Abstract
Potassium channels are membrane proteins that allow the passage of potassium ions at near diffusion rates while severely limiting the flux of the slightly smaller sodium ions. Although studies thus far have focused on the narrowest part of the channel, known as the selectivity filter, channels are long pores with multiple ions that traverse the selectivity filter, the water-filled central cavity, and the rest of the pore formed by cytoplasmic domains. Here, we present experimental analyses on Kir3.2 (GIRK2), a G protein-activated inwardly rectifying potassium (Kir) channel, showing that a negative charge introduced at a pore-facing position in the cavity (N184) below the selectivity filter restores both K+ selectivity and inward rectification properties to the nonselective S177W mutant channel. Molecular modeling demonstrates that the negative residue has no effect on the geometry of the selectivity filter, suggesting that it has a local effect on the cavity ion. Moreover, restoration of selectivity does not depend on the exact location of the charge in the central cavity as long as this residue faces the pore, where it is in close contact with permeant ions. Our results indicate that interactions between permeant ions and the channel cavity can influence ion selectivity and channel block by means of an electrostatic effect.
Keywords: inward rectification, permeation, permeability, GIRK modeling, Kir3
Potassium channels are found in all kingdoms and domains and serve a wide range of important physiological functions, such as volume regulation and potassium transport in plants (1, 2) and the control of insulin release, blood flow, heart rate, neuronal signaling, and potassium secretion in the kidney and inner ear of animals (3–5). These physiological functions rely critically on the channel's ability to permeate K+ and not Na+, which is smaller and more abundant in nature. To understand how K+ channels fulfill their physiological functions, and to gain some appreciation as to how their remarkable selectivity for K+ permeation might have evolved, it is important to characterize all of the features of the K+ channel pore that contribute to its ability to discriminate between potassium and other ions.
Our understanding of K+ channel selectivity has gained considerable insight from the KcsA potassium channel crystal structure, which revealed that four identical subunits come together to form a central pore (6). The pore is most constricted over a narrow span, termed the selectivity filter, which is formed by four loops bearing the amino acid sequence GYG near the extracellular side of the membrane. This sequence is highly conserved throughout the K+ channel superfamily and is thought to underlie the basis of K+ selectivity; therefore, it is called the K+ channel signature sequence (7). Although it is clear that this sequence plays a key role in selectivity, it makes up only a fraction of the channel's entire permeation pathway, and the relative contributions to K+ selectivity from the selectivity filter and the rest of the channel pore are still not known.
This gap in our knowledge is highlighted by the existence of channels that bear the K+ channel signature sequence yet are poor at discriminating K+ from Na+, such as the hyperpolarization-activated cyclic nucleotide-gated cation channel (8, 9), the Slick channel (2) and the rSK2 channel (10). As for those channels that we call K+ channels, selectivity ranges from a 1,000- to 10-fold preference for K+ over Na+ (4). Even for the same K+ channel, ion selectivity may vary with the extent of channel activation and/or inactivation (11–13). It is not yet understood whether these wide-ranging differences in ion selectivity could be accounted for by differences in the conformation of the selectivity filter and/or by structural features elsewhere in the channel.
Mutations outside the selectivity filter, including positions in the transmembrane helix (14, 15) and the cytoplasmic domain (16), have been shown to affect selectivity, although the underlying mechanism is unknown. In a previous study, we identified mutations in the pore-lining inner M2 segment of Kir3.2 [a G protein-activated inwardly rectifying potassium (Kir) channel], such as the S177W mutation, that greatly reduce the channel's ability to discriminate between K+ and Na+ (15). With this nonselective channel as a starting point, we sought to define determinants of K+ selectivity by using molecular evolution and searched for second-site suppressors that restore K+ selectivity to the S177W mutant channel. We identified several second-site suppressors, all outside the selectivity filter, revealing previously unidentified structural elements that are important for K+ selectivity and suggesting that selectivity may not be controlled solely by the selectivity filter (15). By following up on a particular second-site suppressor in the channel central cavity, we now demonstrate that K+ selectivity can be regulated through judicious engineering of cavity residues in Kir3.2 channels. We show that introducing a negative charge at pore-facing positions in the channel's central cavity dramatically increases the selectivity for K+ over Na+ and improves channel block by intracellular cations and polyamines, a feature known as inward rectification. The experimental data presented here and the theoretical analyses presented in our companion paper (17) strongly support an electrostatic mechanism. These findings then raise the possibility that the long pore of K+ channels not only allows channel modulation by physiologically relevant blocking ions but also enables incremental improvements of K+ selectivity during evolution before the perfection of the selectivity filter.
Results
Mutations in the Pore-Lining Inner Transmembrane Helix (M2) of Kir3.2 Affect K+ Selectivity.
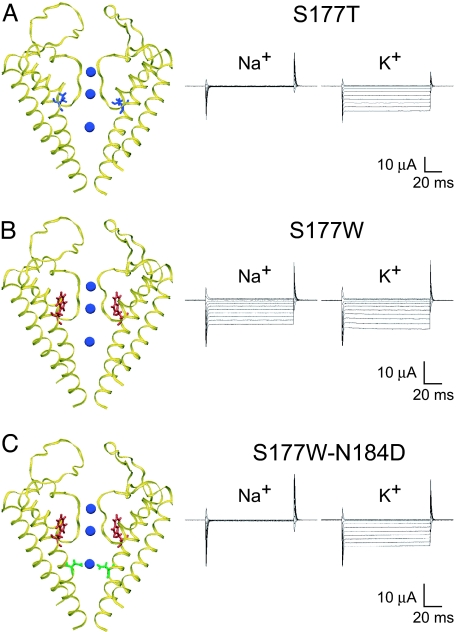
A previous study of Kir3.2 channels identified a conservative mutation in the M2 inner helix, S177T, that causes the channel to open in the absence of regulatory Gβγ proteins while leaving other properties unchanged (14) (Fig. 1A). Meanwhile, a less conservative substitution at this position, S177W, nearly abolishes the channel's ability to discriminate between Na+ and K+ ions (Fig. 1B), even though these channels still bear the intact K+ channel signature sequence. Starting from this nonselective channel, we used the molecular evolution approach to identify several mutations, all within the transmembrane domains, that restore selectivity to the double mutant channel bearing the S177W mutation (15). In particular, the N184D mutation of the M2 inner helix completely restores K+ selectivity (Fig. 1C). To begin our analysis of how these mutations affect K+ selectivity, we first explore the possibility that the S177W mutation compromises K+ selectivity by causing some distortion of the selectivity filter and then ask how the N184D mutation might restore K+ selectivity.
Fig. 1.
Mutations of the M2 inner transmembrane helix residues control K+ selectivity. (A) (Left) Homology model of the Kir3.2 channel with the S177T (blue) mutation on the M2 inner helix, behind the selectivity filter. (Right) Representative current traces obtained from Xenopus oocytes expressing S177T channels bathed in 90 mM Na+ or 90 mM K+ show that S177T channels carry K+ current but not Na+ current. (B) Kir3.2 channel with an S177W (red) mutation that fills the space behind the selectivity filter. Current traces illustrate large inward fluxes for both K+ and Na+ for this nonselective S177W mutant. (C) Kir3.2 double mutant channel with a S177W (red) mutation and a N184D (green) mutation in the center of the central cavity near a K+ ion (blue). The double mutant channel has regained K+ selectivity and exhibits similar current traces to the K+-selective S177T channel. Currents were recorded at membrane potentials ranging from +40 to −150 mV from a holding potential of 0 mV and are shown in 20-mV increments.
Effect of the M2 Helix Mutations on the Geometry of the Selectivity Filter.
Homology models of the Kir3.2 channel that are based on the crystal structure of the closely related bacterial Kir channel KirBac1.1 (18) provide guidance on how these M2 mutations might affect the selectivity filter (Figs. 1 and 2). S177T (blue residue in Fig. 1A) is situated between the pore helices, where it causes only minor perturbations to the selectivity filter (blue curve in Fig. 2). In contrast, Trp (W) at this position (red residue in Fig. 1 B and C) fills the space behind the selectivity filter and causes systematic deviations to the filter (red curve in Fig. 2). Thus, selectivity filter distortions by S177W, but not by S177T, likely contribute to the differences in selectivity between these two mutant channels. It is possible that S177W decreases filter flexibility, consistent with a theory that a rigid filter cannot discriminate between ions (19). Alternatively, S177W could have reduced K+ selectivity by displacing Y157, a critical site for KcsA selectivity (19). N184D (green residue in Fig. 1C), however, is >9 Å away and protrudes into the central pore, adjacent to the cavity ion. N184D alone does not produce systematic rearrangements in the selectivity filter (dashed green curve); the S177W-N184D double mutant channel has a pattern of distortion (green curve) similar to that of the S177W single mutant channel (Fig. 2). Thus, it appears that N184D restores K+ selectivity without readjusting the selectivity filter.
Fig. 2.
Average distortions in the selectivity filter introduced by mutant residues. The relative displacement of amino acids from the K+ signature sequence, TTIGYG, was deduced from Kir3.2 mutant models. The filters of S177T (blue) and N184D (green dashed) mutant channels are similar to WT models (not shown). However, S177W (red) introduces systematic distortions into the filter (up to 1.5 Å). Consistent with this observation, there is greater variation in the filter geometry for S177W models compared with WT models. The N184D mutant combined with S177W (green) does not restore the pattern of distortion created by S177W (red). The S177W-N184D and N184D distributions are significantly different at all positions as analyzed with nonparametric statistics.
Only Negative Charges in the Cavity Enhance K+ Selectivity.
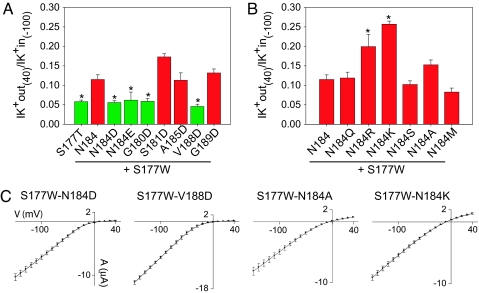
To explore how this cavity residue affects selectivity, we replaced N184 with amino acids comprising a range of sizes and chemistries (Q, E, R, K, S, A, and M, in addition to D), all in the background of the nonselective S177W channel (Fig. 3). We then examined the currents generated by these double mutant channels in Xenopus oocytes by using two-electrode voltage clamp in either Na+- or K+-containing solutions (Fig. 6, which is published as supporting information on the PNAS web site). Selectivity is quantified by the relative permeability ratio of Na+ to K+ (PNa/PK) and determined under biionic conditions from reversal potential values of the toxin-sensitive Kir3.2 current (20) (Table 1). Only N184E restored K+ selectivity in a manner similar to N184D, whereas the other substitutions resulted in nonselective channels. The double mutant channels, S177W-N184D and S177W-N184E, did not produced significant inward current in 90 mM Na+ bath solution compared with the S177W single mutant channel and various S177W-N184X (where X is A, M, Q, S, K, or R) double mutant channels (Fig. 3). Acidic substitution reduced the PNa/PK ratio to values comparable with WT Kir3.2 channels, whereas hydrophobic, hydrophilic, and basic substitution resulted in nonselective channels (Table 1) (21). These experiments reveal that only acidic residues at this pore-facing site in the channel cavity are capable of reinstating K+ selectivity to the nonselective S177W channel.
Fig. 3.
Negatively charged amino acids in the cavity restore K+ selectivity. Only acidic substitutions at the N184 position of Kir3.2 restore K+ selectivity to the nonselective S177W mutant channel. Current traces were elicited with voltage pulses from +40 to −150 (in 10-mV increments) from a holding potential of 0 mV in 90 mM K+ (green line) and 90 mM Na+ (red line). Only traces at −150 mV are shown. All N184 substitutions are in the background of the S177W mutation. Baseline indicates 0 μA current value. (Scale bars: vertical, 10 μA; horizontal, 20 ms.)
Table 1.
Relative permeabilities for Na+ and K+ ions of mutant Kir3.2 channels
| Mutation(s) | ΔE | PNa/PK | n | After TPNQ subtraction |
||
|---|---|---|---|---|---|---|
| ΔE | PNa/PK | n | ||||
| S177T | −74.6 ± 1.9 | 0.06 ± 0.004 | 10 | −131.4 ± 3.6 | 0.006 ± 0.0009 | 4 |
| S177W | −15.3 ± 0.8 | 0.56 ± 0.02 | 23 | −16.3 ± 2.3 | 0.54 ± 0.04 | 4 |
| S177W-N184Q | −20.3 ± 1.4 | 0.47 ± 0.02 | 18 | — | — | — |
| S177W-N184D | −78 ± 7.1 | 0.06 ± 0.01 | 6 | −126.4 ± 4.1 | 0.008 ± 0.001 | 8 |
| S177W-N184E | −82.1 ± 3.1 | 0.04 ± 0.006 | 10 | −95.8 ± 7.5 | 0.03 ± 0.006 | 3 |
| S177W-N184R | −14.8 ± 0.8 | 0.56 ± 0.02 | 7 | — | — | — |
| S177W-N184K | −14.1 ± 0.9 | 0.58 ± 0.02 | 9 | — | — | — |
| S177W-N184S | −11.9 ± 1 | 0.64 ± 0.02 | 19 | — | — | — |
| S177W-N184A | −14.4 ± 0.8 | 0.57 ± 0.02 | 10 | — | — | — |
| S177W-N184M | −14.8 ± 0.9 | 0.57 ± 0.02 | 10 | — | — | — |
| S177W-G180D | −76.8 ± 5 | 0.06 ± 0.01 | 10 | −119.9 ± 5.3 | 0.01 ± 0.002 | 6 |
| S177W-S181D | −9.5 ± 1.2 | 0.69 ± 0.03 | 7 | — | — | — |
| S177W-A185D | −15.6 ± 2.5 | 0.56 ± 0.05 | 5 | — | — | — |
| S177W-V188D | −43.7 ± 1.8 | 0.18 ± 0.01 | 5 | −106.2 ± 6.3 | 0.02 ± 0.007 | 10 |
| S177W-G189D | −19.3 ± 2.3 | 0.5 ± 0.04 | 11 | — | — | — |
The difference in reversal potential (ΔE) that occurred with 90 mM Na+ and 90 mM K+ solutions was measured from the current–voltage relations obtained from recording Xenopus oocytes expressing Kir3.2 channels and corrected for junction potential (4 mV). The relative permeability ratios of Na+ and K+ (PNa/PK) were then deduced by using a form of the Goldman–Hodgkin–Katz voltage equation (see Materials and Methods). For TPNQ-subtracted values, currents were obtained in Na+ before and after addition of 1 μM TPNQ(a mutant blocker), and the difference current was used to determine the reversal potential. n indicates the number of oocytes. Values are mean ± SEM. —, not determined.
It is believed that the channel interacts electrostatically with cavity ions to further stabilize their presence in the center of the channel/bilayer complex. This interaction is enhanced by charged and polar residues lining the cavity (22) and by the charge dipoles of the pore helices (6). We believe that the primary effect of charged residues at the N184 position is an electrostatic stabilization of the cavity ion, and, in our companion paper (17), we use electrostatic calculations to show that N184D/E stabilize the cavity ion by 10 kBT relative to the WT channel. Such a large energy is biologically significant and can evoke dramatic changes in conduction properties.
All Cavity-Facing Residues Affect Selectivity.
To further test the dependence of K+ selectivity on negative charges in the channel cavity, we systematically introduced Asp (D) residues at several other cavity positions (Fig. 4), with the hypothesis that long-range electrostatic interactions should restore K+ selectivity regardless of the exact position of the mutation. Examination of the inner M2 transmembrane segment revealed two helix faces in close apposition to the conduction pore (Fig. 4A). Thus, we placed Asp (D) on face 1 to substitute for G180, N184, or V188 (in green) and on face 2 to replace S181, A185, or G189 (in red) (Fig. 4A). Interestingly, a striking pattern of selectivity emerges: Asp (D) restores K+ selectivity at all mutated positions along face 1 but at none of the positions on face 2 (Fig. 4B and Table 1; see also Fig. 7, which is published as supporting information on the PNAS web site). This all-or-nothing change in selectivity may be understood from the homology model in Fig. 4C, which shows residues along face 1 (green) residing closer to the cavity ion (blue). Moreover, residues along face 2 (red) have less aqueous exposure; the more hydrophobic environment is likely to favor the protonation of the Asp (D) residues so as to reduce their electrostatic effect.
Fig. 4.
Aspartate (D) scan of the central cavity supports an electrostatic mechanism for K+ selectivity. (A) Side view of the Kir3.2 channel with the S177W (red) mutation on the M2 helix. In the cavity, three residues from two faces, face 1 (green) and face 2 (red), are shown. Residues that were subjected to Asp (D) substitution are as follows: on face 1, G180, N184, and V188; on face 2, S181, A185, and G189. (B) Asp (D) substitution of residues at every position on face 1 (green), but at none of the positions on face 2 (red), restores K+ selectivity, as shown by values of the Na+ and K+ permeability ratio deduced from the difference in reversal potential measured in 90 mM Na+ and 90 mM K+ solutions. Values are mean ± SEM; values for selective channels are TPNQ-corrected. ∗, P < 0.001. (C) Molecular surface representation of the Kir3.2 cavity viewed from the extracellular side. Residues on face 1 (green) are closer to the cavity ion (blue) and have greater solvent exposure than the residues on face 2 (red).
Effects on Rectification.
The effect of charge mutations on the block and rectification of Kir channels is well known (22–29). We see a correlation between loss of K+ selectivity and reduced potency for channel block at depolarized potentials by intracellular Mg2+ and polyamines, as previously reported (26, 30–32). However, the reduction of rectification by S177W is not as acute as what has been previously reported for other mutations, leading us to believe that the selectivity filter remains largely intact. Interestingly, negative charge mutations of cavity residues along face 1, such as N184D/E, G180D, and V188D, not only restore K+ selectivity but also eliminate the reduction of inward rectification due to the S177W mutation (Fig. 5A and C). In contrast, hydrophobic (A, M) and uncharged hydrophilic (Q, S) substitution at N184 as well as Asp (D) substitution along face 2 have no effect on the rectification properties of S177W-containing mutant channels, whereas positive charge mutations at N184 (K, R) produce a further loss of rectification (Fig. 5 B and C). The increased inward rectification due to negative charges in the cavity, illustrated in Fig. 5A and similar to previous findings (28, 33), can be accounted for by increased channel occupancy by Mg2+ and polyamine, as expected from electrostatic considerations. Accordingly, positive charge mutations are expected to decrease the occupancy of these positively charged blocking ions, resulting in reduced inward rectification, which is what we see experimentally (Fig. 5B). Moreover, the rectification data further support the hypothesis that Asp (D) residues placed on face 2 are neutral, whereas acidic residues introduced to face 1 are negatively charged (Fig. 4C).
Fig. 5.
Mutations in the M2 helix of Kir3.2 also affect inward rectification properties of S177W-bearing mutant channels. Histograms show the ratio of outward K+ current recorded at membrane potential +40 mV (K+ out) and the inward K+ current recorded at −100 mV (K+ in) for all Kir3.2 channels used in this study. (A) Rectification values of the S177W-containing channels with single acidic (D or E) substitutions along the M2 helix. Mutations that restore K+ selectivity (in green) also increase inward rectification to values comparable to those of S177T channels (∗, P < 0.01 for N184E; ∗, P < 0.001 for all others), whereas all nonselective K+ channels (in red) have impaired rectification. (B) Rectification values for various nonselective mutant channels with positive and neutral substitutions at position N184. Basic substitutions at N184 (N184R/K) have the strongest effect by further reducing rectification. ∗, P < 0.001. Values are reported as mean ± SEM. (C) Representative averaged current–voltage (I–V) relations obtained from Xenopus oocytes expressing Kir3.2 mutant channels. Currents were recorded at membrane potentials ranging from +40 mV to −150 mV in 10-mV increments in 90 mM K+ bath solution. Points in the I–V curves represent mean ± SEM for n = 8–10 oocytes.
Discussion
The existence of K+ channels rendered nonselective by means of mutations outside the K+ signature sequence, such as the Kir3.2 S177W mutant channel, as well as native hyperpolarization-activated cyclic nucleotide-gated cation channels lacking K+ selectivity, raise the possibility that K+ selectivity might be controlled by other structural features in addition to the conserved K+ signature sequence. However, this knowledge does not predict whether, where, and how mutations would arise to bestow potassium selectivity. Having identified mutations in the central cavity that restored K+ selectivity to a nonselective S177W mutant channel, we sought to define the biophysical constraints of this effect. Our findings reveal important insights that might help us to understand how K+ selectivity can be regulated and how selective K+ channels evolved.
Evidences for Electrostatic Interactions Between Channel Pore-Facing Residues and Cavity Ions.
The nonselective S177W mutant Kir3.2 channel becomes K+ selective when the pore-facing M2 residues in the central cavity are replaced with negatively charged amino acids, Asp (D), or Glu (E). This observation and several other aspects of our Asp (D) scan of the cavity residues of Kir3.2 suggest that the recovery of K+ selectivity is mediated not by an increased selectivity at the filter sites but by electrostatic interactions between pore-facing residues and cavity ions.
First, because Trp (W) is a nonpolar residue, it is unlikely that the Asp (D) residues in the cavity electrostatically interact with S177W (Fig. 1). Second, based on our models of the Kir3.2 mutant channels, the second-site suppressor, N184D, does not produce significant deviation of selectivity filter amino acids compared with WT or a K+-selective mutant (S177T). Also, although N184D restores K+ selectivity to the S177W mutant channel, it does not alter the deviation of the filter amino acids introduced by S177W (Fig. 2). Third, as expected for an electrostatic mechanism, K+ selectivity was reinstated by Asp (D) substitutions at three different pore-facing M2 sites (G180D, N184D, and V188D) but not at partially buried sites where acidic residues are likely to bear no charge (Fig. 4). Fourth, the second-site suppressor V188D is nearly three helical turns away from the bottom of the selectivity filter and S177W, yet its effect on selectivity is as profound as that of N184D (two helical turns away) and G180D (one helical turn away) (Fig. 4). Notably, these positions are nearly equidistant to the cavity ion. Fifth, we generally think of K+ selectivity in terms of the packing geometry and motion of the selectivity filter. In principle, the packing geometry of a folded protein can be altered by mutations in unpredictable ways; mutations at different sites are unlikely to have the same effect on the packing geometry of the selectivity filter. Importantly, substitutions of G180, N184, and V188 with Asp (D) all produced the same, nearly complete gain in K+ selectivity despite their drastically different initial side-chain chemistry (Fig. 4). Finally, we have also found that there is a correlation between the mutations' effects on K+ selectivity and the inward rectification properties, as expected from electrostatic considerations of the interaction of cavity-facing residues with the positively charged blocking ions (Fig. 5).
These observations strongly implicate electrostatic interactions with cavity ions as the cause for enhanced K+ selectivity, but given that such interactions generally have equal effects on K+ and Na+, it is not clear exactly how K+ selectivity is enhanced. In our companion paper (17) in the following pages, we explore this question theoretically and develop a mechanistic explanation. Briefly, we show that favorable interactions of cations with N184D change their rates into and out of the cavity site, shifting the rate-limiting step for conduction away from a transition that is nearly equal for K+ and Na+ to one that is much slower for Na+.
Evolving K+ Selectivity by Mutation of the Residues Lining the Permeation Pathway.
Our findings provide one plausible answer to the question that originally motivated our molecular evolution experiments: How might K+ selectivity have evolved? With the K+ selectivity considerations focused on the precise alignment of the selectivity filter to approximate the hydration shell of K+ but not Na+, which differ by ≈1/10 in diameter (0.4 Å), it follows that K+ selectivity must have emerged during evolution by means of progressive improvements in the placement and dynamics of the backbone carbonyl oxygen atoms of the selectivity filter until they were able to specifically coordinate the partially dehydrated K+: a remarkable feat. Another possibility emerges from our finding of the tremendous gain in K+ selectivity by means of electrostatic stabilization of the cavity ion. In this scenario, a primordial channel could have first gained some structural features that endowed a modest preference for K+ binding in the pore but with little evidence for K+ selectivity in ion permeation (in essence, analogous to the S177W channel) and then accomplished functional K+ selectivity during evolution with simple mutations of pore-lining residues that strengthen ion interaction, likely electrostatic in nature. An already functional (although primitive) K+ channel would stand to be selected for, and improve its pore-lining structures in evolution, resulting in K+ channels displaying a range of features for K+ selectivity. Here, biology may echo engineering in instigating more than one mechanism to ensure the robustness of a desired outcome. Although discussions of evolution by nature are speculative, these considerations are in line with the results from our previous molecular evolution experiments (15) and the experimental verification of predictions presented in this study.
Toward a Concerted Control of K+ Selectivity Along the Entire Channel Permeation Pathway.
Although our results concentrate on the effect of negative charges along the transmembrane inner helix of K+ channels on selectivity and rectification, other residues along the conduction pathway may evoke the same phenomenon. For Kir channels in particular, additional cytoplasmic cation-binding sites act in concert with cavity cation-binding sites to control inward rectification (4, 23–25, 27–29). Although most K+ channels are highly selective, the contribution from the selectivity filter may only be part of the story for K+ selectivity. The presence of multiple ion-binding sites may provide additional amplification of K+ selectivity and could allow an ion channel to achieve a high degree of selectivity in multiple discrete steps during evolution. It also provides a rich forum for potential regulation and opens the possibility for changing the selectivity of K+ channels by adjusting the channel's interaction with ions outside the selectivity filter. The possibility of altering K+ selectivity further raises the question of whether the changes in K+ selectivity during voltage-gated K+ channel activation and inactivation may arise from conformational changes outside the selectivity filter.
Materials and Methods
Constructs and Oocyte Expression.
Kir3.2 (GIRK2) was cloned into the HindIII–XhoI sites of a derivative of pGEM vector (34). Site-directed mutagenesis was performed based on PCR of the full-length plasmid by using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), and the entire cDNA was sequenced. Kir3.2 mutant cDNAs were linearized at the AflII restriction site, and capped cRNA were synthesized by using the Amplicap T7 High Yield Message Maker kit (Epicentre Technologies, Madison, WI). Stage V–VI Xenopus laevis oocytes were collected, injected with 1–30 ng of each cRNA, and maintained at 16°C before recording in ND96 solution (96 mM NaCl/2 mM KCl/1 mM MgCl2/5 mM Hepes, pH 7.4) or analogous solutions in which Na+ was replaced with K+ or N-methyl-d-glucamine.
Electrophysiology.
Macroscopic currents were recorded from oocytes with two-electrode voltage clamp (GeneClamp 500B, Axon Instruments, Foster City, CA). Electrodes were filled with 3 M KCl and had a resistance of 0.4–1 MΩ. A small chamber with a fast perfusion system (ValveLink 16, AutoMate Scientific, San Francisco, CA) was used to change extracellular solutions (90 mM KCl or NaCl/2 mM MgCl2/10 mM Hepes, pH adjusted to 7.4 with KOH or NaOH) and was connected to a ground by a 3 M KCl agarose bridge. All recordings were conducted at room temperature (22–25°C).
Permeability Measurements.
Permeability ratios were determined under biionic conditions with a form of the Goldman–Hodgkin–Katz equation, ΔErev = Erev,Na − Erev,K = RT/zF ln(PNa[Na]o/PK[K]o), where R, T, z, and F have their usual meanings (4), Erev,Na is the reversal potential measured in Na+-containing solution, and Erev,K is the reversal potential in K+-containing solution. To control for the contribution of endogenous channels, the difference between currents obtained before and after addition of a 1 μM concentration of a nonoxidizable mutant blocker of the Kir3.2 channel (tertiapin-Q, TPNQ) was used to determine the reversal potential (20). TPNQ subtraction did not alter the PNa/PK measurement for nonselective channels, but it did decrease the estimate for PNa/PK of selective channels. We therefore applied subtraction only for selective channels (see Table 1). All reversal potential measurements were corrected for junction potential (4 mV). Statistics were carried out with one-way ANOVA followed with Tukey's multiple comparison test, comparing all mutants with S177W.
Modeling.
We used the KirBac1.1 crystal structure as a template to construct 3D homology models of Kir3.2 by using the alignment in ref. 35. A representative structure was chosen based on agreement with known mutational analysis and a fitness score that accounts for biophysical properties and the geometric closeness of the model to the template (36, 37). Subsequent mutations were generated from this original WT model. It is of note that KirBac1.1 lacks two conserved residues that flank the selectivity filter and play a crucial role in selectivity and function, possibly through forming a stabilizing salt bridge (31, 38). This salt bridge was introduced at the initial stage of construction by using artificial restraints. Channels were minimized for 3,000 steps in a vacuum by using the CHARMM27 parameter set with full electrostatics in NAMD (39). Potassium ions were included in the S1 and S3 filter sites and the cavity and held fixed during minimization. This process was repeated 100 times for each mutant to construct the curves in Fig. 2. rmsds of the filter atoms compared with the initial WT model were averaged over the set of models. This averaging analysis was repeated for the corresponding WT-to-WT control runs to account for stochastic variation introduced by homology modeling. The graph in Fig. 2 shows the rms values normalized by the appropriate control pattern.
Supplementary Material
Acknowledgments
We thank B. Cohen for comments on the manuscript, Y. F. Lin for her expertise in Kir3.2 channel electrophysiological properties, F. Lesage (CNRS UMR 6097, Valbonne, France) for Kir3.2 cDNA, and Z. Lu (University of Pennsylvania, Philadelphia, PA) for the gift of TPNQ. This work was supported by a National Science Foundation Interdisciplinary Informatics Fellowship (to M.G.) and National Institute of Mental Health Grant MH065334. L.Y.J. and Y.N.J. are Investigators of the Howard Hughes Medical Institute.
Abbreviations
- Kir
inwardly rectifying potassium
- TPNQ
tertiapin-Q.
Footnotes
The authors declare no conflict of interest.
References
- 1.Brownlee C. Curr Biol. 2002;12:R402–R404. doi: 10.1016/s0960-9822(02)00886-2. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee A, Joiner WJ, Wu M, Yang Y, Sigworth FJ, Kaczmarek LK. J Neurosci. 2003;23:11681–11691. doi: 10.1523/JNEUROSCI.23-37-11681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashcroft FM. Ion Channels and Diseases. London: Academic; 2000. [Google Scholar]
- 4.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 5.Shieh CC, Coghlan M, Sullivan JP, Gopalakrishnan M. Pharmacol Rev. 2000;52:557–594. [PubMed] [Google Scholar]
- 6.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 7.Heginbotham L, Abramson T, MacKinnon R. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 9.Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Cell. 1998;93:717–729. doi: 10.1016/s0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- 10.Shin N, Soh H, Chang S, Kim DH, Park CS. Biophys J. 2005;89:3111–3119. doi: 10.1529/biophysj.105.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starkus JG, Kuschel L, Rayner MD, Heinemann SH. J Gen Physiol. 1997;110:539–550. doi: 10.1085/jgp.110.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J, Sigworth FJ. J Gen Physiol. 1997;110:101–117. doi: 10.1085/jgp.110.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiss L, LoTurco J, Korn SJ. Biophys J. 1999;76:253–263. doi: 10.1016/S0006-3495(99)77194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi BA, Lin YF, Jan YN, Jan LY. Neuron. 2001;29:657–667. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 15.Bichet D, Lin YF, Ibarra CA, Huang CS, Yi BA, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2004;101:4441–4446. doi: 10.1073/pnas.0401195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe H, Sackin H, Palmer LG. J Gen Physiol. 2000;115:391–404. doi: 10.1085/jgp.115.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabe M, Bichet D, Qian X, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2006;103:14361–14366. doi: 10.1073/pnas.0606662103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 19.Noskov SY, Berneche S, Roux B. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 20.Jin W, Lu Z. Biochemistry. 1998;37:13291–13299. doi: 10.1021/bi981178p. [DOI] [PubMed] [Google Scholar]
- 21.Navarro B, Kennedy ME, Velimirovic B, Bhat D, Peterson AS, Clapham DE. Science. 1996;272:1950–1953. doi: 10.1126/science.272.5270.1950. [DOI] [PubMed] [Google Scholar]
- 22.Chatelain FC, Alagem N, Xu Q, Pancaroglu R, Reuveny E, Minor DL., Jr Neuron. 2005;47:833–843. doi: 10.1016/j.neuron.2005.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, MacKinnon R. Nature. 1994;371:243–246. doi: 10.1038/371243a0. [DOI] [PubMed] [Google Scholar]
- 24.Stanfield PR, Davies NW, Shelton PA, Sutcliffe MJ, Khan IA, Brammar WJ, Conley EC. J Physiol. 1994;478(Part 1):1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Jan YN, Jan LY. Neuron. 1995;14:1047–1054. doi: 10.1016/0896-6273(95)90343-7. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara Y, Kubo Y. J Gen Physiol. 2002;120:677–693. doi: 10.1085/jgp.20028663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubo Y, Murata Y. J Physiol. 2001;531:645–660. doi: 10.1111/j.1469-7793.2001.0645h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo D, Ramu Y, Klem AM, Lu Z. J Gen Physiol. 2003;121:261–275. doi: 10.1085/jgp.200208771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie LH, John SA, Weiss JN. J Physiol. 2003;550:67–82. doi: 10.1113/jphysiol.2003.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makary SM, Claydon TW, Enkvetchakul D, Nichols CG, Boyett MR. J Physiol. 2005;568:749–766. doi: 10.1113/jphysiol.2005.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibb KM, Rose T, Makary SY, Claydon TW, Enkvetchakul D, Leach R, Nichols CG, Boyett MR. J Biol Chem. 2003;278:49537–49548. doi: 10.1074/jbc.M307723200. [DOI] [PubMed] [Google Scholar]
- 32.Makary SM, Claydon TW, Dibb KM, Boyett MR. Biophys J. 2006;90:4018–4034. doi: 10.1529/biophysj.105.073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurata HT, Phillips LR, Rose T, Loussouarn G, Herlitze S, Fritzenschaft H, Enkvetchakul D, Nichols CG, Baukrowitz T. J Gen Physiol. 2004;124:541–554. doi: 10.1085/jgp.200409159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesage F, Duprat F, Fink M, Guillemare E, Coppola T, Lazdunski M, Hugnot JP. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- 35.Bichet D, Haass FA, Jan LY. Nat Rev Neurosci. 2003;4:957–967. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 36.Minor DL, Jr, Masseling SJ, Jan YN, Jan LY. Cell. 1999;96:879–891. doi: 10.1016/s0092-8674(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 37.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Yu M, Jan YN, Jan LY. Proc Natl Acad Sci USA. 1997;94:1568–1572. doi: 10.1073/pnas.94.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comput Phys. 1999;151:283–312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.