Abstract
The c-Fes protein tyrosine kinase is implicated in the differentiation of a number of cell types including neuronal, endothelial and myeloid cells. Structurally, Fes consists of a unique N-terminal region, followed by SH2 (Src homology domain 2) and kinase domains. Two coiled-coil (CC) domains (CC1 and CC2) located within the unique N-terminal region are critical regulators of Fes activity in vivo and may function to recruit Fes activators and/or substrates. A yeast two-hybrid screen, utilizing a K-562 cell cDNA library and the Fes CC2 domain as bait, identified an interacting clone encoding the CC domain and B-box motifs (residues 114–357) of the transcriptional co-repressor KRAB-associated protein (KAP)-1. KAP-1114–357 interacted with full-length Fes in yeast, and the KAP-1 CC domain was sufficient to bind the Fes N-terminal region in Sf-9 cells. Co-expression of Fes with full-length KAP-1 in human 293T cells stimulated Fes autophosphorylation and led to KAP-1 tyrosine phosphorylation. Association of endogenous Fes and KAP-1 was also observed in HL-60 myeloid leukaemia cells. Together, these data identify a novel Fes–KAP-1 interaction, and suggest a dual role for KAP-1 as both a Fes activator and downstream effector.
Keywords: c-Fes, coiled-coil, KAP-1, non-receptor protein tyrosine kinase, protein–protein interaction, yeast two-hybrid system
Abbreviations: AD, activation domain; CC, coiled-coil; CML, chronic myelogenous leukaemia; DBD, DNA binding domain; GST, glutathione S-transferase; HA, haemagglutinin; KAP, Kruppel-associated box-associated protein; KRAB, Kruppel-associated box; NBT, Nitro Blue Tetrazolium; PTK, protein tyrosine kinase; RIPA, radio-immunoprecipitation assay; SDS-SB, SDS-sample buffer; SH, Src homology; TMF, TATA element modulatory factor
INTRODUCTION
The human c-Fes locus is the cellular homologue of the transforming oncogenes expressed by several avian and feline retroviruses and encodes a 93 kDa non-receptor PTK (protein tyrosine kinase) [1–8]. The c-Fes kinase is expressed in granulocytic and monocytic cell lineages, vascular endothelial cells and various neuronal cell types where it is activated by a variety of cytokines and growth factors [6–18; reviewed in 19,20]. A number of studies have suggested an important role for Fes in the differentiation, survival and proliferation of these diverse cell types. For example, Fes antisense oligonucleotides blocked PMA-induced differentiation of HL-60 human promyelocytic leukaemia cells and FDC-P1/MAC-11 murine myeloid precursor cells [21]. In addition, previous studies have shown that constitutively active forms of Fes induce macrophage differentiation in human promonocytic U937 cells and promote survival and differentiation of the myeloid leukaemia cell line, TF-1 [22,23]. Furthermore, a human CML (chronic myelogenous leukaemia)-derived cell line (K-562) that lacks Fes expression can be induced to undergo growth arrest and terminal differentiation following the re-introduction of Fes, implicating this kinase as a suppressor of CML progression [24,25]. In brain capillary endothelial cells, Fes over-expression leads to fibroblast growth-factor (FGF)-independent tube formation [14]. More recently, Fes has been demonstrated to accelerate NGF-induced neurite outgrowth in PC12 cells implicating the kinase in neuronal differentiation [11,14]. Although Fes is well documented as a modulator of cell differentiation, the mechanisms by which Fes kinase activity is regulated and the downstream differentiation pathways activated by Fes remain poorly characterized.
Fes consists of a unique N-terminal region and a central SH2 (Src homology 2) domain, both of which contribute to the regulation of the C-terminal kinase domain (reviewed in [26]). Unlike other non-receptor PTKs, Fes lacks an SH3 domain, a negative regulatory tail, and other features that contribute to negative regulation of c-Src, c-Abl and other cytoplasmic PTKs. Within the unique N-terminal region are two putative coiled-coil (CC) homology domains, termed CC1 and CC2 [24,27]. CCs are well-characterized protein–protein interaction motifs consisting of amphipathic α-helices with a heptad repeat pattern in which hydrophobic residues in the first and fourth positions align to form an interface between the interacting helices [28]. Interestingly, CC domains are present in Fes (and its close homologue Fer), but they are not found in other non-receptor PTKs, suggesting that they are responsible for the unique regulatory and signalling properties of Fes.
Several studies have identified important roles for the CC domains in the regulation of Fes kinase and biological activities. For example, mutations within CC1 increase Fes kinase and differentiation-inducing activities, suggesting an inhibitory role for CC1 in vivo [22,24]. On the other hand, mutation of CC2 partially reverses the increase in Fes activity that follows mutation of CC1, implicating the CC2 domain as a positive regulator [22,24]. Gel-filtration studies suggest a role for the CC domains in the inter-conversion of Fes between inactive monomeric and active oligomeric states [22]. These motifs however, may also mediate novel heterotypic interactions with upstream Fes activators and/or downstream Fes substrates.
In the present study, to identify novel Fes CC binding partners, a yeast two-hybrid screen was initiated, with the Fes CC2 domain serving as bait. A clone encoding the B boxes and CC domain of KAP (Kruppel-associated box-associated protein)-1 was identified as a Fes CC2 interacting partner from a K-562 CML cell cDNA library. KAP-1 is a transcriptional co-repressor that is recruited to DNA through association with KRAB (Kruppel-associated box) domains found in a variety of Kruppel-class Cys2-His2 zinc finger proteins [29–31]. Once recruited to DNA, KAP-1 is thought to repress transcription through interactions with heterochromatin proteins, histone deacetylases and/or histone methyltransferases [32–36]. Full-length Fes and KAP-1 formed stable complexes in yeast and mammalian cells, including differentiated HL-60 myeloid leukaemia cells. The KAP-1 CC domain was sufficient to interact with the Fes N-terminus (which includes the Fes CC domains) in insect cells. Co-expression of full-length wild-type KAP-1 and Fes in human 293T cells resulted in KAP-1 tyrosine phosphorylation as well as increased Fes kinase activity, suggesting a role for KAP-1 as both a Fes activator and substrate.
MATERIALS AND METHODS
Yeast expression plasmids
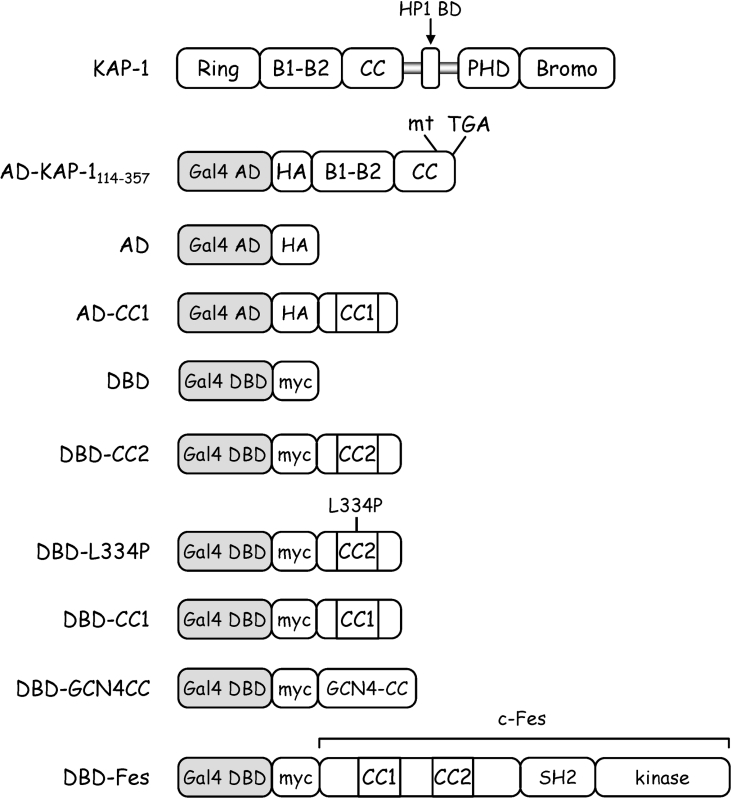
The yeast two-hybrid fusion proteins used in this study are illustrated in Figure 1. The pGBKT7 vector (Clontech) encodes the Gal4 DBD (DNA binding domain) and expresses a leucine-selectable marker, whereas the pGADT7 vector (Clontech) expresses the Gal4 AD (activation domain) and confers tryptophan selection. A human K-562 cDNA library, (Clontech) fused in frame to an N-terminal Gal4 AD and a HA (haemagglutinin) epitope tag, was amplified according to the manufacturer's instructions. Coding regions for the Fes CC1 domain (amino acids Met1–Leu176), CC2 domain (amino acids Leu291–Gly392) and CC2 domain harbouring an L334P mutation [22] were PCR-amplified and subcloned into pGBKT7 for expression of DBD–CC1, DBD–CC2 and DBD–L334P respectively. To express DBD–GCN4CC, the coding sequence for the CC domain of the yeast transcription factor GCN4 was PCR-amplified and inserted into pGBKT7. The coding region for full-length wild-type Fes was also PCR-amplified and inserted into pGBKT7 to express DBD–Fes. The Fes CC1 domain coding region was also subcloned into pGADT7 to express AD–CC1. All clones expressed from pGBKT7 included a Myc tag, while clones expressed from pGADT7 included the HA epitope. Nucleotide sequences of all PCR-derived cDNA clones were confirmed by DNA sequence analysis.
Figure 1. Expression constructs used in the yeast two-hybrid system.
Wild-type Kap-1 is shown at the top for reference. AD–KAP-1114–357 was isolated in a yeast two-hybrid screen using the c-Fes CC2 domain as bait. This clone includes amino acids 114–357 of KAP-1 which encompass the B boxes and CC domain. A frameshift mutation (mt) at residue 357 results in a stop codon (TGA) ten amino acids downstream. The AD construct consists of the Gal4 activation domain plus a C-terminal HA epitope tag, while the DBD protein includes the Gal4 DNA binding domain followed by a Myc epitope tag. All AD or DBD fusion proteins were generated by in-frame fusion of the indicated proteins downstream of the HA or Myc tag, respectively. CC1 includes the Fes CC1 domain (Met1–Leu176), CC2 includes the Fes CC2 domain (Leu291–Gly392), whereas GCN4–CC encompasses the CC domain of the yeast transcription factor, GCN4. L334P includes the Fes CC2 domain with a proline substitution for Leu334. Full-length wild-type Fes was also fused to the DBD to generate DBD–Fes.
Yeast two-hybrid analysis
The yeast two-hybrid screen employed the Matchmaker Gal4 Two-Hybrid System 3 (Clontech) in which Saccharomyces cerevisiae strain AH109 was co-transformed with the bait plasmid DBD-CC2 and the K-562 cell cDNA library (AD-library) according to the manufacturer's protocol. For the initial library screen, yeast co-transformed with DBD–CC2 and AD-library plasmids were plated on minimal synthetic dropout medium lacking leucine, tryptophan, adenine and histidine (SD−L−W−A−H) to select for interacting clones. Fifty putative Fes CC2 interacting clones were initially identified out of approximately 4.5×106 co-transformed yeast cells. Following three additional rounds of selection, CC2-interacting clones were recovered and re-tested to eliminate false-positive interactions with the Gal4 DBD alone. One of these clones was sequenced and identified as a partial KAP-1 clone (AD–KAP-1114–357). All subsequent yeast two-hybrid analyses were initially plated onto solid minimal synthetic dropout medium lacking leucine and tryptophan (SD−L−W) to select for the presence of both plasmids. Three or four independent colonies were then replated onto SD−L−W medium and SD−L−W−A−H medium. All yeast were grown at 30 °C.
Yeast protein extraction
Yeast co-transformations were performed as described for the yeast two-hybrid analyses and protein extraction was performed as described previously [37] with minor modification. Briefly, independent colonies exhibiting growth on solid SD−L−W medium were used to inoculate liquid SD−L−W medium and grown overnight. The resulting culture was diluted to an D600 of 0.2 in liquid YPAD (1% yeast extract, 2% peptone, 0.004% adenine and 2% dextrose) medium and grown until the D600 reached 0.6. Yeast (7.5 D600 units) were collected by centrifugation, resuspended in 300 μl of water and combined with 300μl of 0.2M NaOH. Following incubation at room temperature (22 °C) for 5 min, yeast cells were pelleted and resuspended in 150 μl of 2× SDS-SB (SDS-sample buffer).
Recombinant baculovirus generation and protein expression in Sf-9 insect cells
Recombinant baculoviruses for the expression of the Fes N-terminal region (Fes N; amino acids 1–450), the SH2 and kinase domains (Fes ΔN; amino acids 451–822), an SH2 deletion mutant (Fes ΔSH2; lacks residues 451–540), the Fes kinase domain (Fes kinase; amino acids 541–822), as well as full-length Fes and GST (glutathione S-transferase) have been described previously [27,38]. All Fes constructs included a C-terminal FLAG epitope tag. To express GST–KAP-1114–357, the corresponding coding region was PCR-amplified from AD–KAP-1114–357 and subcloned into the baculovirus transfer vector pVL-GST [38]. The coding sequence for the KAP-1 CC domain (Phe220–Thr359) was PCR-amplified from pC3-FLAG-KAP (full-length human KAP-1 expression plasmid provided by Dr Frank Rauscher III, The Wistar Institute Cancer Center, Philadelphia, PA, U.S.A.) and inserted into pVL-GST for expression of GST–KAPCC.
Recombinant baculoviruses were generated from the pVL-GST constructs by co-transfection of Sf-9 cells with Baculogold DNA (Pharmingen) utilizing CellFectin® Reagent (Invitrogen) according to the manufacturer's protocol. Following 4 days in culture, the primary viral supernatant was clarified by centrifugation and this primary baculovirus stock was amplified according to methods described previously [38]. For protein expression, 2.5×106 Sf-9 cells were infected with baculoviruses for 1 h at room temperature (22 °C). The virus was replaced with fresh medium and cells were incubated at 27 °C for 2–3 days.
Sf-9 cell lysis and GST-pull-down analyses
Infected Sf-9 cells were washed twice with ice-cold PBS and pelleted by centrifugation. Cell pellets were re-suspended in RIPA (radio-immunoprecipitation assay) buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 1 mM EDTA, 2 mM PMSF, 25 μg/ml aprotinin, 25 μg/ml leupeptin, 2 mM sodium orthovanadate and 25 mM sodium fluoride] and sonicated for 10 s. The lysate was clarified by micro-centrifugation and a portion was combined with an equal volume of 2× SDS-SB, heated to 95 °C for 5 min and stored at −80 °C. The remaining lysate was incubated with 20 μl of GSH (glutathione)–agarose beads (50% slurry) for 2 h at 4 °C. The GSH–agarose precipitate was washed three times with RIPA buffer, re-suspended in 2× SDS-SB, heated to 95 °C for 5 min and stored at −80 °C.
293T cell transfection, lysis and immunoprecipitation
Mammalian cell expression vectors for Fes, Fes-L145P and Fes-KE include C-terminal FLAG epitope tags and have been described previously [22]. KAP-1 was expressed from the pC3 FLAG–KAP-1 expression vector, which encodes KAP-1 amino acids Pro20–Pro835 plus an N-terminal FLAG epitope tag. For transient transfections, 293T cells were plated in 60 mm (1.0×106 cells) or 100 mm (2.5×106 cells) plates and transfected 1 day later according to the calcium phosphate method as described previously [24]. At 2–4 days post-transfection, cells were lysed in Fes lysis buffer [50 mM Tris/HCl (pH 7.4), 50 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 2 mM PMSF, 25 μg/ml aprotinin, 25 μg/ml leupeptin, 2 mM sodium orthovanadate and 25 mM sodium fluoride] and sonicated for 10 s. The lysate was clarified by micro-centrifugation and a portion was combined with an equal volume of 2× SDS-SB, heated to 95 °C for 5 min and stored at −80 °C. For immunoprecipitation, the remaining lysate was incubated with 5 μl of a Fes-specific rabbit polyclonal antiserum (provided by Dr Peter Greer, Queen's University Cancer Research Institute, Ontario, Canada) or Fes-specific goat polyclonal antiserum at 1 μg/ml (Fes N-19, Santa Cruz Biotechnology) with 20 μl of protein G–Sepharose (1:1 w/v slurry) for 2 h at 4 °C. Alternatively, FLAG-tagged Fes was immunoprecipitated with 20 μl of the anti-FLAG-M2 agarose affinity gel (Sigma). KAP-1 was also immunoprecipitated with the anti-FLAG-M2 agarose affinity gel. All immunoprecipitates were washed three times with RIPA buffer, re-suspended in 2× SDS-SB, heated to 95 °C for 5 min and stored at −80 °C.
Immunoblot analyses
Antibodies anti-Myc (9E10), anti-GST (B-14) and anti-phosphotyrosine (PY99) were obtained from Santa Cruz Biotechnology. Antibodies anti-HA (HA-7) and anti-FLAG (M2) were obtained from Sigma. All antibodies were used at 1 μg/ml except for the anti-HA antibody that was used at a 1:10000 dilution. Aliquots of cell lysates or immunoprecipitates were separated by discontinuous SDS/PAGE (10% or 12% gels). Proteins were transferred to PVDF membranes, blocked overnight at 4 °C or for 1 h at room temperature (22 °C) in TBST–BSA [10 mM Tris/HCl (pH 7.4), 150 mM NaCl, 0.1% Tween 20 and 1.5% BSA], and probed for 1 h at room temperature (22 °C) or overnight at 4 °C with primary antibodies diluted in TBST–BSA.
Following incubation with primary antibodies, membranes were probed for 1 h at room temperature (22 °C) with the appropriate alkaline phosphatase-conjugated secondary antibodies (Southern Biotechnology) diluted 1:10000 in TBST–BSA. All blocking and antibody incubations were followed by three TBST washes for 5 min at room temperature (22 °C). Detection was performed using the CDP-STAR® Chemiluminescence Reagent detection system (PerkinElmer) or with BCIP (5-bromo-4-chloro-3-indolyl phosphate) and NBT (Nitro Blue Tetrazolium). In some instances, blots were stripped by a 30 min incubation at 50 °C in strip buffer (62.5 mM Tris/HCl, 100 mM 2-mercaptoethanol and 2% SDS) and re-probed.
Detection of Fes–KAP-1 complexes in HL-60 cells
HL-60 cells (a gift of Dr Richard Steinman, University of Pittsburgh Cancer Institute, Pittsburgh, PA, U.S.A.) were grown in RPMI-1640 medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biological) and 50 μg/ml gentamicin (Gibco). Cells were maintained at a density of 105–106 cells/ml. To induce granulocytic differentiation, cells were seeded at a density of 5.0×105 cells/ml and treated with DMSO (Sigma) to a final concentration of 1.25% or all-trans retinoic acid (1 μM; a gift of Dr Neil Hukriede, University of Pittsburgh School of Medicine, Pittsburgh, PA, U.S.A.). Differentiation was assessed after 4 days as the percentage of cells able to reduce NBT. To detect Fes–KAP-1 complexes, cells (2.5×107) were sonicated in 0.5 ml of 50 mM Tris/HCl (pH 7.4), 1 mM EDTA, 50 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 2 mM PMSF, 2 mM sodium orthovanadate, 25 mM sodium fluoride and Protease Inhibitor Cocktail Set III (Calbiochem). Lysates were clarified and aliquots were incubated with 2 μg of a Fes-specific goat polyclonal antiserum (Fes C-19; Santa Cruz Biotechnology) for 1 h at 4 °C, followed by an additional hour with 30 μl protein G–Sepharose beads (AP Biotech; 1:1 w/v slurry) at 4 °C. The beads were then washed three times with lysis buffer and protein complexes were eluted by heating in SDS-SB. Immunoprecipitates and clarified lysates were subjected to immunoblot analysis with antibodies to Fes (C-19; 1 μg/ml) and KAP-1 (Bethyl Laboratories, A300-274A; 1:1000 dilution). Lysates were also blotted with actin antibodies (Chemicon MAB1501; 1:10000 dilution) as a loading control.
RESULTS
Identification of KAP-1 as a Fes CC domain binding protein
The Fes CC2 domain has been identified as an important modulator of Fes kinase and biological activities, suggesting that this motif may recruit binding partners involved in the regulation of kinase activity and downstream signalling [22,24]. To identify Fes CC2-interacting proteins, the CC2 domain was used as bait to screen a K-562 cell cDNA library by yeast two-hybrid analysis. K-562 cells were originally derived from a patient with CML [39] and are devoid of detectable Fes expression [6,25]. Re-expression of Fes, however, promotes growth arrest and terminal differentiation of K-562 cells, suggesting that upstream activators and downstream effectors for Fes are present in this cell line [24,25]. Sequence analysis identified one of the CC2-interacting K562 library clones as the transcriptional co-repressor KAP-1. This clone (KAP-1114–357) lacked N-terminal amino acids 1–113 of KAP-1 and contained a base deletion within codon 357, resulting in a frame-shift and the generation of a stop codon 10 amino acids downstream. As illustrated in Figure 1, KAP-1114–357 retains the B boxes and CC domain but lacks the RING domain, heterochromatin protein-1 binding domain, plant homeodomain and bromo-domain.
KAP-1114–357 selectively interacts with the Fes CC2 domain
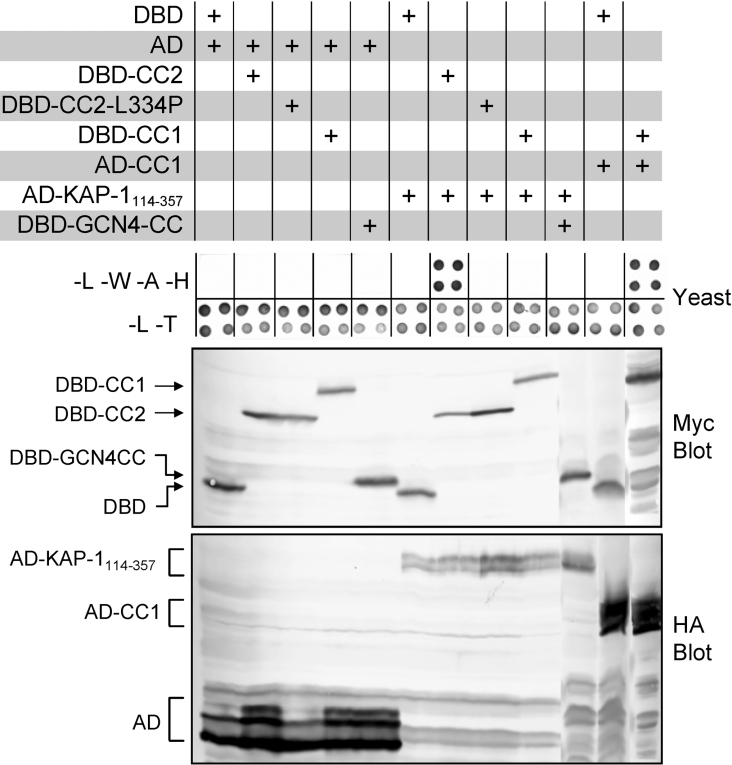
To determine whether the KAP-1 clone has selectivity towards the Fes CC2 domain, the ability of KAP-1114–357 to interact with additional CC domains was tested in the yeast two-hybrid system. Various CC domains were fused to the yeast Gal4 AD or to the Gal4 DBD as described in the Materials and Methods section. The CC fusion proteins that were used in this experiment are illustrated in Figure 1 and the results are shown in Figure 2. The AD–KAP-1114–357 fusion, which includes the KAP-1 CC domain, interacted with the Fes DBD–CC2 fusion protein as indicated by growth on SD-L-W-A-H medium. Interactions were not observed between AD–KAP-1114–357 and the Gal4 DBD alone or between the Fes DBD–CC2 and the Gal4 AD alone. Although AD–KAP-1114–357 interacted with the Fes CC2 domain, it failed to interact with Fes CC1–DBD or an unrelated CC domain from the yeast transcription factor GCN4 (DBD–GCN4CC). These observations suggest that KAP-1114–357 is selective for the Fes CC2 motif and is not a general CC interacting protein. Despite the lack of interaction between Fes DBD–CC1 and AD–KAP-1114–357, DBD–CC1 did interact with a Fes AD–CC1 fusion protein, indicating that the Fes CC1 domain is functional when expressed in yeast. Mutation of a Leu334 to proline within the Fes CC2 domain (DBD–CC2-L334P) eliminated interaction with KAP-1114–357. Previous studies have established that Leu334 occupies a critical position within the heptad repeat predicted to form the CC structure [22,28,40]. Clarified yeast cell lysates were immunoblotted with antibodies against the Myc and HA epitopes to verify expression of the DBD and AD fusion proteins, respectively (Figure 2, lower panels).
Figure 2. KAP-1 selectively interacts with the Fes CC2 domain.
Yeast strain AH109 was co-transformed with the indicated pairs of plasmids and grown either on plates lacking leucine and tryptophan (−L−W) or on plates lacking leucine, tryptophan, adenine and histidine (−L−W−A−H; upper panel). Growth on −L−W plates selects for the presence of both plasmids and growth on −L−W−A−H plates is indicative of protein–protein interaction. Results from four independent transformants are shown for each combination. Yeast colonies from −L−W plates in the upper panel were grown in liquid medium and cell lysates were immunoblotted with Myc (middle panel) or HA antibodies (lower panel) to detect the DBD and AD fusions respectively. The positions of the fusion proteins are indicated on the left. Data are representative of three independent experiments.
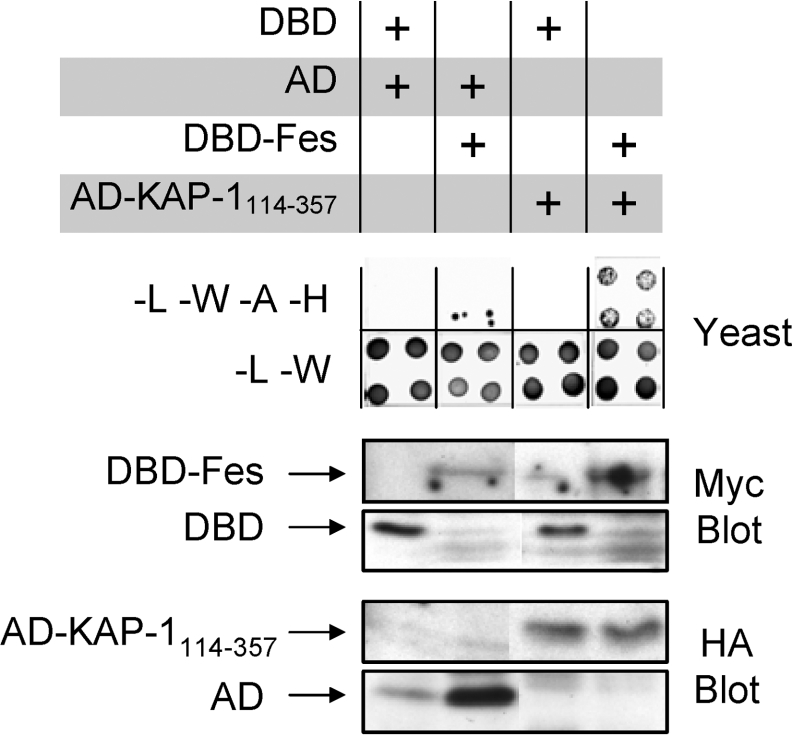
In a final series of experiments, we investigated the interaction of full-length Fes with KAP-1114–357 in the yeast system. Fes was fused to the Gal4 DBD (DBD–Fes) and assessed for association with AD–KAP-1114–357. AD–KAP-1114–357 interacted with DBD–Fes, permitting growth on SD-L-W-A-H medium (Figure 3). Immunoblots of yeast cell lysates demonstrate expression of the DBD–Fes and AD–KAP-1114–357 fusion proteins as well as the DBD and AD controls.
Figure 3. KAP-1 interacts with full-length Fes.
Yeast strain AH109 was co-transformed with the indicated pairs of plasmids. Colonies were replated on −L−W plates to verify transformation of both plasmids and on −L−W−A−H plates to test for protein–protein interaction (upper panel). Results from four independent transformants are shown for each combination. Colonies from −L−W plates were grown in liquid medium and clarified lysates were subjected to immunoblot analyses utilizing Myc antibodies to detect DBD-fusion proteins and HA antibodies to visualize AD-fusion proteins as indicated (lower panels). The positions of the AD and DBD proteins are indicated on the left. Data are representative of three independent experiments.
The KAP-1 CC domain directly interacts with the Fes N-terminal region
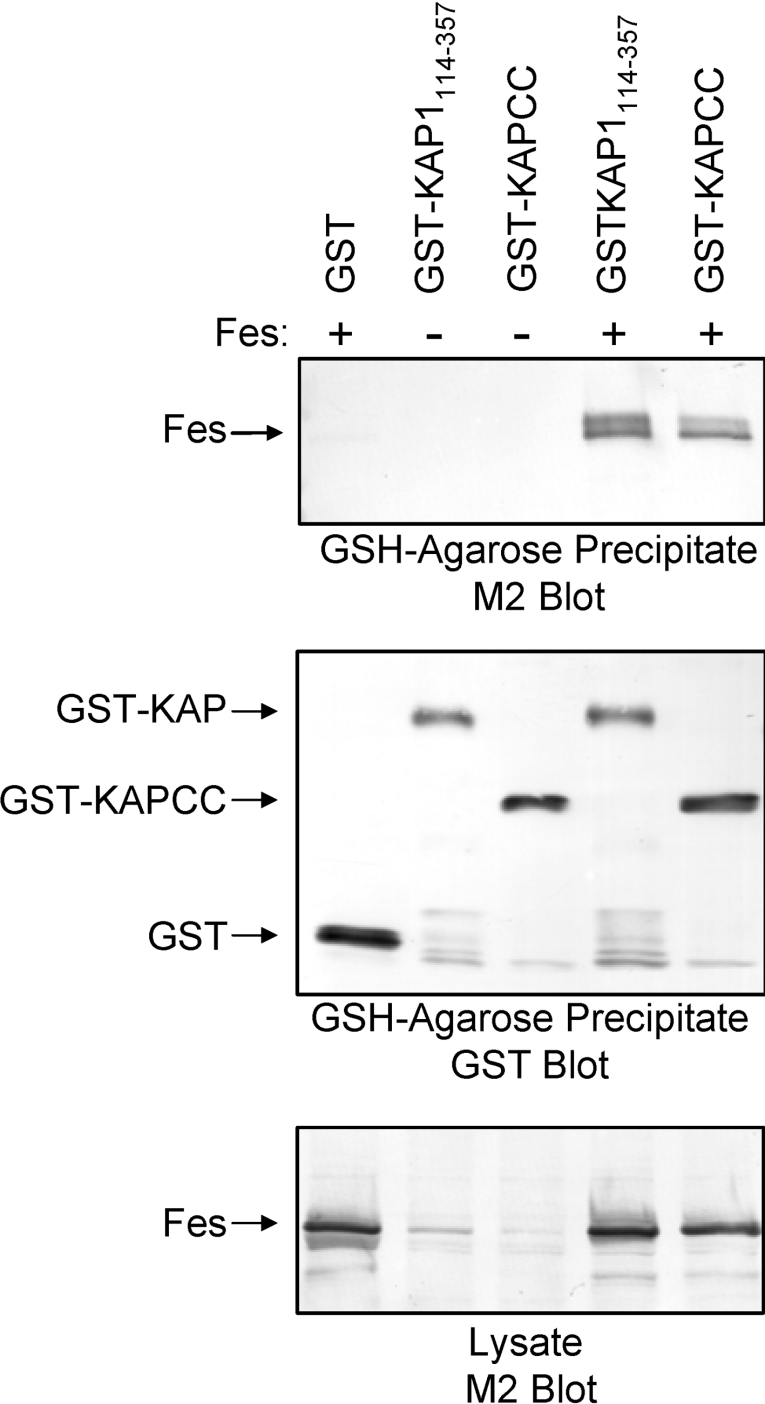
We next investigated whether the results observed in the yeast twohybrid assay were due to direct interaction between the KAP-1 and Fes proteins. In these studies, Fes and KAP-1 proteins were co-expressed in Sf-9 insect cells using recombinant baculoviruses. The baculoviruses used for these experiments are illustrated in Figure 4. In the first experiment, GST–KAP-1114–357 was co-expressed with FLAG-tagged full-length Fes. GST–KAP-1114–357 was then precipitated with glutathione–agarose beads and associated Fes was visualized by immunoblotting with FLAG-specific antibodies. GST–KAP-1114–357 bound full-length wild-type Fes, whereas no association with Fes was observed with GST alone (Figure 5). Because KAP-1114–357 contains a CC domain that interacted with the Fes CC2 domain in the yeast two-hybrid assay (Figure 2), we also tested whether the KAP-1 CC domain alone was sufficient to interact with Fes. As shown in Figure 5, a GST–KAP-1 CC domain fusion protein also associated with Fes at levels comparable to those observed for GST–KAP-1114–357. This result shows that the KAP-1 CC domain is sufficient for recognition of c-Fes.
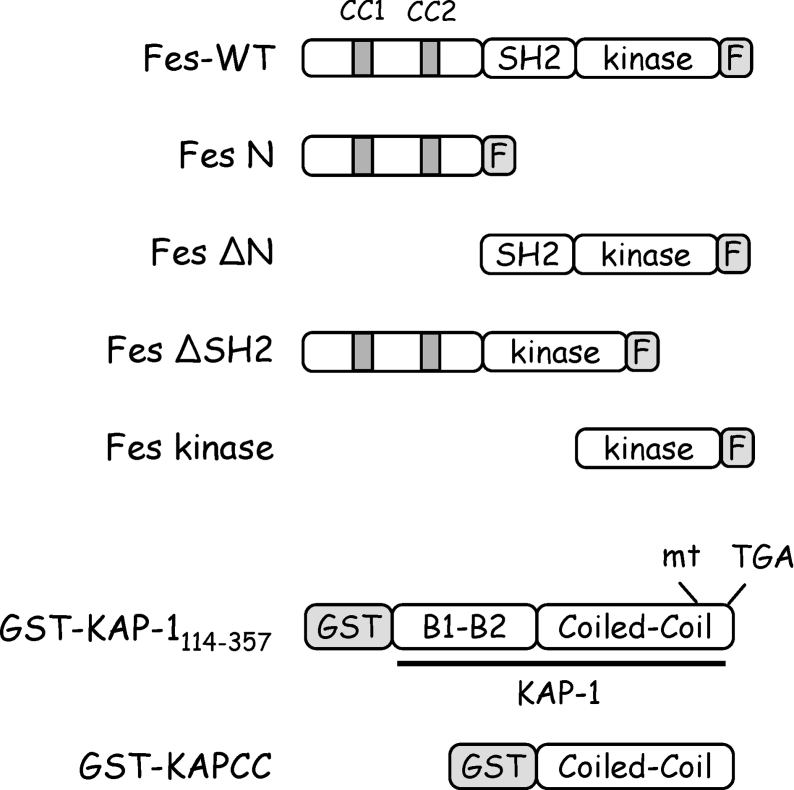
Figure 4. Baculoviruses expression constructs.
Recombinant baculoviruses were created for expression of wild-type Fes and a series of deletion mutants in Sf-9 insect cells. The Fes deletion mutants include the Fes N-terminal region alone (Fes N), the Fes SH2 domain plus the kinase domain (Fes ΔN), the Fes N-terminal domain plus the Fes kinase domain (Fes ΔSH2) and the Fes kinase domain alone (Fes kinase). All Fes constructs include a C-terminal FLAG epitope tag (F). Baculoviruses were also created for expression of GST fused to the KAP-1 CC domain (GST–KAPCC) or to the B boxes plus the CC domain (GST–KAP-1114–357).
Figure 5. The KAP-1 CC domain interacts with full-length Fes.
GST, GST–KAP-1114–357 and GST–KAPCC were co-expressed with full-length Fes in Sf-9 cells. GST fusion proteins were precipitated with GSH–agarose beads followed by immunoblot analysis with antibodies raised against the FLAG epitope (M2) to detect associated Fes (upper panel) or against GST to verify protein recovery (middle panel). In the lower panel, clarified lysates were immunoblotted with the FLAG (M2) antibody to control for Fes expression levels. The positions of the Fes and GST-fusion proteins are indicated on the left. Data are representative of two independent experiments.
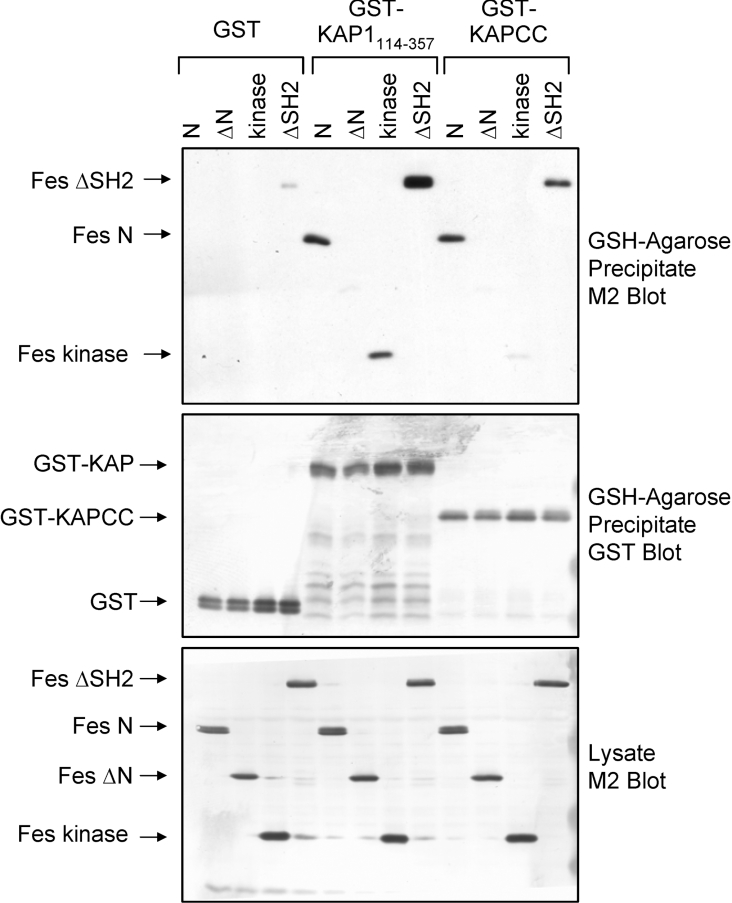
To identify the Fes domains required for KAP-1 binding, the Fes deletion mutants illustrated in Figure 4 were co-expressed with GST–KAP114–357 or GST and assessed for association as described in Figure 5 for full-length Fes. Figure 6 (upper panel) demonstrates that the Fes N-terminal region alone (N) and a Fes deletion mutant lacking the SH2 domain (ΔSH2) exhibited strong interaction with KAP-1114–357. Neither Fes deletion construct interacted with GST, demonstrating a dependence on the KAP-1 sequence. In contrast, a construct lacking the N-terminal domain (ΔN) did not bind to KAP-1114–357. These results demonstrate that KAP-1114–357 interacts with the unique N-terminal region of Fes, which contains the CC2 domain used in the original two-hybrid screen (Figure 2). Surprisingly, a Fes construct containing only the Fes kinase domain also interacted with KAP-1114–357. This result suggests the presence of a second KAP-1114–357 interacting motif that becomes accessible following deletion of the Fes N-terminal and SH2 domains (see the Discussion section). The blots in the upper panel of Figure 6 were stripped and re-probed with antibodies against GST (Figure 6, middle panel) to demonstrate equivalent levels of GST-fusion proteins. Clarified lysates were blotted for FLAG to verify equal expression of the Fes deletion mutants (Figure 6, lower panel).
Figure 6. The KAP-1 CC domain associates with the Fes N-terminal region.
Sf-9 insect cells were infected with recombinant baculoviruses carrying GST, GST–KAP-1114–357 or GST–KAPCC together with the Fes N, ΔN, ΔSH2 or kinase constructs as indicated. GST-fusion proteins were precipitated with GSH–agarose beads followed by immunoblot analysis with antibodies raised against the FLAG epitope (M2) to detect associated Fes proteins (upper panel). The blot in the upper panel was stripped and reprobed with GST antibodies to verify fusion protein recovery (middle panel). Cell lysates were immunoblotted with the FLAG antibody (M2) to detect the Fes deletion mutants (lower panel). The positions of the Fes and GST-fusion proteins are indicated on the left. Data are representative of three independent experiments.
To determine whether the KAP-1 CC domain alone was sufficient to interact with the Fes N-terminal region and kinase domain, GST–KAPCC was co-expressed with the Fes deletion constructs and assessed for association. As shown in Figure 6 (upper panel), the KAP CC domain efficiently interacted with Fes mutants that retained the N-terminal region (N and ΔSH2), whereas deletion mutants lacking this region (ΔN and kinase) exhibited low or undetectable association with KAPCC (upper panel). Together, these data suggest that KAP114–357 interaction with the Fes N-terminal region is dependent on the KAP-1 CC domain whereas a secondary contact between KAP114–357 and the Fes kinase domain is independent of the KAP CC domain and may reside in the B-box motifs.
Fes interaction with full-length KAP-1 in mammalian cells requires an activating mutation in the Fes CC1 domain
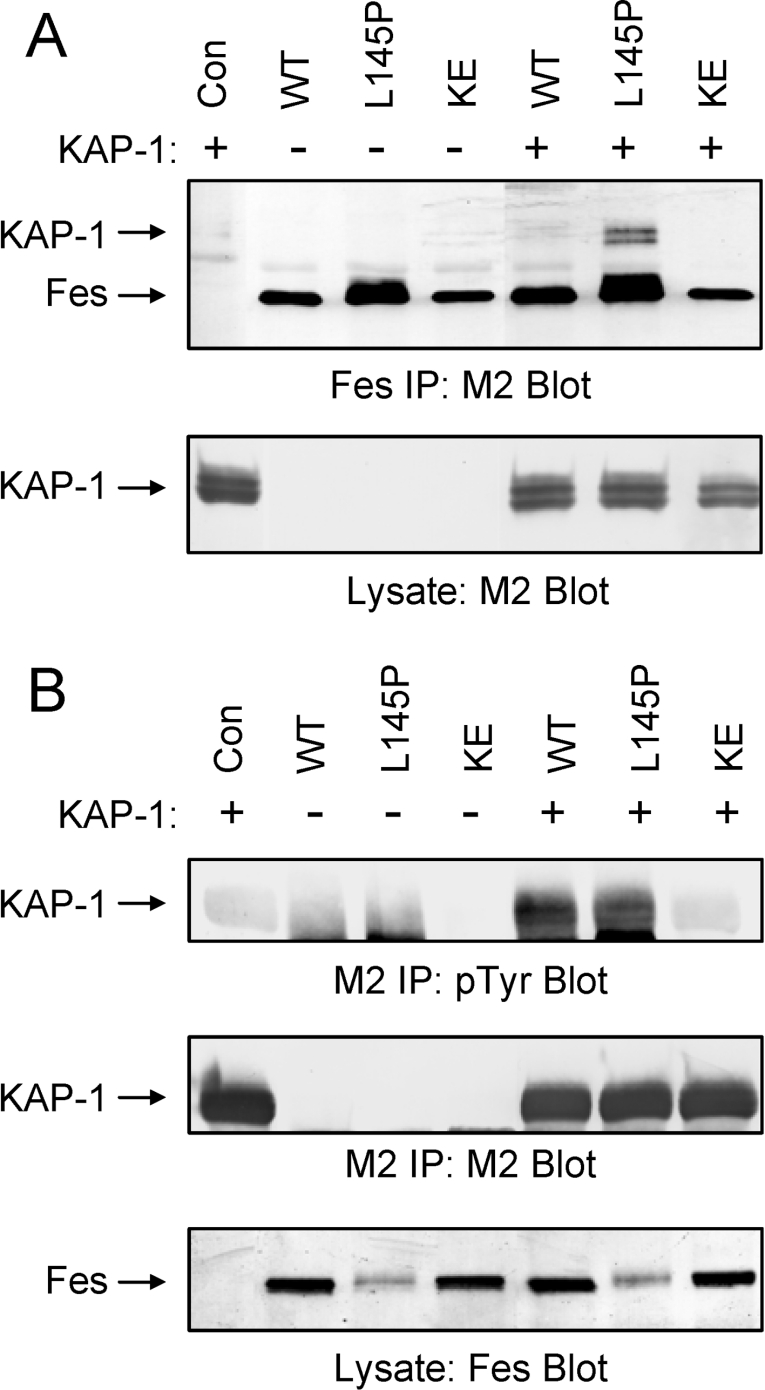
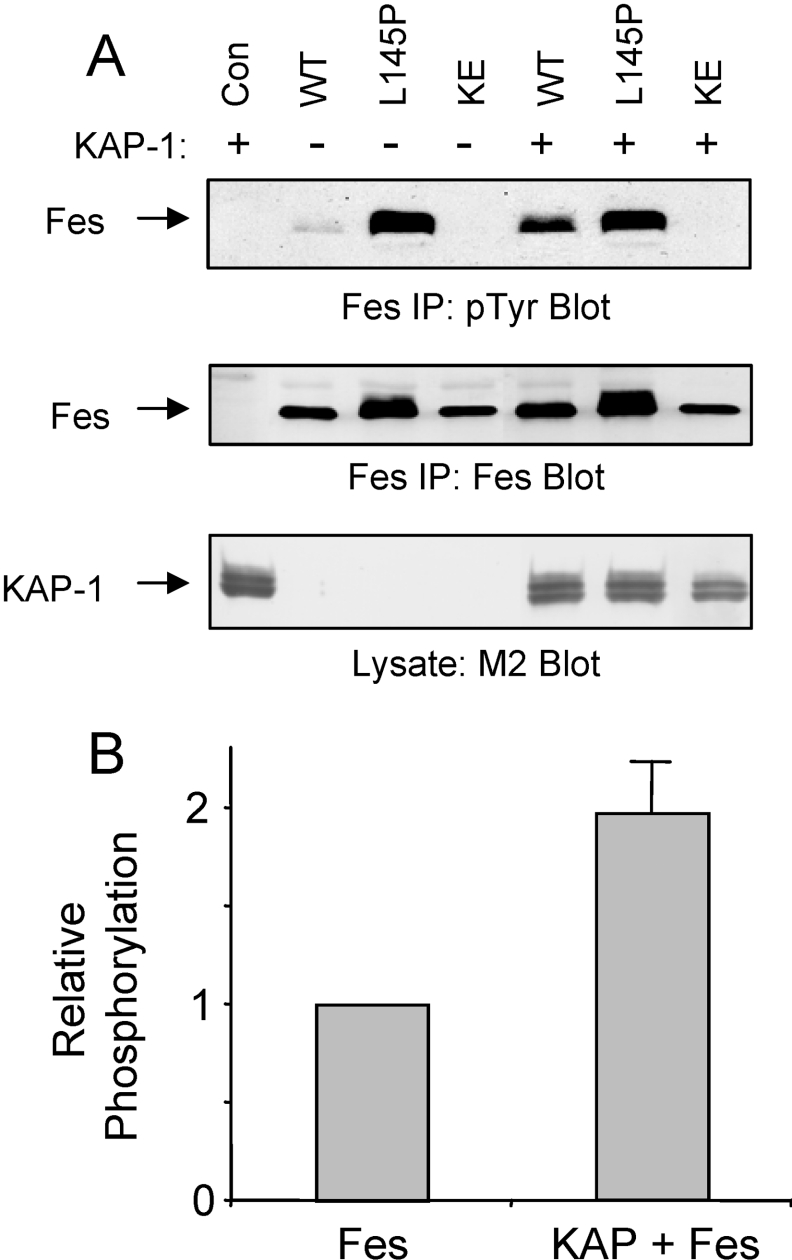
To determine whether Fes interacts with KAP-1 in mammalian cells, co-immunoprecipitation analyses were performed. For these studies, 293T cells were co-transfected with a KAP-1 expression plasmid as well as expression vectors for wild-type Fes (WT), a kinase dead form (KE) of Fes containing a glutamate to lysine mutation at residue 590 within the ATP binding site, or a CC1 point mutant (L145P) shown previously to induce strong activation of Fes kinase activity [22]. The KAP-1 and Fes constructs were tagged with the FLAG epitope. Fes was then immunoprecipitated from clarified cell extracts with an antibody directed against the N-terminal region, and associated KAP-1 was identified via immunoblotting with a FLAG-specific antibody (M2; Figure 7A, upper panel). Surprisingly, full-length KAP-1 interaction with wild-type Fes was low or undetectable. The introduction of a point mutation within the CC1 domain significantly enhanced association of Fes with KAP-1. This mutation has been shown to enhance Fes kinase and biological activities [22] through a mechanism that may expose the CC2 domain for substrate recruitment (see the Discussion section). Control FLAG (M2) immunoblots show equivalent recovery of Fes protein in each immunoprecipitate as well as equivalent expression of KAP-1 under all conditions.
Figure 7. Full-length KAP-1 is a Fes binding partner and substrate in 293T cells.
Full-length KAP-1 was transiently expressed in 293T cells in the absence (Con) or presence of full-length wild-type Fes (WT), a Fes mutant containing a proline substitution at Leu145 within the CC1 domain (L145P) or a kinase-inactive Fes mutant (KE) in which conserved Lys590 was replaced with glutamate. Fes constructs include C-terminal FLAG epitope tags whilst KAP-1 contains an N-terminal FLAG tag. (A) Fes proteins were immunoprecipitated with an antibody directed against the Fes N-terminal region and immunoblotted with the FLAG antibody to detect associated KAP-1 (upper panel). M2 also detects immunoprecipitated Fes. Lysates were blotted with the M2 antibody to verify equivalent expression of KAP-1 in each case (lower panel). (B) KAP-1 was immunoprecipitated with the FLAG antibody (M2) and immuoblotted with anti-phosphotyrosine antibodies (pTyr; upper panel). This blot was then stripped and reprobed with the M2 antibody to verify KAP-1 recovery (middle panel). Lysates were probed with the Fes antibody to verify Fes protein expression (lower panel). The positions of Fes and KAP-1 are indicated on the left. Data are representative of three independent experiments.
KAP-1 is a Fes substrate
The direct interactions between Fes and KAP-1 observed in yeast and mammalian cells suggested that Fes binding may induce KAP-1 tyrosine phosphorylation. To test this hypothesis, 293T cells were co-transfected with Fes and KAP-1 expression plasmids as described for Figure 7(A). Following immunoprecipitation of KAP-1 with FLAG-specific antibodies, phosphotyrosine content was analysed by Western blotting with the anti-phosphotyrosine antibody PY99 (Figure 7B, upper panel). Co-expression of KAP-1 with either wild-type Fes or the active L145P mutant induced strong KAP-1 tyrosine phosphorylation. In contrast, KAP-1 tyrosine phosphorylation was undetectable in the absence of Fes or upon co-expression of a kinase-defective Fes mutant (KE). The blot in the upper panel of Figure 7(B) was stripped and re-probed with a FLAG-specific antibody to demonstrate equivalent immunoprecipitation of KAP-1. Immunoblots of the clarified lysates demonstrate equivalent expression of the wild-type and L145P forms of Fes in the presence and absence of KAP-1.
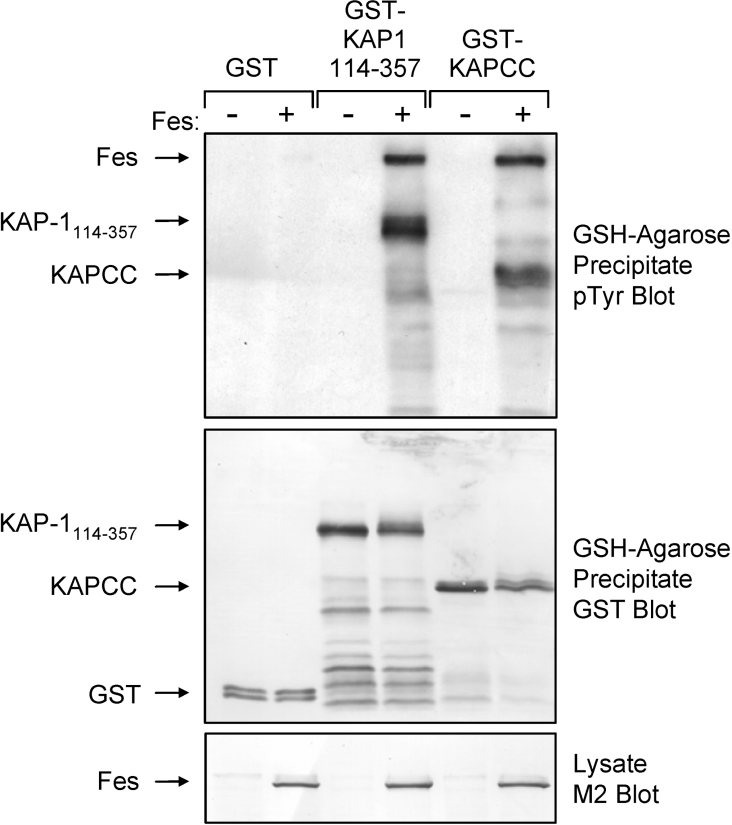
To provide further evidence that KAP-1 is a direct substrate for Fes and to map the region of KAP-1 that is phosphorylated, Sf-9 insect cells were infected with the GST–KAP-1114–357 and GST–KAPCC baculoviruses in the absence or presence of a c-Fes baculovirus. GST fusion proteins were precipitated with glutathione–agarose beads and KAP phosphotyrosine content was assessed by immunoblotting (Figure 8). Both GST–KAP-1114–357 and GST–KAPCC were strongly tyrosine phosphorylated upon co-expression with Fes (Figure 8, upper panel). Fes did not induce phosphorylation of GST alone. The blot in Figure 8, upper panel was stripped and re-probed with anti-GST antibodies to verify equivalent recovery of the GST fusion proteins (Figure 8, middle panel), while Fes expression was confirmed in the cell lysates by anti-FLAG immunoblotting (Figure 8, lower panel). Together these data indicate that KAP-1 is a Fes substrate and that at least one site of KAP tyrosine phosphorylation occurs within the KAP-1 CC domain.
Figure 8. Fes phosphorylates the KAP-1 CC domain.
Sf-9 cells were infected with baculoviruses carrying GST, GST–KAP-1114–357 or GST fused to the KAP-1 CC domain (GST–KAPCC) in the presence or absence of a Fes baculovirus. GST fusion proteins were precipitated with glutathione–agarose beads and immunoblotted with anti-phosphotyrosine antibodies (pTyr; upper panel). This blot was stripped and reprobed with a GST antibody to verify GST fusion protein recovery (middle panel). Cell lysates were immunoblotted with FLAG antibodies (M2) to demonstrate equivalent Fes protein expression (lower panel). Data are representative of three independent experiments.
A slower migrating tyrosine phosphorylated protein was also observed in the GSH-agarose precipitates when Fes was co-expressed with GST–KAP-1114–357 and GST–KAPCC but not with GST alone (Figure 8, upper panel). This protein co-migrated with the Fes band identified in the clarified lysates, suggesting that active Fes associates with the KAP-1 CC domain. Note that unlike 293T cells, high-level expression of wild-type Fes in Sf-9 insect cells results in constitutive kinase activity [38].
KAP-1 is a Fes kinase activator
Previous studies suggest that Fes is maintained in an inactive state in mammalian cells [22,24]. Fes activation may result from CC-mediated oligomerization via N-terminal CC domains and subsequent trans-autophosphorylation of the activation loop tyrosine [22,24,27,38]. Because KAP-1 binds to the Fes CC2 domain in the N-terminal region, we reasoned that this interaction could affect Fes kinase activity. To test this hypothesis, full-length KAP-1 was co-expressed in 293T cells with wild-type (WT), active (L145P) or kinase-dead (KE) forms of Fes. Fes proteins were immunoprecipitated and probed for kinase activity via immunoblotting with anti-phosphotyrosine antibodies (Figure 9A, upper panel). In agreement with previous studies, autophosphorylation of wild-type Fes was barely detectable in 293T cells [24,41]. Co-expression with KAP-1 enhanced wild-type Fes autophosphorylation nearly 2-fold (Figure 9B). Disruption of the CC1 domain (Fes L145P) released Fes kinase activity and KAP-1 co-expression had no further effect. KAP-1 did not affect the kinase-dead form of Fes, suggesting that the observed increase in wild-type Fes phosphotyrosine content results from direct KAP-1 binding and does not involve another kinase. The blot in the upper panel of Figure 9(A) was stripped and re-probed with a Fes-specific antibody to demonstrate equivalent recovery of Fes proteins (Figure 9A, middle panel), and clarified lysates were probed with a FLAG-specific antibody to verify KAP-1 expression (Figure 9A, lower panel).
Figure 9. KAP-1 induces Fes kinase activity.
(A) 293T cells were transfected with full-length KAP-1 in the absence or presence of wild-type Fes (WT), Fes containing a Leu145 to proline mutation within the CC1 domain (L145P), or a kinase-inactive form of Fes (KE) containing a Glu590 to lysine mutation within the ATP binding pocket. Fes was then immunoprecipitated from clarified lysates with an N-terminal-specific antibody and immunoblotted for phosphotyrosine content (pTyr; upper panel). This blot was stripped and reprobed with the Fes antibody to verify expression (middle panel). Clarified lysates were immunoblotted with the FLAG antibody (M2) to verify equivalent KAP-1 expression (lower panel). (B) Wild-type Fes phosphotyrosine content was quantified using a BioRad GS710 Scanning Densitometer and Quantity One software. Relative phosphotyrosine content was normalized for protein level and is expressed as a fold induction in the presence of KAP-1. Results are mean±S.E.M., n=6.
Association of endogenous Fes with KAP-1 in HL-60 leukaemia cells
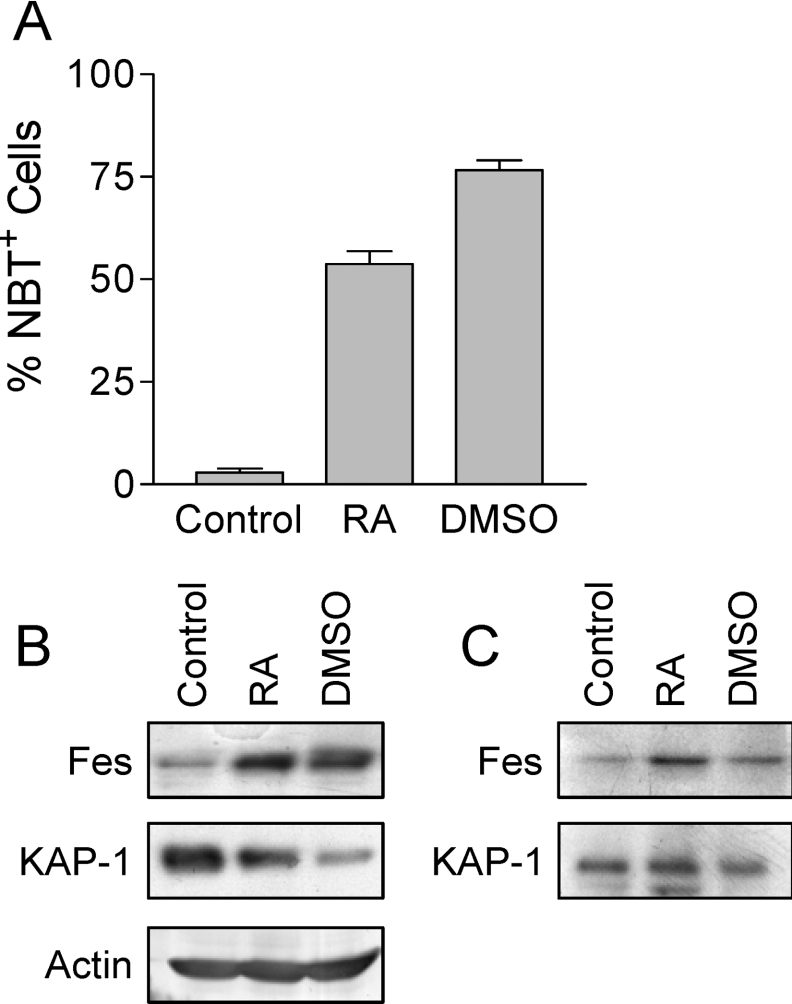
To determine whether Fes and KAP-1 interact in a physiological setting, we looked for Fes–KAP-1 complexes in HL-60 cells that were induced to differentiate along the granulocytic pathway by treatment with retinoic acid or DMSO. Treatment of HL-60 cells with either of these agents led to the induction of differentiation as well as an increase in the level of Fes protein (Figure 10), as has been reported previously [6]. KAP-1 expression was easily detected in lysates of HL-60 cells, although the overall levels of KAP-1 appeared to decline in the differentiated cell lysates. To determine whether endogenous Fes and KAP-1 associate in HL-60 cells, Fes was immunoprecipitated from the cell lysates and probed for the presence of associated KAP-1 by immunoblotting. Figure 10(C) shows the presence of KAP-1 in the Fes immunoprecipitates, supporting the presence of Fes–KAP-1 complexes in differentiated HL-60 cells. This result identifies KAP-1 as a binding partner for Fes in a cell context where Fes is normally expressed and is biologically active [6,20].
Figure 10. Association of endogenous Fes with KAP-1 in HL-60 cells.
(A) HL-60 myeloid leukaemia cells were treated with retinoic acid (RA) or DMSO to induce terminal differentiation or left untreated (Control). The extent of differentiation was assessed 4 days later as the percentage of cells able to reduce NBT. Results are mean±S.D., n=6. (B) Cell lysates from control and differentiated cells in (A) were probed for Fes and KAP-1 protein levels by immunoblotting as described in the Materials and Methods section. An actin blot was also performed as a loading control. (C) Fes was immunoprecipitated from HL-60 cell lysates and probed for associated KAP-1 as well as Fes by immunoblotting. This experiment was repeated five times, and the level of Fes–KAP-1 interaction shown was observed in two of these experiments. KAP-1 recovery was weaker or absent in the other trials, which is likely to reflect heterogeneity in the level of Fes protein available for immunoprecipitation.
DISCUSSION
In the present paper, the transcriptional co-repressor KAP-1 was identified as an interacting partner for the non-receptor PTK, Fes. Molecular recognition of KAP-1 by Fes was mediated by mutual CC domain interactions, although other domains may contribute to the association (see below). KAP-1 represents the first protein identified to interact with the Fes CCs, and defines a new mode of substrate recognition for non-receptor protein tyrosine kinases. In addition, KAP-1 binding stimulated Fes autophosphorylation and led to KAP-1 tyrosine phosphorylation, suggesting that KAP-1 is an activator as well as a downstream substrate for Fes.
The KAP-1 clone (KAP-1114–357) isolated from the K562 library screen exhibited selectivity toward the Fes CC2 domain, as it did not interact with the Fes CC1 domain or the CC domain from the yeast transcription factor GCN4 (Figure 2) [42]. Interaction of KAP-1 with the Fes CC2 domain was eliminated upon mutation of the CC2 Leu334 to a proline residue, a substitution previously shown to destabilize CC2 function [22,43]. Moreover, the KAP-1 CC domain alone was sufficient to interact with the Fes N-terminal region, which contains the CC domains. In contrast, the KAP-1 CC domain failed to associate with Fes deletion mutants lacking the N-terminal sequences. Together with the yeast data, these studies strongly suggest that the KAP-1 CC domain interacts directly with the Fes CC2 domain.
Although the Fes CC2 domain is sufficient for KAP-1 binding, experiments in Sf-9 insect cells suggest additional points of contact. This second KAP-1 interaction motif may lie in the Fes kinase domain as this domain alone interacted with KAP-1114–357 (Figure 6). In contrast, the KAP-1 CC domain alone did not interact with the Fes kinase domain. As KAP-1114–357 contains only the B-boxes and CC domain of KAP-1, this result suggests that the KAP-1 B-boxes are most likely to be responsible for interaction with the Fes kinase domain. Accessibility to this second KAP-1 interaction domain may be important for stable interaction of full-length KAP-1 and Fes in mammalian cells as well, as association was only detected following the introduction of a destabilizing leucine to proline mutation in the Fes CC1 domain (Figure 7A). Interestingly, this same mutation released Fes transforming activity in Rat-2 cells and promoted Fes-induced differentiation and survival of TF-1 myeloid leukaemia cells [22]. In light of these observations, it will be interesting to determine whether KAP-1 or other CC2-binding proteins are required for the biological effects of Fes.
Co-expression of full-length Fes with KAP-1 in mammalian cells resulted in KAP-1 tyrosine phosphorylation (Figure 7B). Phosphorylation was not detected in the absence of Fes or following co-expression with a kinase inactive form of Fes (Fes-KE), suggesting that KAP-1 is a direct substrate for the Fes kinase domain. Similar studies in Sf-9 cells demonstrated strong tyrosine phosphorylation of KAP-1114–357 and the KAP-1 CC domain following co-expression with Fes (Figure 8). Our results also identify KAP-1 Tyr242 as a phosphorylation site for Fes, as this is the only tyrosine residue present in the KAP-1 CC domain. Tyrosine phosphorylation within the CC domain could potentially alter KAP-1 oligomerization and interactions with other transcription factors.
In addition to serving as a Fes substrate, KAP-1 also stimulated Fes autophosphorylation (Figure 9). The mechanism by which KAP-1 association activates Fes tyrosine kinase activity is not entirely clear. KAP-1 is oligomeric [44], suggesting a mechanism in which KAP-1 oligomers recruit multiple Fes monomers, promoting Fes oligomerization and autophosphorylation. Interestingly, the breakpoint cluster region protein has also been identified as a Fes N-terminal domain binding protein and Fes activator, suggesting that Fes activation via substrate association with its N-terminal region may represent a general activation mechanism [45]. Like KAP-1, the breakpoint cluster region protein has an N-terminal CC domain that mediates tetramerization [46].
KAP-1 represents a new member of a growing list of transcription factors that are modulated by Fes and the closely related Fer kinase. Fes has been shown to activate STAT3 (signal transducer and activator of transcription 3) by direct tyrosine phosphorylation in human 293T cells. Macrophages derived from mice in which the endogenous fes gene was replaced with a kinase inactive mutant exhibited reduced STAT3 activation in response to GM–CSF (granulocyte/macrophage colony-stimulating factor) [47,48]. More recently, Fes has been shown to enhance PU.1 and C/EBPα activity in myeloid cell lines [23,49]. STAT3, PU.1 and C/EBPα together with Fes have been implicated in myeloid cell differentiation suggesting that these transcription factors may act downstream of Fes [21,22,24,25,50–52]. In the present study we show that Fes stably associates with KAP-1 in both control and terminally differentiated HL-60 cells (Figure 10), supporting a connection between these signalling proteins in myeloid differentiation. Other work has shown that differentiation of HL-60 cells promotes localization of Fes to the nucleus [53], where it may come into contact with KAP-1 and other transcription factors that regulate the differentiation programme. This possibility will be the subject of future investigation. Fer associates with and phosphorylates the nuclear transcription factor TMF (TATA element modulatory factor) [54]. TMF binds to the TATA element of RNA polymerase II promoters where it represses transcription by competing for TATA binding protein recruitment to the TATA element. Binding and phosphorylation of transcriptional regulators such as TMF and KAP-1 by the Fes and Fer kinases suggests a more general function for this kinase family in the regulation of transcription.
Acknowledgments
We thank Dr Frank Rauscher III of the The Wistar Institute Cancer Center, Philadelphia, PA, U.S.A. for the KAP-1 mammalian expression vector, and Dr Peter Greer of Queen's University Cancer Research Institute, Ontario, Canada for the Fes-specific rabbit polyclonal antiserum. This work was supported by NIH (National Institutes of Health) Grants CA58667 and CA101828.
References
- 1.Lee W. H., Bister K., Pawson A., Robins T., Moscovici C., Duesberg P. H. Fujinami sarcoma virus: an avian RNA tumor virus with a unique transforming gene. Proc. Natl. Acad. Sci. U.S.A. 1980;77:2018–2022. doi: 10.1073/pnas.77.4.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groffen J., Heisterkamp N., Shibuya M., Hanafusa H., Stephenson J. R. Transforming genes of avian (v-fps) and mammalian (v-fes) retroviruses correspond to a common cellular locus. Virology. 1983;125:480–486. doi: 10.1016/0042-6822(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 3.Hampe A., Laprevotte I., Galibert F., Fedele L. A., Sherr C. J. Nucleotide sequences of feline retroviral oncogenes (v-fes) provide evidence for a family of tyrosine-specific protein kinase genes. Cell. 1982;30:775–785. doi: 10.1016/0092-8674(82)90282-3. [DOI] [PubMed] [Google Scholar]
- 4.Hanafusa T., Wang L. H., Anderson S. M., Karess R. E., Hayward W. S., Hanafusa H. Characterization of the transforming gene of Fujinami sarcoma virus. Proc. Natl. Acad. Sci. U.S.A. 1980;77:3009–3013. doi: 10.1073/pnas.77.5.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C. C., Hammond C., Bishop J. M. Nucleotide sequence of v-fps in the PRCII strain of avian sarcoma virus. J. Virol. 1984;50:125–131. doi: 10.1128/jvi.50.1.125-131.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smithgall T. E., Yu G., Glazer R. I. Identification of the differentiation-associated p93 tyrosine protein kinase of HL-60 leukaemia cells as the product of the human c-fes locus and its expression in myelomonocytic cells. J. Biol. Chem. 1988;263:15050–15055. [PubMed] [Google Scholar]
- 7.MacDonald I., Levy J., Pawson T. Expression of the mammalian c-fes protein in hematopoietic cells and identification of a distinct fes-related protein. Mol. Cell. Biol. 1985;5:2543–2551. doi: 10.1128/mcb.5.10.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman R. A., Gabrilove J. L., Tam J. P., Moore M. A., Hanafusa H. Specific expression of the human cellular fps/fes-encoded protein NCP92 in normal and leukemic myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2379–2383. doi: 10.1073/pnas.82.8.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greer P., Haigh J., Mbamalu G., Khoo W., Bernstein A., Pawson T. The Fps/Fes protein-tyrosine kinase promotes angiogenesis in transgenic mice. Mol. Cell. Biol. 1994;14:6755–6763. doi: 10.1128/mcb.14.10.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigh J., McVeigh J., Greer P. The fps/fes tyrosine kinase is expressed in myeloid, vascular endothelial, epithelial and neuronal cells and is localized in the trans-golgi network. Cell Growth Differ. 1996;7:931–944. [PubMed] [Google Scholar]
- 11.Shibata A., Laurent C. E., Smithgall T. E. The c-Fes protein-tyrosine kinase accelerates NGF-induced differentiation of PC12 cells through a PI3K-dependent mechanism. Cell. Signal. 2003;15:279–288. doi: 10.1016/s0898-6568(02)00089-x. [DOI] [PubMed] [Google Scholar]
- 12.Park W. Y., Ahn J. H., Feldman R. A., Seo J. S. c-Fes tyrosine kinase binds to and activates STAT3 after granulocyte-macrophage colony-stimulating factor stimulation. Cancer Lett. 1998;129:29–37. doi: 10.1016/s0304-3835(98)00077-9. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda T., Fukada T., Takahashi-Tezuka M., Okuyama Y., Fujitani Y., Hanazono Y., Hirai H., Hirano T. Activation of Fes tyrosine kinase by gp130, an interleukin-6 family cytokine signal transducer, and their association. J. Biol. Chem. 1995;270:11037–11039. doi: 10.1074/jbc.270.19.11037. [DOI] [PubMed] [Google Scholar]
- 14.Kanda S., Lerner E. C., Tsuda S., Shono T., Kanetake H., Smithgall T. E. The nonreceptor protein-tyrosine kinase c-Fes is involved in fibroblast growth factor-2-induced chemotaxis of murine brain capillary endothelial cells. J. Biol. Chem. 2000;275:10105–10111. doi: 10.1074/jbc.275.14.10105. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H., Foltenyi K., Kashiwada M., Donahue L., Vuong B., Hehn B., Rothman P. Fes mediates the IL-4 activation of insulin receptor substrate-2 and cellular proliferation. J. Immunol. 2001;166:2627–2634. doi: 10.4049/jimmunol.166.4.2627. [DOI] [PubMed] [Google Scholar]
- 16.Izuhara K., Feldman R. A., Greer P., Harada N. Interaction of the c-fes proto-oncogene product with the interleukin-4 receptor. J. Biol. Chem. 1994;269:18623–18629. [PubMed] [Google Scholar]
- 17.Hanazono Y., Chiba S., Sasaki K., Mano H., Miyajima A., Arai K., Yazaki Y., Hirai H. c-fps/fes protein-tyrosine kinase is implicated in a signalling pathway triggered by granulocyte-macrophage colony-stimulating factor and interleukin-3. EMBO J. 1993;12:1641–1646. doi: 10.1002/j.1460-2075.1993.tb05809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brizzi M. F., Aronica M. G., Rosso A., Bagnara G. P., Yarden Y., Pegoraro L. Granulocyte-macrophage colony-stimulating factor stimulates JAK2 signalling pathway and rapidly activates p93fes, STAT1 p91, and STAT3 p92 in polymorphonuclear leukocytes. J. Biol. Chem. 1996;271:3562–3567. doi: 10.1074/jbc.271.7.3562. [DOI] [PubMed] [Google Scholar]
- 19.Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat. Rev. Mol. Cell. Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- 20.Smithgall T. E., Rogers J. A., Peters K. L., Li J., Briggs S. D., Lionberger J. M., Cheng H., Shibata A., Scholtz B., Schreiner S., Dunham N. The c-Fes family of protein-tyrosine kinases. Crit. Rev. Oncog. 1998;9:43–62. doi: 10.1615/critrevoncog.v9.i1.40. [DOI] [PubMed] [Google Scholar]
- 21.Manfredini R., Balestri R., Tagliafico E., Trevisan F., Pizzanelli M., Grande A., Barbieri D., Zucchini P., Citro G., Franceschi C., Ferrari S. Antisense inhibition of c-fes proto-oncogene blocks PMA-induced macrophage differentiation in HL60 and in FDC-P1/MAC-11 cells. Blood. 1997;89:135–145. [PubMed] [Google Scholar]
- 22.Cheng H. Y., Schiavone A. P., Smithgall T. E. A point mutation in the N-terminal CC domain releases c-Fes tyrosine kinase activity and survival signalling in myeloid leukemia cells. Mol. Cell. Biol. 2001;21:6170–6180. doi: 10.1128/MCB.21.18.6170-6180.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J., Feldman R. A. Activated Fes protein tyrosine kinase induces terminal macrophage differentiation of myeloid progenitors (U937 cells) and activation of the transcription factor PU.1. Mol. Cell. Biol. 2002;22:1903–1918. doi: 10.1128/MCB.22.6.1903-1918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H., Rogers J. A., Dunham N. A., Smithgall T. E. Regulation of c-Fes tyrosine kinase and biological activities by N-terminal coiled-coil oligomerization domains. Mol. Cell. Biol. 1999;19:8335–8343. doi: 10.1128/mcb.19.12.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Smithgall T. E., Glazer R. I. K562 leukemia cells transfected with the human c-fes gene acquire the ability to undergo myeloid differentiation. J. Biol. Chem. 1989;264:10276–10281. [PubMed] [Google Scholar]
- 26.Smithgall T. E. Signal transduction pathways regulating hematopoietic differentiation. Pharmacol. Rev. 1998;50:1–19. [PubMed] [Google Scholar]
- 27.Read R. D., Lionberger J. M., Smithgall T. E. Oligomerization of the Fes tyrosine kinase. Evidence for a coiled-coil domain in the unique N-terminal region. J. Biol. Chem. 1997;272:18498–18503. doi: 10.1074/jbc.272.29.18498. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A. Coiled coils: new structures and new functions. Trends Biochem. Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 29.Kim S. S., Chen Y. M., O'Leary E., Witzgall R., Vidal M., Bonventre J. V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moosmann P., Georgiev O., Le Douarin B., Bourquin J. P., Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechner M. S., Begg G. E., Speicher D. W., Rauscher F. J., III Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill C., Qutob M. S., Yee S. P., Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 35.Schultz D. C., Friedman J. R., Rauscher F. J., III Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Rogers J. A., Read R. D., Li J., Peters K. L., Smithgall T. E. Autophosphorylation of the Fes tyrosine kinase. Evidence for an intermolecular mechanism involving two kinase domain tyrosine residues. J. Biol. Chem. 1996;271:17519–17525. doi: 10.1074/jbc.271.29.17519. [DOI] [PubMed] [Google Scholar]
- 39.Lozzio B. B., Lozzio C. B., Bamberger E. G., Feliu A. S. A multipotential leukemia cell line (K-562) of human origin. Proc. Soc. Exp. Biol. Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- 40.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Smithgall T. E. Co-expression with BCR induces activation of the FES tyrosine kinase and phosphorylation of specific N-terminal BCR tyrosine residues. J. Biol. Chem. 1996;271:32930–32936. doi: 10.1074/jbc.271.51.32930. [DOI] [PubMed] [Google Scholar]
- 42.O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 43.Takashima Y., Delfino F. J., Engen J. R., Superti-Furga G., Smithgall T. E. Regulation of c-Fes tyrosine kinase activity by coiled-coil and SH2 domains: analysis with Saccharomyces cerevisiae. Biochemistry. 2003;42:3567–3574. doi: 10.1021/bi0272499. [DOI] [PubMed] [Google Scholar]
- 44.Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W., Rauscher F. J., III Reconstitution of the KRAB–KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 45.Lionberger J. M., Smithgall T. E. The c-Fes protein-tyrosine kinase suppresses cytokine-independent outgrowth of myeloid leukemia cells induced by Bcr-Abl. Cancer Res. 2000;60:1097–1103. [PubMed] [Google Scholar]
- 46.Zhao X., Ghaffari S., Lodish H., Malashkevich V. N., Kim P. S. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat. Struct. Biol. 2002;9:117–120. doi: 10.1038/nsb747. [DOI] [PubMed] [Google Scholar]
- 47.Nelson K. L., Rogers J. A., Bowman T. L., Jove R., Smithgall T. E. Activation of STAT3 by the c-Fes protein-tyrosine kinase. J. Biol. Chem. 1998;273:7072–7077. doi: 10.1074/jbc.273.12.7072. [DOI] [PubMed] [Google Scholar]
- 48.Senis Y., Zirngibl R., McVeigh J., Haman A., Hoang T., Greer P. A. Targeted disruption of the murine fps/fes proto-oncogene reveals that Fps/Fes kinase activity is dispensable for hematopoiesis. Mol. Cell. Biol. 1999;19:7436–7446. doi: 10.1128/mcb.19.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J., Ogata Y., Ali H., Feldman R. A. The Fes tyrosine kinase: a signal transducer that regulates myeloid-specific gene expression through transcriptional activation. Blood Cells Mol. Dis. 2004;32:302–308. doi: 10.1016/j.bcmd.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 50.McLemore M. L., Grewal S., Liu F., Archambault A., Poursine-Laurent J., Haug J., Link D. C. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 2001;14:193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D. E., Zhang P., Wang N. D., Hetherington C. J., Darlington G. J., Tenen D. G. Absence of granulocyte colony-stimulating factor signalling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henkel G. W., McKercher S. R., Yamamoto H., Anderson K. L., Oshima R. G., Maki R. A. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- 53.Tagliafico E., Siena M., Zanocco-Marani T., Manfredini R., Tenedini E., Montanari M., Grande A., Ferrari S. Requirement of the coiled-coil domains of p92 c-Fes for nuclear localization in myeloid cells upon induction of differentiation. Oncogene. 2003;22:1712–1723. doi: 10.1038/sj.onc.1206279. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz Y., Ben-Dor I., Navon A., Motro B., Nir U. Tyrosine phosphorylation of the TATA element modulatory factor by the FER nuclear tyrosine kinases. FEBS Lett. 1998;434:339–345. doi: 10.1016/s0014-5793(98)01003-5. [DOI] [PubMed] [Google Scholar]