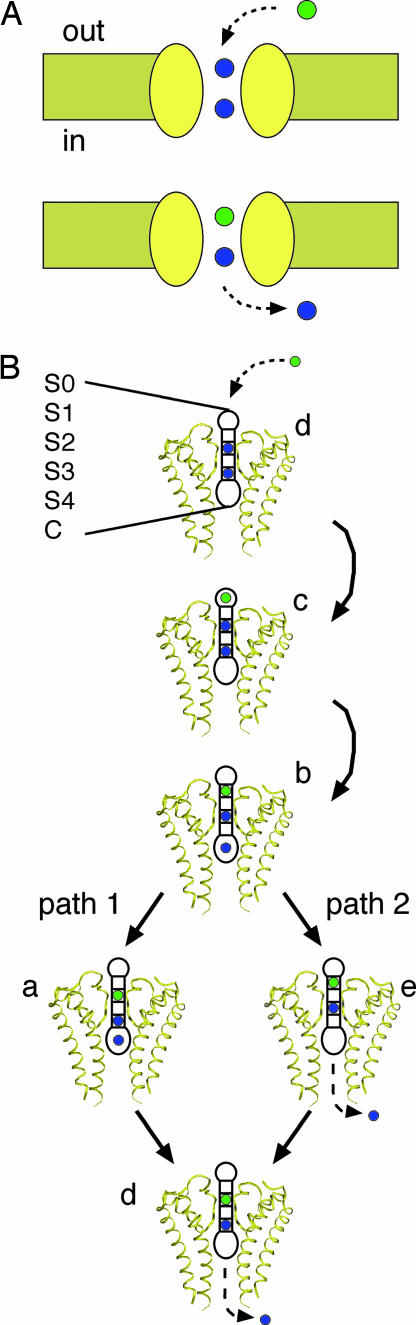
Our analysis thus far suggests that the primary effect of the charge mutations in the cavity is to change the cavity-binding energetics of K+ and Na+ equally, while leaving the selectivity filter geometry largely unchanged (21). We set out to explore how stabilizing K+ and Na+ equally in the cavity could enhance K+ selectivity by using realistic kinetic models in which the rate constants reflect the underlying physics of ion movement through the channel. We begin with a simplified scenario and imagine a channel harboring two ions (blue) with an ion (green) poised to enter the channel from the extracellular solution (Fig. 2A). According to the “knock-on model,” ion movement in the multiion, single-file channel is tightly coupled, and the entry of the green ion displaces an ion already in the channel, forcing it into the intracellular space (23). Repeating this process results in the translocation of ions from outside to inside the cell. Detailed models of this sequence of events in a more realistic channel pore have been developed and tested based on physiological measurements and high-resolution structures of KcsA (20). We have adapted this scheme to Kir3.2, and the inward translocation of an ion can be seen in Fig. 2B where each step along the pathway is represented by a ribbon diagram of the full channel. Ions reside in one of six distinct positions, in the extracellular site (S0), the filter (S1–S4), or the central cavity site (C). Inward transport occurs in two distinct modes via path 1 (d → c → b → a → d) or path 2 (d → c → b → e → d). Note that, the cavity site remains occupied along path 1 but not path 2. This difference immediately suggests a nontrivial role for the cavity ion-binding site in determining the kinetics of transport.
Fig. 2.
Single species models of ion permeation. (A) An abstract representation of a channel spanning the membrane to illustrate ion movement through a multiion pore. On the top, an extracellular ion (green) enters the channel causing the bottom ion in the channel to enter the intracellular space. This process moves a single ion from the outside of the cell to the inside. (B) A realistic model of ion movement through a potassium channel is specified by five states, a–e. The channel has six discrete ion binding sites labeled according to standard nomenclature: S0, extracellular site; S1–S4, filter sites; C, cavity site. Explicit representations of the channel illustrate how these binding sites are positioned within the channel. Inward flow is initiated by an extracellular ion entering the channel, d → c, and permeation proceeds via path 1 (b → a → d) or path 2 (b → e → d). Just as in A, this process moves a single ion from the outside of the cell to the inside.
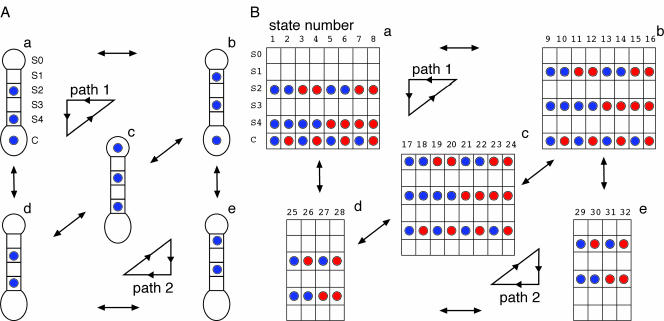
Next, we discuss the extension of this model to allow for both inward and outward movements of ions (Fig. 3A) and the possibility of multiple ion species in the channel at once (Fig. 3B). Whereas the scheme in Fig. 2B depicts unidirectional movements of ions entering the cell, a realistic model for ion permeation has to incorporate the equally important outward flow of ions through the channel. In this more complete model, the equivalent kinetic scheme can be redrawn as in Fig. 3A. Here, inward current is depicted by counterclockwise transitions along path 1 or clockwise transitions along path 2. This model, although accurate, only allows for one kind of ion in the channel at a time. Experimentally, we determine permeability ratios from reversal potential measurements under bi-ionic conditions with both K+ and Na+ present. Therefore, we must consider one final generalization of the scheme, which allows for mixing of both K+ (blue) and Na+ (red) ions in the channel (Fig. 3B). In this case, the total number of configurations jumps from 5 to 32, but the spatial organization of the ions in the channel remains the same. We therefore assigned each of the new states to a subgroup corresponding to one of the states (a to e) in Fig. 3A, so that there are eight states each in subgroups a–c and four states each in subgroups d and e (Fig. 3B). We refer generically to rates from states in subgroup a to those in subgroup b by kab. Realize that this is a shorthand notation and not all rates between subgroups are equal. We now want to motivate the choice of kinetic rate constants through the channel for both the K+ and Na+ ions.
Fig. 3.
Multispecies model of ion permeation. (A) A model of reversible ion permeation. The backward transitions, not illustrated in Fig. 2B, are required for microscopic reversibility. Rewriting the state diagram, we produce this equivalent abstract model connected by 12 rate constants (2 per arrow). (B) A multiion model of reversible ion permeation. This model allows for potassium (blue) or sodium (red) at each position. It has 32 states that are grouped into five subgroups, a–e, corresponding to the five states in A.
Model Rates Predicted from Molecular Calculations and Experiment.
As suggested by our molecular calculations with K+ or Na+ ions in the cavity, we require that the energies of both ions in the cavity and their rates of movements between the cavity and the cytoplasm are equal. Most importantly, all cavity rates depend on mutations at position 184, such that enhanced cation binding in the cavity (N184D/E) increases, to the same extent, the in rates for moving K+ or Na+ into the cavity (kda, keb, and kcb) and decreases, to the same extent, the out rates for moving K+ or Na+ out of the cavity (kad, kbe, and kbc) (details in Supporting Text, which is published as supporting information on the PNAS web site). Rates were determined subject to these constraints by simultaneously fitting the model to the single-channel conductance values measured in isotonic KCl or isotonic NaCl solutions (Fig. 7, which is published as supporting information on the PNAS web site) and the permeability ratios determined under biionic conditions for the S177W mutant channel (see Supporting Text).
A Kinetic Mechanism Explains the Gain in K+ Selectivity.
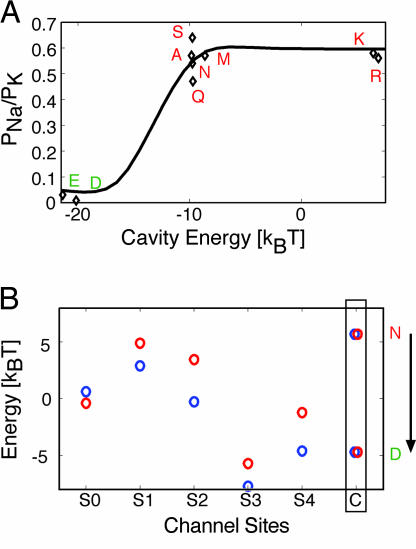
Using this realistic model for K+ and Na+ permeation through the S177W channel, we numerically determined the reversal potential and PNa/PK ratio. Remarkably, this kinetic model predicts that electrostatic stabilization of the cavity ion makes the channel extremely selective for K+ (solid line in Fig. 4A). Moreover, this result is robust and can be reproduced with fewer parameters with a minimal model (see Fig. 8, which is published as supporting information on the PNAS web site). This finding supports the dependence of the experimentally measured permeability ratios of S177W–N184X on these mutants' electrostatic binding energy predicted by continuum calculations (diamonds in Fig. 4A). Interestingly, significant gain of selectivity requires a 10-kBT change in cavity binding, which is the incremental change elicited by introducing a negative charge at position 184 (Fig. 1B). This finding explains the binary response of selectivity recovery in Fig. 1A.
Fig. 4.
The kinetic model predicts experimentally measured PNa/PK values. (A) Predicted permeability ratios versus the electrostatic component of the cavity binding energy for the S177W channel. The multiion model (solid curve), solved in Supporting Text, corresponds well with the experimental PNa/PK values (diamonds). Single-letter amino acid codes indicate mutations at N184; in red are the nonselective mutant channels (N, Q, S, A, M, R, and K) and in green are the K+-selective ones (D and E). (B) Single ion energies for sodium (red circles) and potassium (blue circles) at all six binding sites along the channel pore (S0, S1, S2, S3, S4, and C). The energy of both ions is identical in the cavity site (C) and is altered by the same amount by charge mutations in the cavity; however, K+ is favored over Na+ at all positions except the extracellular site S0. For comparison to calculations on a selective channel, KcsA is predicted to be K+-selective by −2.2, +4.3, +8.8, +3.0, and −2.0 kBT for sites S0–S4 (24). In the present simulations, filter binding energies, S0–S4, were determined from kinetic modeling as described in Supporting Text.
Some insight into the channel's ability to select K+ over Na+ may be gleaned from the energetic predictions of this model for K+ (blue circles in Fig. 4B) and Na+ (red circles in Fig. 4B). The free energies for K+ and Na+ in the cavity are equal and altered to the same extent by charge mutations (Fig. 4B), as expected from continuum-electrostatic calculations (Fig. 1B). Without negatively charged cavity-facing residues to stabilize the cavity ion, state e is stabilized relative to state a forcing conduction through path 2 (see Figs. 2B and 3). Along this pathway, both K+ and Na+ are rate-limited by the exit of the cavity ion to the cytoplasm (b → e) because of low occupancy of state b, because the transition from c to b is also energetically unfavorable in the absence of electrostatic stabilization of the cavity ion. This step is comparable for K+ and Na+ because kbe is equivalent for both ions, and the filter sites S1 and S3 are only slightly selective for K+ over Na+, ≈2 kBT each (Fig. 4B), which is about half the selectivity predicted for these sites in WT KcsA (24). With negative charges in the cavity, however, state a is stabilized and conduction occurs via path 1, but the cavity ion dwell time is lengthened because of electrostatic interactions, thus making it difficult for the cavity ion to exit to the cytoplasm (a → d). This increased dwell time provides more opportunity for the filter to select for the filter ions, particularly in light of the nearly doubled selectivity at sites S2 and S4 compared with S1 and S3 (Fig. 4B), thereby bringing about the dramatic changes to the permeability ratio seen in Fig. 4A. We will return to this theme in Discussion and use a simplified scenario to illustrate the basis of kinetic regulation of K+ selectivity (see Fig. 6).
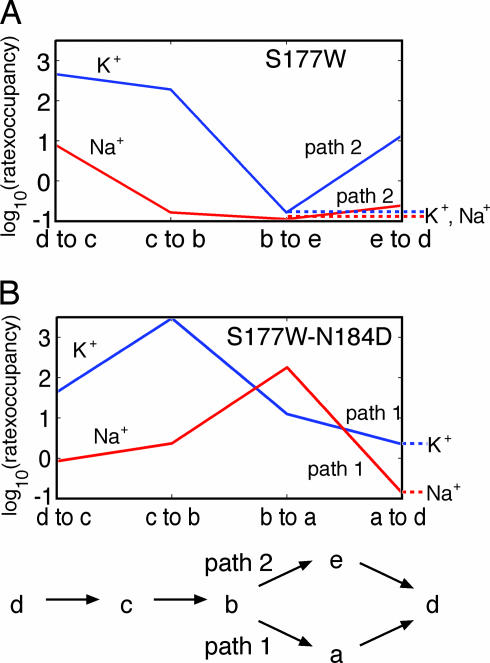
Fig. 6.
Stabilizing the cavity ion affects K+ and Na+ rate-limiting steps differently. We used the multiion model to analyze the rate-limiting steps for both K+ and Na+ movement under conditions of inward flow (100 mM outside, 10 mM inside). Different species were considered separately for illustrative purposes. The units of the y axis are log of picoamps, and only the dominant pathway is shown for each case. (A) Inward, unidirectional flux for the S177W nonselective channel. Exit of the cavity ion to the cytoplasm is rate-limiting for K+ and Na+ because of the reduced occupancy of state b. (B) Inward, unidirectional flux for the S177W–N184D K+-selective channel. The rate-limiting step for both K+ and Na+ is an exit of the cavity ion to the cytoplasm (this rate constant is assumed equal for both ions). Na+ flux through this pathway is reduced because of greater selectivity at the S2 and S4 filter sites, which reduces the steady-state occupancy of state a relative to K+. We note that the net flux (inward − outward) is constant along the permeation pathway, but here we show only the inward component of the flux.
Recovery of K+ Selectivity Is Independent of the Mutation Causing the Initial Loss of Selectivity.
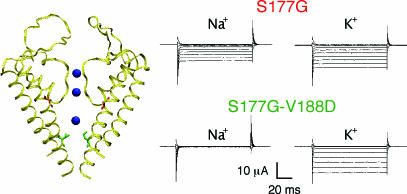
It is crucial to our analysis that the mutations at positions 177 and 184 independently affect the channel as suggested by our theoretical analysis (21). To address this concern, we discovered that S177G, like S177W, abolishes channel selectivity (Fig. 5, PNa/PK = 0.4 ± 0.1 SEM). The marked size difference between Gly (G) and Trp (W) suggests that the details of selectivity loss are different in these cases. Our next step was to determine whether a negative charge introduced to face 1 of M2 could recover this loss of selectivity. Indeed, the S177G–V188D double mutant channels are highly selective (PNa/PK = 0.03 ± 0.01 SEM). These findings further buttress the notion that the enhanced K+ selectivity is a general feature of cavity ion stabilization as suggested by our theory.
Fig. 5.
The nonselective S177G mutant Kir3.2 channel is also suppressed by aspartate (D) mutation in the channel cavity (V188D). (Left) Homology model of Kir3.2 channel with the S177G (red) mutation on the M2 helix, behind the selectivity filter, and V188D (green) at a pore-facing position in the cavity. (Right) Current traces for S177G (Upper) illustrate that the channel is nonselective and permeates both Na+ and K+. The V188D mutation restores K+ selectivity to S177G-bearing channels as shown by the S177G–V188D (Lower) current recorded in Na+ or K+ solutions. Current traces were recorded at membrane potentials ranging from +40 to −140 mV in 20-mV increments from Xenopus oocyte expressing mutant channels and bathed in 90 mM Na+ or K+ solutions.
Discussion
Charge mutations have a powerful effect on the interaction of permeant ions with the channel (Fig. 1B), and they have been shown to have an equally large impact on the channel conduction properties (14, 15, 17, 18). It is not surprising then that single-charge mutations should also affect the K+ selectivity of a channel (Fig. 1A); in fact, basic substitution of a pore-lining acidic residue switches the cation-anion selectivity of a cyclic nucleotide-gated channel (25). What is surprising is that this change in selectivity is accomplished in a completely nonselective way at a site that is not the selectivity filter. Using a standard model of potassium channel ion transport, we show that our experimental results can be entirely explained by nonselective stabilization of cations in the cavity site without appealing to the possibility that charge mutations in the cavity affect the geometry of the selectivity filter (see Fig. 4B).
We further illustrate this mechanism by considering the simplified scenarios of a channel under a strong inward concentration gradient of only K+ or Na+ (Fig. 6); the same conclusions hold for the multiion model used to compute Fig. 4A. The ion energies in Fig. 4B influence the probability of occupancy of each state depicted in Fig. 3A and the transition rates between states. Without electrostatic stabilization of the cavity ion by negatively charged residues, it is more energetically costly (and less likely) to occupy state b in which the cavity site is filled. In this case path 2 is preferred by both ions, and the rate-limiting steps for K+ or Na+ permeation are similar (Fig. 6A). Because the model predicts that filter sites S1 and S3 have very small selectivity (Fig. 4B), the steady-state populations at the bottleneck (state b) are nearly equal, resulting in roughly equal flux of both ions. In contrast, with negative charges to stabilize the cavity ion, states b and a are readily occupied by both K+ and Na+, and ions move primarily along path 1 (Fig. 6B). K+ ions have no difficulty moving through the selectivity filter and entering state a, whereas Na+ ions experience greater discrimination at sites S2 and S4 (Fig. 4B); hence, conduction along path 1 is suppressed for Na+ despite the stabilization of the cavity ion. As a result, the rate-limiting step for K+ is much less demanding than the rate-limiting step for Na+ (Fig. 6B). It is in this manner that tuning the stabilization of the cavity ion moved the rate-limiting step away from a state that is relatively nonselective (state b) to one that is more selective (state a) (Fig. 4B), and therefore brings about amplification of the K+ selectivity (Fig. 4A).
K+ selectivity is generally discussed in terms of the free energy of K+ compared with other ions at the selectivity filter sites. Although such a distinction is required at some sites or barriers along the channel, it is not sufficient to ensure high selectivity. Here we show with kinetic modeling, that even when some filter sites are selective, by 4–5 kBT, this selectivity does not translate to a selective channel. Adjusting rates elsewhere in the channel brings out this selectivity. This effect is especially important when the channel sites are only mildly selective; in fact, our models predict that the channel is selective for all cavity-binding energies when all filter sites are very selective, >5 kBT. It is most likely that the selectivity of the filter sites was only modest at the earliest points of evolution. Therefore, it is at these nascent stages that the adoption of multiple ion binding sites may have been most important.
Although most K+ channels are highly selective, as we show here with reengineered channels, simply measuring the selectivity does not delineate the contribution to selectivity from the selectivity filter alone versus the cavity. We have focused on the effects of charge mutations in the cavity site, but any binding site is likely to affect the mode of ion conduction and thus channel selectivity according to our model, although the details may be different. Cation-binding sites are found throughout the extracellular, transmembrane, and cytoplasmic domains of various channels, including Kir channels (1, 26–31). The presence of these multiple ion-binding sites increases the complexity of transport in unexpected ways, as shown here, and suggests an expanded role for the different domains of the channel.
Materials and Methods
Constructs and Oocyte Expression, Electrophysiology, and Permeability Measurements.
A description of all experimental techniques can be found in the companion paper (21).
Modeling.
We used Modeller8v0 (32) to construct homology models of Kir3.2 by using KirBac1.1 as a template structure as described (21). A representative S177W mutant structure was chosen, and subsequent mutations at position 184 were generated by using the PSFGEN module in NAMD2.5 (33). Channels were minimized for 3,000 steps in vacuum by using the CHARMM27 parameter set with full electrostatics in NAMD (33). A high dielectric constant of 40.0 was used to account for the lack of water surrounding the cavity ion. The choice of this value did not strongly influence the final free energy values. Potassium ions were included in the S1 and S3 filter sites and the cavity and held fixed during minimization.
Electrostatic calculations were carried out by using the program APBS with the PARSE parameter set (34). All calculations treated protein as dielectric 10.0 and water as dielectric 80.0 and included a symmetric bath of 0.1 M counterions. The effect of the membrane was added as a continuous slab of dielectric 2.0 (35). For free energy calculations for the transfer of ions from bulk water to the cavity, we calculated the energy of the ion in the channel and subtracted the energy of the channel system without the cavity ion and the energy of the ion in bulk water. Estimates of pKa shifts indicate that at most two N184D residues are charged (see Supporting Text), in accord with experiments on pore lining charges (36). Hence, all calculations involving basic or acidic mutations were performed with two charged residues in the pore from diagonal subunits.
The kinetic model was solved in Matlab as detailed in Supporting Text. Rate constants, ion concentrations, and additional details on solving the model can be found in Supporting Text.
Supplementary Material
Acknowledgments
We thank B. Cohen for comments on the manuscript; H. Lecar and H. Y. Wang for assistance in constructing the model; the Pittsburgh Supercomputing Center (MCB030034P) in Pittsburgh, PA, and D. Agard for computer time; F. Lesage (Centre National de la Recherche Scientifique, Unité Mixte de Recerche 6097, Valbonne, France) for Kir3.2 cDNA; and Z. Lu (University of Pennsylvania, Philadelphia, PA) for tertiapin. M.G. was supported by a National Science Foundation Interdisciplinary Informatics Fellowship. L.Y.J. and Y.N.J. are Investigators of the Howard Hughes Medical Institute. This work was supported by National Institute of Mental Health Grant MH065334.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sinauer: Sunderland, MA; 2001. [Google Scholar]
- 2.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Bezanilla F, Armstrong CM. J Gen Physiol. 1972;60:588–608. doi: 10.1085/jgp.60.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 5.Berneche S, Roux B. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 6.Aqvist J, Luzhkov V. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- 7.Allen TW, Bliznyuk A, Rendell AP, Kuyucak S, Chung SH. J Chem Phys. 2000;112:8191–8204. [Google Scholar]
- 8.Nishida M, MacKinnon R. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 9.Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 10.Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, Choe S. Nat Neurosci. 2005;8:279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 11.Hille B, Schwarz W. J Gen Physiol. 1978;72:409–442. doi: 10.1085/jgp.72.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopatin AN, Makhina EN, Nichols CG. J Gen Physiol. 1995;106:923–955. doi: 10.1085/jgp.106.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spassova M, Lu Z. J Gen Physiol. 1998;112:211–221. doi: 10.1085/jgp.112.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haug T, Sigg D, Ciani S, Toro L, Stefani E, Olcese R. J Gen Physiol. 2004;124:173–184. doi: 10.1085/jgp.200308949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brelidze TI, Niu X, Magleby KL. Proc Natl Acad Sci USA. 2003;100:9017–9022. doi: 10.1073/pnas.1532257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Niu X, Brelidze TI, Magleby KL. J Gen Physiol. 2006;128:185–202. doi: 10.1085/jgp.200609493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara Y, Kubo Y. J Gen Physiol. 2006;127:401–419. doi: 10.1085/jgp.200509434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen MF, Chen TY. J Gen Physiol. 2003;122:133–145. doi: 10.1085/jgp.200308844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morais-Cabral JH, Zhou Y, MacKinnon R. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 20.Kutluay E, Roux B, Heginbotham L. Biophys J. 2005;88:1018–1029. doi: 10.1529/biophysj.104.052043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bichet D, Grabe M, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2006;103:14355–14360. doi: 10.1073/pnas.0606660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roux B, MacKinnon R. Science. 1999;285:100–102. doi: 10.1126/science.285.5424.100. [DOI] [PubMed] [Google Scholar]
- 23.Hodgkin AL, Keynes RD. J Physiol (London) 1955;128:28–60. doi: 10.1113/jphysiol.1955.sp005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noskov SY, Berneche S, Roux B. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 25.Qu W, Moorhouse AJ, Chandra M, Pierce KD, Lewis TM, Barry PH. J Gen Physiol. 2006;127:375–389. doi: 10.1085/jgp.200509378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo Y, Murata Y. J Physiol (London) 2001;531:645–660. doi: 10.1111/j.1469-7793.2001.0645h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo D, Ramu Y, Klem AM, Lu Z. J Gen Physiol. 2003;121:261–275. doi: 10.1085/jgp.200208771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanfield PR, Davies NW, Shelton PA, Sutcliffe MJ, Khan IA, Brammar WJ, Conley EC. J Physiol (London) 1994;478:1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Jan YN, Jan LY. Neuron. 1995;14:1047–1054. doi: 10.1016/0896-6273(95)90343-7. [DOI] [PubMed] [Google Scholar]
- 30.Xie LH, John SA, Weiss JN. J Physiol (London) 2003;550:67–82. doi: 10.1113/jphysiol.2003.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Z, MacKinnon R. Nature. 1994;371:243–246. doi: 10.1038/371243a0. [DOI] [PubMed] [Google Scholar]
- 32.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comp Phys. 1999;151:283–312. [Google Scholar]
- 34.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabe M, Lecar H, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2004;101:17640–17645. doi: 10.1073/pnas.0408116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Root MJ, MacKinnon R. Science. 1994;265:1852–1856. doi: 10.1126/science.7522344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.