Abstract
The ubiquitin E3 ligase Cbl has been shown to negatively regulate tyrosine kinase receptors, including the stem cell factor receptor/c-Kit. Impaired recruitment of Cbl to c-Kit results in a deregulated positive signalling that eventually can contribute to carcinogenesis. Here, we present results showing that Cbl is activated by the SFKs (Src family kinases) and recruited to c-Kit in order to trigger receptor ubiquitination. We demonstrate that phosphorylated Tyr568 and Tyr936 in c-Kit are involved in direct binding and activation of Cbl and that binding of the TKB domain (tyrosine kinase binding domain) of Cbl to c-Kit is specified by the presence of an isoleucine or leucine residue in position +3 to the phosphorylated tyrosine residue on c-Kit. Apart from the direct association between Cbl and c-Kit, we show that phosphorylation of Cbl by SFK members is required for activation of Cbl to occur. Moreover, we demonstrate that Cbl mediates monoubiquitination of c-Kit and that the receptor is subsequently targeted for lysosomal degradation. Taken together, our findings reveal novel insights into the mechanisms by which Cbl negatively regulates c-Kit-mediated signalling.
Keywords: Cbl, degradation, Src family kinase, stem cell factor receptor/c-Kit, receptor tyrosine kinase, ubiquitin ligase
Abbreviations: APS, adapter protein with a pleckstrin homology and Src homology domain; CSF-1, colony-stimulating factor-1; EGF, epidermal growth factor; FBS, foetal bovine serum; Grb2, growth-factor-receptor-bound protein 2; GST, glutathione S-transferase; PAE cells, porcine aortic endothelial cells; PDGF, platelet-derived growth factor; PE, phycoerythrin; PI3K, phosphoinositide 3-kinase; pY568, phospho-Tyr568; RING, really interesting new gene; RTK, receptor tyrosine kinase; SCF, stem cell factor; SFK, Src family kinase; SH2 domain, Src homology 2 domain; TKB domain, tyrosine kinase binding domain; VEGF, vascular endothelial growth factor
INTRODUCTION
c-Kit belongs to the family of type III RTKs (receptor tyrosine kinases) and plays a major role in survival, expansion and differentiation of haematopoietic progenitor cells, as well as stem cells involved in reproduction and pigmentation (for a review, see [1]). Moreover, it is essential for the function of mast cells. These c-Kit-dependent processes are triggered by the binding of SCF (stem cell factor) to c-Kit, which results in dimerization of the receptor and subsequent activation of its intrinsic tyrosine kinase activity. This leads to autophosphorylation of the receptors on specific tyrosine residues, which then serve as docking sites for signalling molecules containing either an SH2 (Src homology 2) domain or a PTB (phosphotyrosine-binding) domain respectively (for a review, see [2]). Proteins reported to interact with ligand-stimulated c-Kit include PI3K (phosphoinositide 3-kinase), PLC-γ1 (phospholipase C-γ1), STAT (signal transducer and activator of transcription), SFKs (Src family kinases) and a number of adaptor proteins such as Grb2 (growth-factor-receptor-bound protein 2), Grb7, Grb10, Crk and APS (adapter protein with a pleckstrin homology and Src homology domain). Their activation and recruitment normally constitute the first steps in the signal relay cascade from activated c-Kit to intracellular downstream targets.
Under normal circumstances, the activation of c-Kit is tightly regulated and its inappropriate activation is associated with development of a number of malignancies, including acute myeloid leukaemia, small cell lung cancer, gastrointestinal stromal tumours and testicular cancer (for a review, see [3]). Negative regulation of RTKs occurs through the action of protein tyrosine phosphatases as well as through proteolytic degradation.
Recent studies have shown that a number of RTKs undergo ubiquitination, targeting them for degradation. Ubiquitination serves as a general key regulatory mechanism of signalling in normal cells and is mediated by ubiquitin ligases that attach single or multiple ubiquitin moieties to their targets, resulting in mono- or poly-ubiquitinated proteins respectively. Members of the Cbl family of ubiquitin ligases, namely Cbl, Cbl-b and Cbl-c, play a major role in the ligand-dependent ubiquitination of many RTKs [4]. They all possess a highly conserved N-terminal domain consisting of a TKB domain (tyrosine kinase binding domain) and a RING (really interesting new gene) finger domain, which are both necessary for ubiquitin ligase activity. The TKB domain is required for Cbl recruitment to tyrosine-phosphorylated proteins [5]. The RING finger domain associates with the E2 ubiquitin-conjugating enzyme, UbcH7 (ubiquitin-conjugating enzyme H7) [6], leading to the transfer of ubiquitin molecules to lysine residues on the target protein. The C-terminal half of Cbl contains a proline-rich region and potential phosphorylation sites, which in turn can allow interaction of Cbl with proteins possessing SH3 or SH2 domains respectively and confer adaptor functions to Cbl proteins (reviewed in [7]).
Examples of RTKs known to be targeted and negatively regulated by Cbl family members include the receptors for EGF (epidermal growth factor), CSF-1 (colony-stimulating factor-1), PDGF (platelet-derived growth factor), VEGF (vascular endothelial growth factor) and hepatocyte growth factor (c-Met) [8–12]. Upon mono- or poly-ubiquitination, the proteins are destined for degradation via the lysosome or the proteasomes respectively [13], although in some cases receptors are recycled to the plasma membrane. Dysfunction or impaired recruitment of Cbl to RTKs results in deregulated positive signalling that eventually can lead and contribute to carcinogenesis [14].
Several reports claim that internalization and degradation of c-Kit are mediated by Cbl in an Src-dependent manner. The current opinion suggests that upon SCF stimulation, SFKs are recruited to autophosphorylated Tyr568 of c-Kit and thereby activated. Active Src kinases then phosphorylate Cbl, which in turn tags the receptor for proteasomal degradation [15]. The interaction of Cbl and c-Kit during these processes has been studied by some groups and so far it is assumed to be primarily mediated indirectly by the adaptor proteins APS or Grb2 [16–18].
In this paper, we present results suggesting that Cbl needs not only to be activated by Src but also to be recruited to c-Kit upon SCF stimulation in order to elicit receptor ubiquitination. We show that the N-terminal TKB domain of Cbl can directly interact with c-Kit at phosphorylated Tyr568 and Tyr936. The binding specificity of Cbl for these tyrosine residues is determined by Ile571 and Leu939 of c-Kit respectively. The specific Cbl loss-of-binding mutant of c-Kit, namely c-Kit I571A/L939A, accordingly failed to bind Cbl, while phosphorylation of Cbl appeared to be intact. Consequently, ubiquitination and degradation of I571A/L939A-c-Kit was retarded despite intact ligand-stimulated kinase activities of c-Kit and Src. Wild-type c-Kit was further found to be primarily monoubiquitinated and targeted to the lysosome for degradation. Thus the present paper adds a novel perspective on the dynamics and regulation of c-Kit signalling with a sequential recruitment of signal transduction molecules such as Src and Cbl to Tyr568 of c-Kit.
EXPERIMENTAL
Antibodies, ligands and inhibitors
Human recombinant SCF and murine recombinant IL-3 (interleukin-3) were obtained from ProSpec-Tany Technogene (Rehovot, Israel). The rabbit antiserum Kit-C1, recognizing the C-terminal domain of c-Kit, was affinity-purified as described in [19]. The anti-phosphotyrosine antibody 4G10 was from Upstate Biotechnology (Charlottesville, VA, U.S.A.), and anti-GST (glutathione S-transferase) and anti-Cbl (c-15) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). The 9E10 antibody recognizing Myc tag was from Roche Diagnostics, anti-ubiquitin P4D1 from Covance Research Products (Denver, PA, U.S.A.) and anti-ubiquitin FK1 from Biomol (Plymouth Meeting, PA, U.S.A.). The Src antibody Ab-1 was from Oncogene Sciences. Redivue [γ-32P]ATP was purchased from Amersham Biosciences. The Src inhibitor SU6656 was from SUGEN/Calbiochem (San Diego, CA, U.S.A.). PE (phycoerythrin)-conjugated monoclonal YB5.B8 against human c-Kit was from BD Pharmingen. Clasto-lactacystin (Biomol) or chloroquine (Sigma, St. Louis, MI, U.S.A.) was used for proteasomal or lysosomal inhibition respectively. GST fusion proteins of N- and C-terminal parts of Cbl have previously been described [20].
Cell culture
PAE (porcine aortic endothelial) cells were grown in Ham's F-12 medium with 10% (v/v) FBS (foetal bovine serum), 100 units/ml penicillin and 100 μg/ml streptomycin. COS-1 cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were cultured in a humidified incubator at 37 °C with 5% CO2. Cells were starved in 0.1% FBS overnight and stimulated with 100 ng/ml SCF for the indicated periods of time unless stated otherwise.
Transient and stable transfection
Transient transfection of COS-1 and PAE cells was conducted by employing Lipofectamine™/Plus reagents (Invitrogen) according to the manufacturer's instructions. The mutants of c-Kit in pcDNA3 used in the present paper as well as PAE cells stably expressing wild-type and mutant human c-Kit have previously been described [16,21,22]. In all experiments, the GNNK+ (the alternative splice form of human c-Kit containing the peptide sequence GNNK in the extracellular juxtamembrane region) isoform of c-Kit was used [23,24].
Immunoprecipitation, pulldown and Western blotting
Experimental procedures were conducted as described in [24]. Immunodetection was performed by enhanced chemiluminescence using the Super Signal Dura from Pierce (Rockford, IL, U.S.A.) and a CCD camera (charge-coupled-device camera; LAS-3000; Fujifilm).
Affinity fishing of GST fusion proteins with immobilized peptides
The following peptides were synthesized: CEEINGNNY(p)-VYIDPTQ [pY568 (phospho-Tyr568)], CEEINGNNY(p)AYIDPTQ (pY568, Ala569), CEEINGNYY(p)VAIDPTQ (pY568, Ala570), CEEINGNYY(p)VYADPTQ (pY568, Ala571), STNHIY-(p)SNLANCS (pY936), STNHIY(p)ANLANCS (pY936, Ala937), STNHIY(p)SALANCS (pY936, Ala938), STNHIY(p)SNAANCS (pY936, Ala939) and CSDSTNEY(p)MDMKPGV (pY721). Peptides were dissolved at 1 mg/ml in 50 mM Tris buffer (50 mM Tris/HCl, pH 7.4) and 5 mM EDTA (pH 8) and immobilized on Affigel 10 beads (Bio-Rad, Hercules, CA, U.S.A.; 1 mg of peptide/ml of Affigel) according to the manufacturer's instructions. A 1:1 slurry (50 μl) was incubated for 3 h at 4 °C with 1 μg of purified GST–Cbl fusion protein and rolled end-over-end for 3 h in the cold with cell lysates. After thorough washing of the beads, bound protein was eluted by boiling SDS sample buffer, run out on an SDS/10% polyacrylamide gel, electrotransferred to Immobilon-P and probed for GST. For competition experiments, the respective soluble peptide (100 μM) was added to the incubation mixture of fusion protein and immobilized peptides.
In vitro kinase assay
Kinase activity of c-Src was assessed essentially as described in [22].
Internalization and degradation experiments
Internalization and degradation experiments using 125I-SCF were performed as previously described [25].
Internalization of c-Kit was determined after stimulation with SCF (100 ng/ml) for the indicated periods of time, using a PE-anti-human CD117 antibody (BD Biosciences Pharmingen) and analysis by flow cytometry using a FACSort instrument (BD Biosciences).
RESULTS
The N-terminal domain of Cbl interacts with c-Kit in a phospho-dependent manner
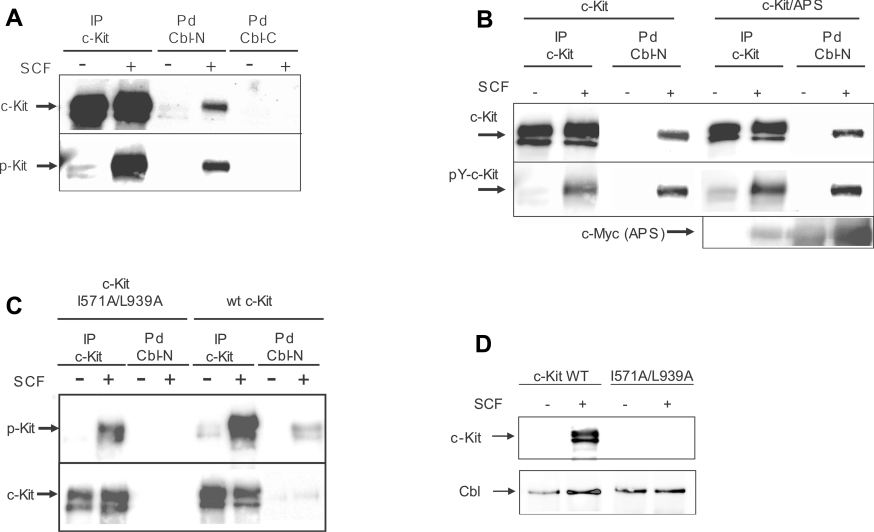
Several reports indicate that Cbl and tyrosine kinase receptors can interact both directly and indirectly through adaptor proteins such as Grb2, APS or the p85 subunit of PI3K [17,26–28]. To address the issue of whether the interaction between Cbl and c-Kit occurs in a direct or indirect manner, COS-1 cells were transfected with c-Kit and stimulated with 100 ng/ml SCF for 5 min. Lysates were incubated with immobilized GST fusion proteins of either the N-terminal or the C-terminal half of Cbl respectively. Pulled down proteins were separated on an SDS/8% polyacrylamide gel, followed by electrotransfer to Immobilon-P. The filter was first probed with a phosphotyrosine antibody (PY-99), followed by stripping and reprobing with Kit-C1 antibody. A phosphorylation-dependent interaction between the N-terminal region of Cbl and c-Kit could be detected, whereas the C-terminal region of Cbl did not interact with c-Kit (Figure 1A). Efficient transfection and activation of c-Kit in COS-1 cells were confirmed by a parallel immunoprecipitation of c-Kit from the same lysates and probing sequentially for phosphotyrosine and c-Kit.
Figure 1. Interaction of the N-terminal half of Cbl with c-Kit at position Tyr568 and Tyr936.
(A) COS-1 cells were transiently transfected with wild-type c-Kit expression plasmids and stimulated with SCF as indicated. After lysis, cell lysates were subjected to either an immunoprecipitation (IP) with c-Kit antibody or a pulldown (Pd) with GST fusion proteins of either the N-terminal or C-terminal half of Cbl. Immunocomplexes were separated on an SDS/8% polyacrylamide gel, blotted and sequentially probed for c-Kit (upper panel) or anti-phosphotyrosine (lower panel). (B) COS-1 cells were co-transfected with expression plasmid for c-Kit and Myc-tagged APS and stimulated with SCF as indicated. After lysis, cell lysates were subjected to immunoprecipitation with anti-c-Kit as well to a pulldown with GST–Cbl (N-term) fusion protein. Complexes were analysed by Western blotting and the membranes were sequentially probed for c-Kit, phosphorylated c-Kit and APS. (C) COS-1 cells were transfected with the c-Kit mutant I571A/L939A and samples were treated as described above. (D) Cbl was immunoprecipitated from COS-1 cells transfected with either wild-type c-Kit or the I571A/L939A mutant and samples were analysed by Western blotting.
It has previously been suggested that the adaptor protein APS mediates an indirect interaction between Cbl and c-Kit [16]. In order to rule out the contribution of APS to the observed interaction between c-Kit and the N-terminal part of Cbl, we co-transfected COS-1 cells with Myc-tagged APS and c-Kit and performed an analogous pulldown experiment with GST–Cbl (N-term) as described above. Surprisingly, we did not detect an expected enhancement of Cbl–c-Kit interaction by co-transfection of APS (Figure 1B) despite substantial amounts of exogenously expressed APS, while the interaction between Cbl and c-Kit appeared equally strong regardless of the presence or absence of APS. The interaction between APS and Cbl is to some extent SCF-independent, which can be explained by the overexpression of APS in this case. Similar experiments were performed using SH2B and Lnk (lymphocyte-specific adapter protein), members of the same family of adaptor proteins as APS, where no binding of Cbl could be detected (results not shown). We observed that mutation of Ile571 and Leu939 in c-Kit (previously reported to be specific binding determinants for binding of APS to Tyr568 and Tyr936 respectively [16]) abolished the observed interaction of c-Kit and Cbl-N. This finding suggests that Cbl interacts directly with c-Kit with the same binding determinants as APS (Figure 1C). To verify this interaction in vivo with endogenous levels of Cbl, we performed co-immunoprecipitation between Cbl and c-Kit using cells transfected with either wild-type c-Kit or the I571A/L939A mutant (Figure 1D).
The interaction between Cbl and c-Kit is direct and specifically determined by the +3 positions to Tyr568 and Tyr936
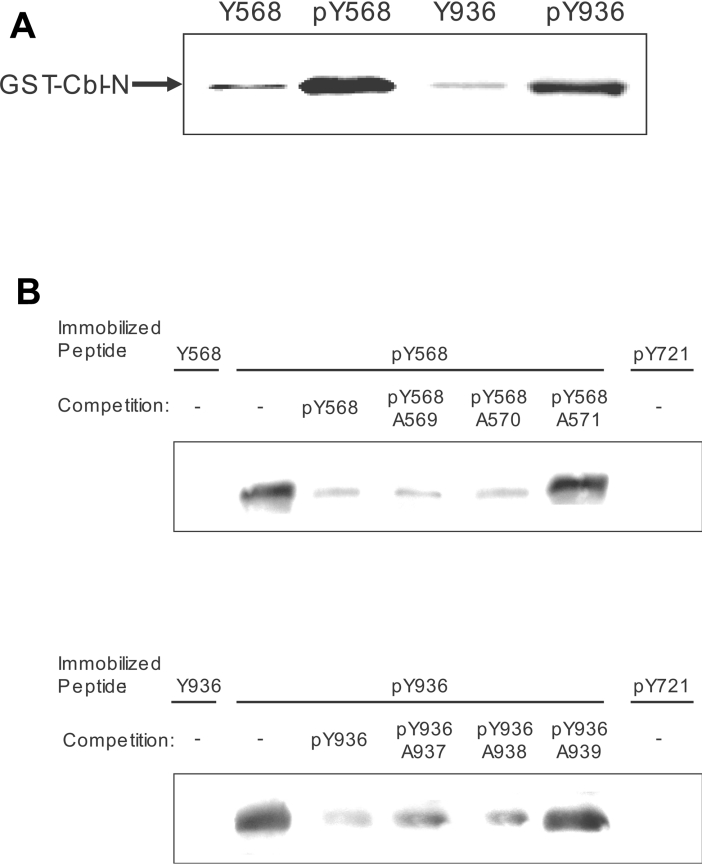
To prove that neither any other molecule besides APS nor endogenous APS is mediating the interaction between Cbl and c-Kit at positions Tyr568 and Tyr936 of c-Kit, we incubated purified GST–Cbl (N-term) fusion protein with immobilized phosphopeptides containing pY568 and pY936. As positive controls for interaction with pY568 or pY936, GST fusion proteins with the SH2 domain of Src or Grb2 were employed respectively (results not shown). Bound proteins were eluted and immunoblotted for GST, revealing that GST–Cbl (N-term) interacts with Tyr568 and Tyr936 in a phosphotyrosine-dependent manner (Figure 2A). Furthermore, the observed interaction could be competed by an excess of soluble phosphopeptide. The competing effect was abolished when peptides with I571A or L939A mutations were used, further supporting the importance of isoleucine/leucine in position +3 to the tyrosine as an important determinant for binding of Cbl to c-Kit (Figure 2B). As a negative control for interaction with Cbl, a peptide based on the peptide sequence around pY721 in c-Kit was used.
Figure 2. Cbl binds directly to phosphorylated Tyr568 and Tyr936 and the specificity is determined by the amino acids at the +3 positions to the tyrosine residues respectively.
(A) Phosphopeptides pY568 and pY936 (sequences in the Experimental section) immobilized to Affigel 10 beads were incubated (3 h at 4 °C) with the indicated purified GST fusion proteins in the presence of either unphosphorylated or phosphorylated peptides in solution as indicated. Samples were analysed by SDS/PAGE and Western blotting, and membranes were probed with an anti-GST antibody. (B) Pulldown experiment performed as described above with competition by soluble peptides (100 μM) as indicated.
Our results provide evidence that the N-terminal part of Cbl interacts with c-Kit at position pY568 and pY936 in a direct manner. As we furthermore determined Ile571 and Leu939 as specific determinants for this interaction, we chose the I571/L939A mutant of c-Kit as a specific Cbl loss-of-binding mutant of c-Kit for further phenotypic investigations.
The c-Kit double mutant I571A/L939A possesses intact intrinsic kinase activity and is able to activate Src upon ligand stimulation
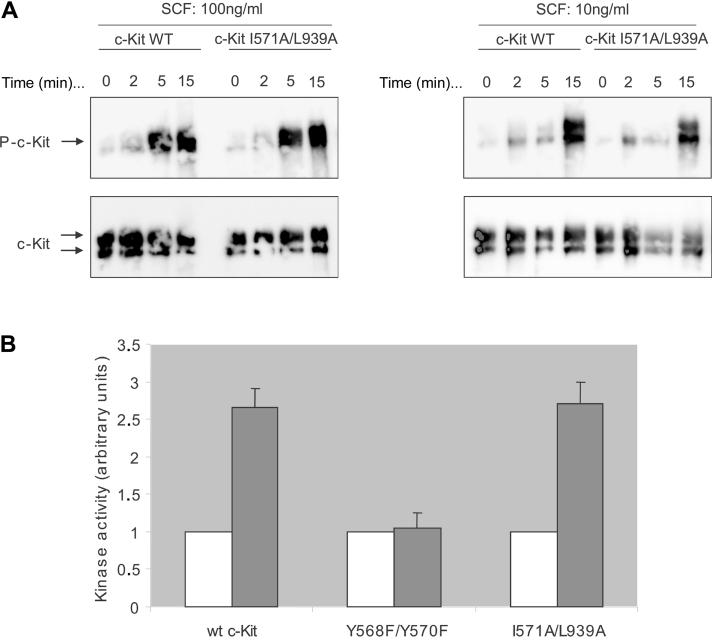
In order to exclude the possibility that the I571A/L939A double mutant of c-Kit blocks Cbl interaction because of its potential loss of intrinsic kinase activity, we examined ligand-stimulated autophosphorylation of the receptor. COS-1 cells transfected with either wild-type c-Kit or double mutant c-Kit I571A/L939A were stimulated with SCF and lysates were immunoprecipitated with a c-Kit antibody, Kit-C1. The samples were analysed by Western blotting, probing with a phosphotyrosine antibody (4G10) and reprobed with Kit-C1 for confirmation of equal amount of protein. The kinase activity of the mutant receptor proved to be intact (Figure 3A). Phosphorylated Tyr568 and Tyr936 of c-Kit are known to bind c-Src and Grb2 respectively and both Src and Grb2 have been reported to be involved in Cbl-mediated regulation of RTK signalling via either phosphorylation of Cbl [29] or indirect recruitment of Cbl to the receptors [30] respectively. Thus, to exclude that the observed Cbl phenotype of the I571A/L939A double mutant might be a consequence of impaired Src activation, we performed an in vitro Src kinase assay. For this purpose, Src was immunoprecipitated from lysates of cells expressing wild-type c-Kit, the I571/L939A mutant of c-Kit or the Y568F/Y570F mutant, known to be deficient in Src activation [22], and therefore serving as a negative control respectively. Immunocomplexes were incubated for 15 min at room temperature with [γ-32P]ATP and acid-denatured enolase as an exogenous substrate for Src. Reactions were terminated after 15 min, proteins were separated on an SDS/polyacrylamide gel and electroblotted on to PVDF membranes. Phosphorylated enolase was detected using PhosphoImager (LAS3000; Fujfilm, Tokyo, Japan) and densitometrically analysed. The membranes were reprobed with an Src antibody to ensure equal levels of Src (results not shown). In parallel, the same samples were analysed using Western blotting and probed for anti-phosphotyrosine as well as c-Kit to verify equal amount of expressed protein and activation (results not shown). We could demonstrate that the wild-type c-Kit and the I571A/L939A double mutant show an increased Src kinase activity upon ligand stimulation, whereas the Y568F/Y570F mutant displayed an Src-negative phenotype as expected (Figure 3B). Intact interaction of Grb2 with the I571A/L939A double mutant of c-Kit has previously been demonstrated by Wollberg et al. [16].
Figure 3. Intrinsic kinase activity and ability to phosphorylate Src of the c-Kit double mutant I571A/L939A.
(A) COS-1 cells were transfected with wild-type c-Kit (lanes 1 and 2) as well as the I571A/L939A (lanes 3 and 4) double mutant and stimulated with SCF for the indicated periods of time (min; values above lanes) and concentrations. Lysates were immunoprecipitated with a c-Kit antibody and analysed by Western blotting. The membrane was probed for tyrosine-phosphorylated residues (4G10) and reprobed for c-Kit as loading control. (B) Densitometric analysis of an Src in vitro kinase assay. Src was immunoprecipitated from an aliquot of cell lysate prepared as described above, followed by incubation (3 h at 4 °C) with 50 μM [γ-32P]ATP in kinase buffer (described in detail in the subsection ‘In vitro kinase assay’ in the Experimental section). Samples were separated on an SDS/8% polyacrylamide gel and blotted on to Immobilon. The filter was incubated with 1 M KOH for 1 h at 55 °C to hydrolyse non-specific phosphoserine present in the proteins, exposed to the PhosphoImager and densitometrically analysed. Src activity upon stimulation in the respective mutant (grey bars) is expressed as fold activity in unstimulated cells (white bars). Mean values and standard deviations for three independent experiments are shown.
These results demonstrate that the I571A/L939A double mutant of c-Kit is impaired in its ligand-induced binding of Cbl to the c-Kit receptor without abolishing kinase activities of either c-Kit or Src respectively, nor does it inhibit binding of Grb2 to c-Kit.
Phosphorylation and recruitment of Cbl to c-Kit are separable events that both are necessary for receptor ubiquitination
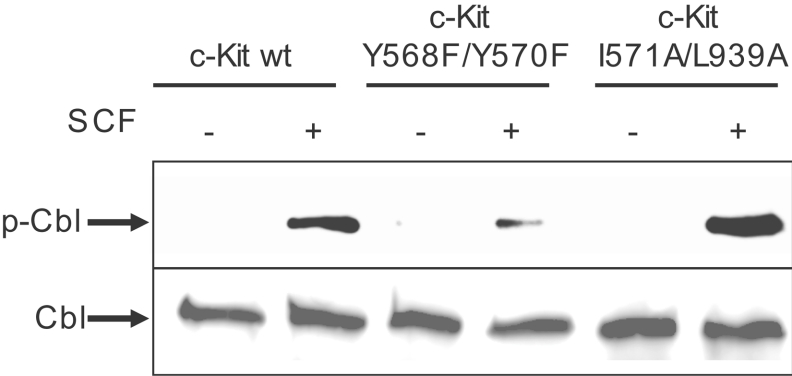
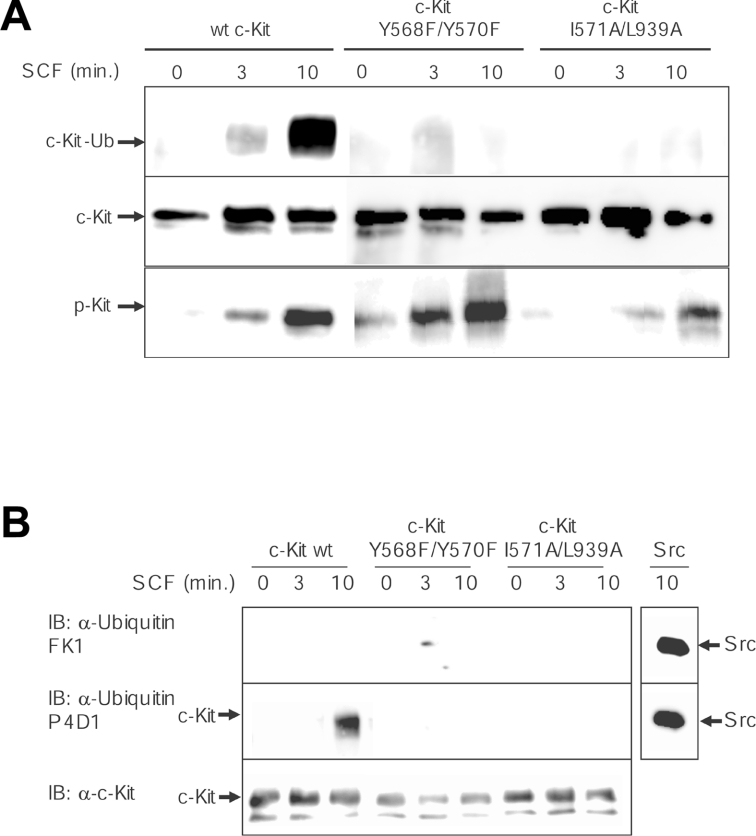
So far we have shown that the I571A/L939A mutant of c-Kit is unable to bind Cbl, but is still capable of activating Src. Next, we were interested to see whether Cbl needs to be recruited to c-Kit in order to be phosphorylated and activated by SFKs. To address this question, COS-1 cells were transfected with wild-type c-Kit as well as the double mutants Y568F/Y570F and I571A/L939A respectively, stimulated with SCF and lysed. Cbl was immunoprecipitated from the lysates and subjected to immunoblotting with anti-phosphotyrosine antibodies. Equal Cbl protein levels were confirmed by reprobing the membrane with an anti-Cbl antibody. Hereby we were able to reveal that phosphorylation of Cbl is deficient in the Y568F/Y570F mutant, but unaffected in the Cbl-I571A/L939A mutant, further confirming that Src kinase activity is intact downstream of I571A/L939A c-Kit (Figure 4). From this, we conclude that Src-dependent phosphorylation of Cbl occurs independently from Cbl interaction with c-Kit and that both events can be temporally and spatially separated. Next, we aimed to examine the ubiquitination phenotype of I571A/L939A c-Kit. The question whether the receptor gets polyubiquitinated or monoubiquitinated following ligand stimulation has not been completely resolved yet in the case of c-Kit. It has been claimed that c-Kit is polyubiquitinated following ligand stimulation and degraded in the proteasomes [15,31], whereas others state that RTKs in general become monoubiquitinated through the action of Cbl, which targets them for degradation in the lysosomes [32,33]. To escape the problem of using an antibody potentially raised against an inappropriate antigen, we chose an anti-ubiquitin antibody recognizing both mono- and poly-ubiquitin moieties (P4D1) for a first experiment. For this, c-Kit was immunoprecipitated from PAE cells that were transiently transfected with the wild-type form, the I571/L939A Cbl-binding mutant or the Y568F/Y570F Src-binding mutant of c-Kit. Immunocomplexes were run on a gel, electrotransferred and probed for ubiquitin. Upon ligand stimulation for 10 min, wild-type c-Kit was markedly ubiquitinated, while none of the c-Kit double mutants displayed receptor ubiquitination despite comparable expression and activation levels of each receptor isoform (Figure 5A). This finding is in accordance with our hypothesis that ubiquitination of c-Kit requires both activation of Cbl by Src and recruitment of phosphorylated Cbl to c-Kit. Thus binding of Cbl to I571/L939 in c-Kit appears to be a specific occurrence secondary to its phosphorylation.
Figure 4. The effect of the wild-type c-Kit versus the two c-Kit double mutants Y568F/Y570F and I571A/L939A on Cbl phosphorylation.
Transiently transfected COS-1 cells expressing wild-type c-Kit or one of the two double mutants Y568F/Y570F or I571A/L939A respectively were stimulated with SCF for the indicated periods of time, immunoprecipitated with a Cbl antibody and subjected to Western blotting. The membrane was probed for tyrosine phosphorylation and Cbl respectively.
Figure 5. Monoubiquitination of c-Kit upon phosphorylation of Tyr568 and Tyr936 and recruitment of Cbl.
(A) PAE cells expressing wild-type, Y568F/Y570F or I571A/L939A c-Kit were stimulated with SCF for the indicated time points, followed by lysis and immunoprecipitation with c-Kit antibody. Samples were subjected to a Western-blot analysis for ubiquitin (P4D1) and c-Kit. (B) An identical experiment was performed, with the exception that the membrane was probed with a selective polyubiquitin antibody (FK1) as well as the P4D1 ubiquitin antibody recognizing both poly- and mono-ubiquitin. Immunoprecipitated Src was included as a positive control for the polyubiquitin antibody. IB, immunoblot.
c-Kit is selectively monoubiquitinated by Cbl
To address the issue of mono- versus poly-ubiquitination of c-Kit, a similar experiment to Figure 5(A) was conducted. This time, membranes containing immunoprecipitated c-Kit proteins were first probed with an antibody specifically reacting with polyubiquitin moieties (FK1), stripped and reprobed with the anti-mono/poly-ubiquitin antibody. As a positive control for a known polyubiquitinated protein, we included immunoprecipitated activated Src [34]. It was consistently observed that only wild-type c-Kit with intact Cbl and Src binding displayed ubiquitination. Furthermore, the ubiquitination signal on c-Kit only became apparent when employing the antibody generated against both mono- and poly-ubiquitin, whereas the specific polyubiquitin antibody did not react with wild-type c-Kit (Figure 5B). Thus it appears that c-Kit is primarily monoubiquitinated upon ligand stimulation in a process that requires Src-dependent phosphorylation and receptor association of Cbl.
Internalization and degradation of c-Kit are dependent on activation of Src and intact Cbl-binding sites
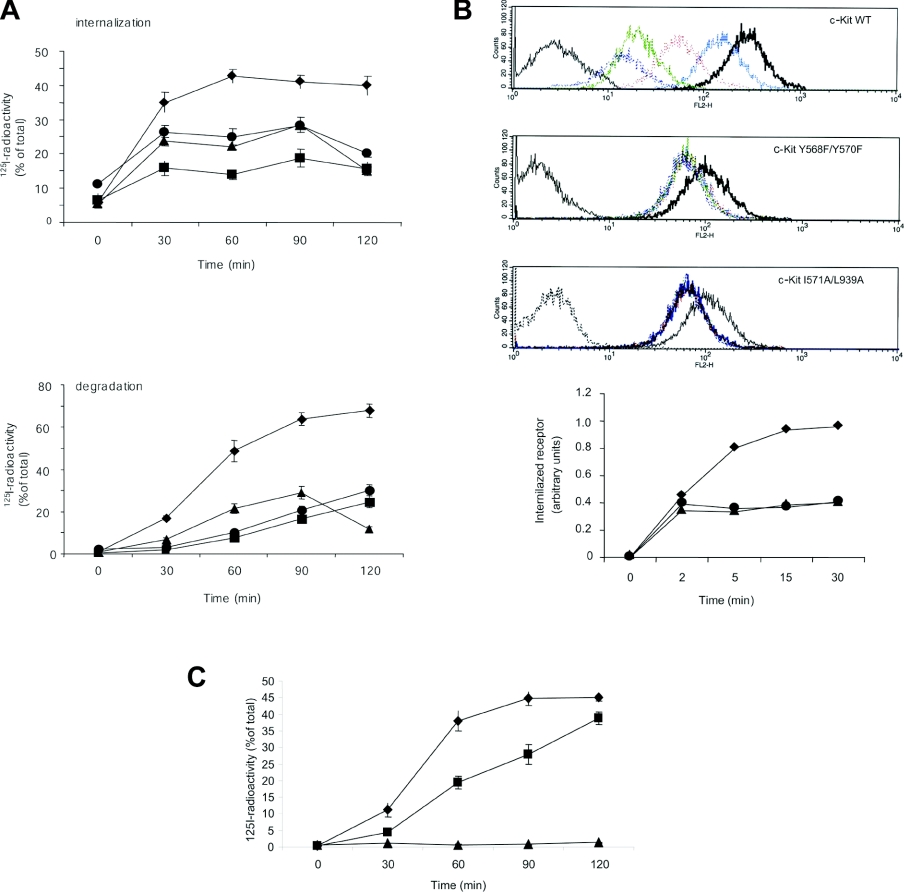
Since it has been demonstrated that ligand-induced monoubiquitination regulates internalization and degradation of several RTKs, we wanted to investigate the effect of inhibition of monoubiquitination on c-Kit internalization and degradation. PAE cells expressing either Y568F/Y570F or I571/L939A mutant forms of c-Kit, which we have shown to inhibit Src-dependent phosphorylation of Cbl or Cbl association respectively, displayed impaired ligand internalization and degradation when compared with wild-type c-Kit (Figure 6A). In addition, inhibition of Src kinase activity by 2 μM SU6656 interfered with degradation of wild-type receptor, which is in agreement with previous reports [24]. We also analysed receptor internalization in Ba/F3 cells expressing wild-type, Y568F/570F or I571A/L939A mutant c-Kit by flow cytometry using an antibody against the extracellular part of c-Kit. Ba/F3 cells were used since they grow in suspension culture and thus do not need to be treated with trypsin in order to do the flow cytometry analysis, which could potentially interfere with receptor detection. In both the Y568F/Y570F and the I571A/L939A mutant c-Kit, internalization of the receptor was decreased compared with wild-type receptor-expressing cells (Figure 6B).
Figure 6. Analysis of degradation of internalized SCF in cells expressing wild-type c-Kit versus I571A/L939A mutant c-Kit and their intracellular processing.
(A) PAE cells expressing wild-type (◆), Y568F/Y570F (▲) or I571A/L939A (●) c-Kit were incubated with 125I-SCF for 60 min on ice, followed by incubation at 37 °C for the indicated periods of time. Cells were washed with an acidic buffer (see the subsection ‘Internalization and degradation experiments’ in the Experimental section) and surface-released radioactivity was determined. Loss of surface-bound radioactivity was taken as a measure of internalization. The medium was collected and subjected to precipitation with 10% (w/v) trichloroacetic acid. Trichloroacetic acidsoluble radioactivity was taken as a measure of degradation of 125I-SCF. As a negative control, wild-type c-Kit-expressing PAE cells were treated with 2 μM SU6656 (■) to inhibit Src activity before examining internalization and SCF degradation. (B) Flow cytometry analysis of internalization. Ba/F3 cells transfected with either wild-type (◆), Y568F/Y570F (▲) or I571A/L939A (●) mutant c-Kit were incubated with 100 ng/ml SCF for the indicated periods of time, followed by flow cytometry analysis using an antibody against the extracellular part of c-Kit. The following colour coding of the histograms was used: grey: negative control; black; 0 min; blue: 2 min; red: 5 min; green: 15 min; purple: 30 min. (C) PAE cells expressing wild-type c-Kit were pre-incubated in the absence of inhibitor (◆) or in the presence of either 100 μM lactacystin (■) or 20 μM chloroquine (▲) for 4 h, followed by the assessment of binding, internalization and degradation of 125I-SCF as described above. Mean values and standard deviations for three independent experiments are shown.
Internalized c-Kit is degraded via the lysosomal pathway
It has recently been shown that the EGF receptor is sorted to lysosomes upon EGF stimulation in a Cbl-dependent manner [35]. Moreover, lysosomal degradation has been suggested to be the general degradation pathway following monoubiquitination of proteins (reviewed in [36]). To test whether c-Kit is degraded in the lysosomes, we examined receptor degradation indirectly by assessing degradation of bound 125I-labelled ligand in the presence or absence of the specific proteasomal or lysosomal inhibitors, lactacystin or chloroquine respectively. Measurement of ligand degradation showed that inhibition of lysosomal action by 20 μM chloroquine completely abolished receptor degradation, whereas inhibition of the proteasome by 100 μM lactacystin only retarded receptor degradation slightly (Figure 6C). Therefore we propose that degradation of c-Kit mainly occurs via monoubiquitination and subsequent targeting to the lysosomes.
DISCUSSION
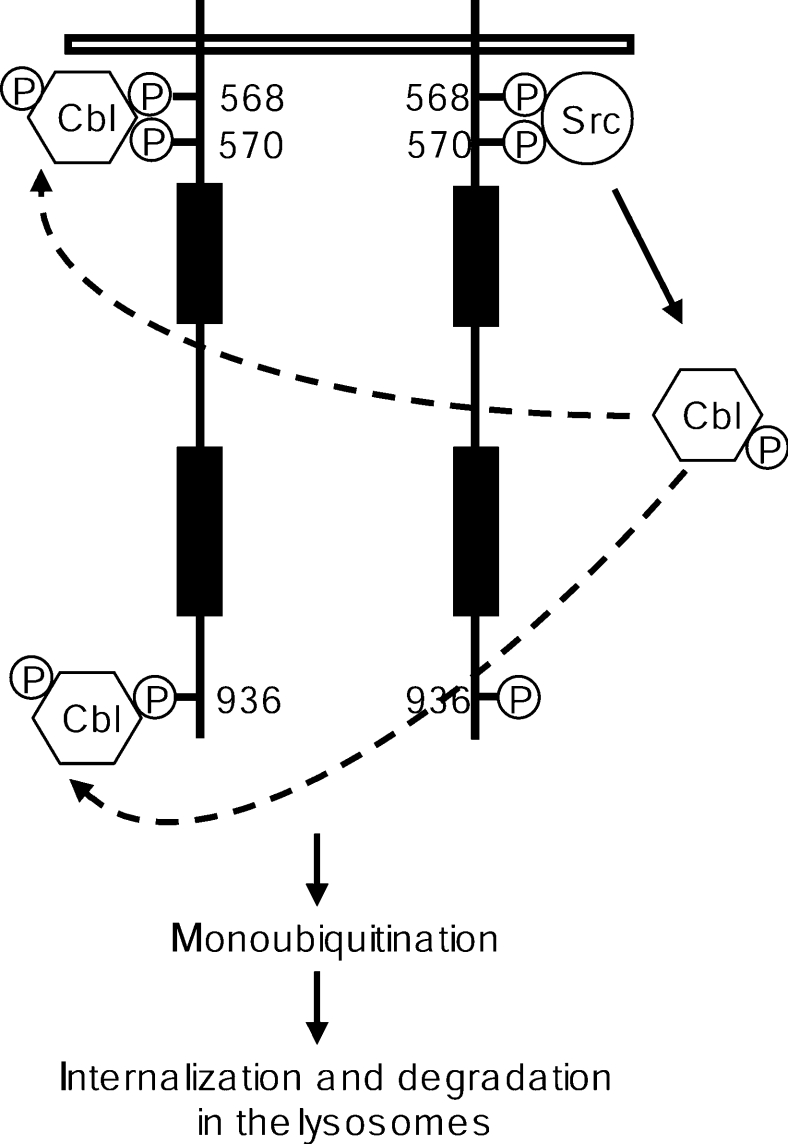
The TKB domain of Cbl mediates binding to target proteins containing phosphorylated tyrosine residues and is indispensable for Cbl to induce ubiquitination of target RTKs [5]. Cbl can associate with the activated RTK either directly or through adaptor proteins. The adaptor protein Grb2 is able to associate with Cbl through proline-rich regions in Cbl interacting with the SH3 domain of Grb2 [27]. It was recently suggested that direct versus indirect association of Cbl with the EGF receptor has distinct function [35]. This further suggests that the sites of ubiquitination might differ depending on a direct or indirect interaction with Cbl, which in turn might differentially regulate association with downstream signal transduction molecules. Here, we present evidence that Cbl can interact with c-Kit in a direct manner at position Tyr568 and Tyr936, and that this interaction is dependent on a leucine or isoleucine residue in position +3. Since Tyr568 of c-Kit is the binding site for several signal transduction molecules besides Cbl, including SFKs, SHP-2 (SH2-domain-containing protein tyrosine phosphatase 2) and the adaptor protein APS, we made the I571A mutant of c-Kit to specifically knock out binding of Cbl without affecting Src kinase activity. This also enabled us to study the role of Src kinase activity versus Cbl association with Tyr568 for the activation of Cbl following c-Kit activation. Hereby we could reveal that Cbl phosphorylation by Src and Cbl recruitment to c-Kit are separate events that are both pivotal for Cbl-mediated ubiquitination of c-Kit. This also includes a sequential binding of Src and Cbl to pY568 of c-Kit (see Figure 7).
Figure 7. Proposed model for the sequential Src- and Cbl-dependent steps during the ubiquitination of c-Kit.
Activation of SFKs, initiated by Src binding to pY568 of ligand-stimulated c-Kit, leads to phosphorylation of Cbl. Phosphorylated Cbl is then recruited to pY568 and pY936 of c-Kit, which in turn results in monoubiquitination of the receptor and its subsequent internalization and degradation in the lysosomes.
A number of tyrosine kinases and RTKs have been shown to bind to Cbl through the TKB of Cbl, including the EGF receptor [28], VEGF receptor [Flt-1 (Fms-like tyrosine kinase-1)] [37], macrophage CSF-1 receptor [38], Met receptor [39], the p75 neurotrophin receptor [40], the PDGF α-receptor [11], Zap-70 (zeta-chain associated protein 70) [41], Syk [42], Src [29] and Fyn [43]. Studies on the consensus sequence for binding of the Cbl TKB using degenerate peptide libraries suggested a consensus of NXpY(S/T)XXP [41]. Many Cbl-binding proteins fulfil this consensus sequence requirements, but there are exceptions (Table 1).
Table 1. Sequence comparison of Cbl-binding sites in various proteins.
(a) The sequences of Cbl-binding sites that conform with the previously described traditional consensus sequences for the binding of Cbl to phosphorylated tyrosine residues [46]. (b) The recently reported sequences of Cbl-binding sites that do not match the previously proposed consensus (APS, Met and c-Kit). c-Fms is the cellular homologue of the transforming oncogene of viral oncogene of McDonough feline sarcoma virus. EGFR, EGR receptor; Let-23, Caenorhabditis elegans Let-23 encoded receptor tyrosine kinase; NTR, neurotrophin receptor; VEGFR, VEGF receptor.
| (a) | |
|---|---|
| Cbl-binding site sequence | Protein |
| LLQPNNYQFC | c-Fms |
| DSFLQRYSSDPT | EGFR |
| SVNSSRYKTEPF | Let-23 |
| TLNSDGYTPEPA | ZAP-70 |
| TVSFNPYEPELA | Syk |
| LIEDNEYTARQG | Src/Fyn |
| KGDGNLYSSLPL | P75 NTR |
| YNSVVLYSTPPI | VEGFR |
| IRGSNEYTEGPS | Sprouty2 |
| (b) | |
| Cbl-binding site sequence | Protein |
| SNESVDYRATFP | Met |
| RAVENQYSFY | APS |
| EEINGNYVYIDP | c-Kit Tyr568 |
| ESTNHIYSNLAN | c-Kit Tyr936 |
The Met receptor has been described to associate with Cbl via an atypical sequence motif [39]. In that case, the required sequence was found to be DpYR. Also the amino acid sequence surrounding Tyr568 and Tyr936 in c-Kit does not strictly meet the requirements of the ‘traditional’ consensus sequence for binding of Cbl to phosphotyrosine residues. Tyr936 has a serine residue in position +1, but otherwise it shows little similarity to the typical Cbl association sites. The two Cbl recruitment sites in c-Kit both have isoleucine/leucine residues in position +3. This is also present in the Cbl association site of the p75 neurotrophin receptor. However, there are no striking similarities. This might point to requirements for Cbl binding that are rather determined by specific tertiary structures than by primary amino acid sequences. One should also keep in mind that the results presented on the binding specificity of Cbl to c-Kit arise from the use of synthetic phosphopeptides derived from the c-Kit sequence and might not fully represent the role of c-Kit in a physiological context.
Ubiquitination of protein has for long time been thought exclusively to occur through polyubiquitination and to target proteins for degradation in the proteasomes. However, in the case of RTKs, it was recently shown that at least some of them undergo multiple monoubiquitination, which targets them for endocytosis and degradation in the lysosomes [32,33]. Several studies have suggested that c-Kit is polyubiquitinated upon ligand stimulation [15,31], but the methods to detect ubiquitination in those studies were not able to distinguish between polyubiquitination and monoubiquitination. In the present study, we have used two types of ubiquitin antibodies: one that is able to recognize both monoubiquitin as well as polyubiquitin (P4D1), and one recognizing only polyubiquitin (FK1). Thus we could show that ubiquitination of c-Kit following ligand stimulation occurs exclusively as monoubiquitination.
Other studies have presented evidence that mono- and poly-ubiquitinations serve distinct purposes. In the case of the receptors for PDGF and EGF, several studies have linked monoubiquitination to regulation of receptor internalization and endosomal routing of receptors for lysosomal degradation [32,33]. By the use of a proteasomal inhibitor, lactacystin, and a lysosomal inhibitor, chloroquine, we showed that degradation of c-Kit is to a large extent dependent on the lysosomal pathway and only to a minor extent on the proteasomal pathway. It has been shown that proteasome inhibitors block lysosomal degradation of ligand, which remains bound to their receptors within the endocytic pathway [44].
Monoubiquitination and degradation of RTKs through the lysosomal pathways are important pathways for regulating receptor activity. It is striking that in many cases transforming mutants of RTKs and other tyrosine kinases carry mutations that render them unable to bind to the Cbl family of ubiquitin E3 ligases (reviewed in [14]). Thus escape of lysosomal degradation will allow the receptor to signal for a prolonged period of time and contribute to the transformed phenotype.
In the present paper, we describe a direct interaction between Cbl and c-Kit. Based on our results, we propose a model in which Cbl is activated through Src-mediated phosphorylation and then associates with the receptor. The physical association of Cbl with c-Kit is necessary for c-Kit to become ubiquitinated (Figure 7). The monoubiquitinated receptor is then internalized, sorted to the lysosomes and degraded. In other systems, such as the EGF receptor, direct association versus indirect association via Grb2 has been shown to have distinct functions [35]. One obvious possibility is that direct versus indirect association with Cbl leads to ubiquitination at distinct lysine residues with distinct functional consequences. Future studies aiming at identifying the sites of ubiquitination in wild-type c-Kit, versus the I571A/L939A mutant c-Kit (that is deficient in direct association), versus the Y703F/Y936F mutant that is defective in Grb2 association [45], will shed light on these issues.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Cancer Society, the research funds at Malmö University Hospital, Alfred Österlund Foundation, Gunnar Nilsson Cancer Foundation and the National Institutes of Health (Bethesda, MD, U.S.A.; to H.B.). E.H. was supported by a postdoctoral fellowship from the Wenner-Gren Foundation and the salary of L.R. is partially funded by the Swedish Research Council.
References
- 1.Lennartsson J., Jelacic T., Linnekin D., Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23:16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 2.Rönnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell. Mol. Life Sci. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PubMed] [Google Scholar]
- 3.Lennartsson J., Voytyuk O., Heiss E., Sundberg C., Sun J., Rönnstrand L. c-Kit signal transduction and involvement in cancer. Cancer Ther. 2005;3:5–28. [Google Scholar]
- 4.Thien C. B., Langdon W. Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 5.Lill N. L., Douillard P., Awwad R. A., Ota S., Lupher M. L., Jr, Miyake S., Meissner-Lula N., Hsu V. W., Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 2000;275:367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 6.Yokouchi M., Kondo T., Houghton A., Bartkiewicz M., Horne W. C., Zhang H., Yoshimura A., Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 7.Thien C. B., Langdon W. Y. Negative regulation of PTK signaling by Cbl proteins. Growth Factors. 2005;23:161–167. doi: 10.1080/08977190500153763. [DOI] [PubMed] [Google Scholar]
- 8.Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee P. S., Wang Y., Dominguez M. G., Yeung Y. G., Murphy M. A., Bowild-typeell D. D., Stanley E. R. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duval M., Bedard-Goulet S., Delisle C., Gratton J. P. Vascular endothelial growth factor-dependent down-regulation of Flk-1/KDR involves Cbl-mediated ubiquitination. Consequences on nitric oxide production from endothelial cells. J. Biol. Chem. 2003;278:20091–20097. doi: 10.1074/jbc.M301410200. [DOI] [PubMed] [Google Scholar]
- 11.Miyake S., Mullane-Robinson K. P., Lill N. L., Douillard P., Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. Consequences on nitric oxide production from endothelial cells. J. Biol. Chem. 1999;274:16619–16628. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- 12.Taher T. E., Tjin E. P., Beuling E. A., Borst J., Spaargaren M., Pals S. T. c-Cbl is involved in Met signaling in B cells and mediates hepatocyte growth factor-induced receptor ubiquitination. J. Immunol. 2002;169:3793–3800. doi: 10.4049/jimmunol.169.7.3793. [DOI] [PubMed] [Google Scholar]
- 13.Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 14.Peschard P., Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 15.Zeng S., Xu Z., Lipkowitz S., Longley J. B. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005;105:226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- 16.Wollberg P., Lennartsson J., Gottfridsson E., Yoshimura A., Rönnstrand L. The adaptor protein APS associates with the multifunctional docking sites Tyr-568 and Tyr-936 in c-Kit. Biochem. J. 2003;370:1033–1038. doi: 10.1042/BJ20020716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokouchi M., Wakioka T., Sakamoto H., Yasukawa H., Ohtsuka S., Sasaki A., Ohtsubo M., Valius M., Inoue A., Komiya S., Yoshimura A. APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene. 1999;18:759–767. doi: 10.1038/sj.onc.1202326. [DOI] [PubMed] [Google Scholar]
- 18.Brizzi M. F., Dentelli P., Lanfrancone L., Rosso A., Pelicci P. G., Pegoraro L. Discrete protein interactions with the Grb2/c-Cbl complex in SCF- and TPO-mediated myeloid cell proliferation. Oncogene. 1996;13:2067–2076. [PubMed] [Google Scholar]
- 19.Blume-Jensen P., Siegbahn A., Stabel S., Heldin C. H., Rönnstrand L. Increased Kit/SCF receptor induced mitogenicity but abolished cell motility after inhibition of protein kinase C. EMBO J. 1993;12:4199–4209. doi: 10.1002/j.1460-2075.1993.tb06104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupher M. L., Jr, Reedquist K. A., Miyake S., Langdon W. Y., Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J. Biol. Chem. 1996;271:24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 21.Blume-Jensen P., Claesson-Welsh L., Siegbahn A., Zsebo K. M., Westermark B., Heldin C. H. Activation of the human c-kit product by ligand-induced dimerization mediates circular actin reorganization and chemotaxis. EMBO J. 1991;10:4121–4128. doi: 10.1002/j.1460-2075.1991.tb04989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennartsson J., Blume-Jensen P., Hermanson M., Pontén E., Carlberg M., Rönnstrand L. Phosphorylation of Shc by Src family kinases is necessary for stem cell factor receptor/c-kit mediated activation of the Ras/MAP kinase pathway and c-fos induction. Oncogene. 1999;18:5546–5553. doi: 10.1038/sj.onc.1202929. [DOI] [PubMed] [Google Scholar]
- 23.Reith A. D., Ellis C., Lyman S. D., Anderson D. M., Williams D. E., Bernstein A., Pawson T. Signal transduction by normal isoforms and W mutant variants of the Kit receptor tyrosine kinase. EMBO J. 1991;10:2451–2459. doi: 10.1002/j.1460-2075.1991.tb07784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voytyuk O., Lennartsson J., Mogi A., Caruana G., Courtneidge S., Ashman L. K., Rönnstrand L. Src family kinases are involved in the differential signaling from two splice forms of c-Kit. J. Biol. Chem. 2003;278:9159–9166. doi: 10.1074/jbc.M211726200. [DOI] [PubMed] [Google Scholar]
- 25.Mori S., Rönnstrand L., Claesson-Welsh L., Heldin C. H. A tyrosine residue in the juxtamembrane segment of the platelet-derived growth factor beta-receptor is critical for ligand-mediated endocytosis. J. Biol. Chem. 1994;269:4917–4921. [PubMed] [Google Scholar]
- 26.Sattler M., Salgia R., Shrikhande G., Verma S., Pisick E., Prasad K. V., Griffin J. D. Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-kit, phosphatidylinositol 3-kinase, and p120(CBL) J. Biol. Chem. 1997;272:10248–10253. doi: 10.1074/jbc.272.15.10248. [DOI] [PubMed] [Google Scholar]
- 27.Waterman H., Katz M., Rubin C., Shtiegman K., Lavi S., Elson A., Jovin T., Yarden Y. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 29.Yokouchi M., Kondo T., Sanjay A., Houghton A., Yoshimura A., Komiya S., Zhang H., Baron R. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 2001;276:35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- 30.Meisner H., Czech M. P. Coupling of the proto-oncogene product c-Cbl to the epidermal growth factor receptor. J. Biol. Chem. 1995;270:25332–25335. doi: 10.1074/jbc.270.43.25332. [DOI] [PubMed] [Google Scholar]
- 31.Miyazawa K., Toyama K., Gotoh A., Hendrie P. C., Mantel C., Broxmeyer H. E. Ligand-dependent polyubiquitination of c-kit gene product: a possible mechanism of receptor down modulation in M07e cells. Blood. 1994;83:137–145. [PubMed] [Google Scholar]
- 32.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 33.Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 34.Harris K. F., Shoji I., Cooper E. M., Kumar S., Oda H., Howley P. M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grøvdal L. M., Stang E., Sorkin A., Madshus I. H. Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp. Cell Res. 2004;300:388–395. doi: 10.1016/j.yexcr.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Marmor M. D., Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi S., Sawano A., Nojima Y., Shibuya M., Maru Y. The c-Cbl/CD2AP complex regulates VEGF-induced endocytosis and degradation of Flt-1 (VEGFR-1) FASEB J. 2004;18:929–931. doi: 10.1096/fj.03-0767fje. [DOI] [PubMed] [Google Scholar]
- 38.Wilhelmsen K., Burkhalter S., van der Geer P. C-Cbl binds the CSF-1 receptor at tyrosine 973, a novel phosphorylation site in the receptor's carboxy-terminus. Oncogene. 2002;21:1079–1089. doi: 10.1038/sj.onc.1205166. [DOI] [PubMed] [Google Scholar]
- 39.Peschard P., Ishiyama N., Lin T., Lipkowitz S., Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J. Biol. Chem. 2004;279:29565–29571. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 40.Ohrt T., Mancini A., Tamura T., Niedenthal R. c-Cbl binds to tyrosine-phosphorylated neurotrophin receptor p75 and induces its ubiquitination. Cell. Signalling. 2004;16:1291–1298. doi: 10.1016/j.cellsig.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Lupher M. L., Jr, Songyang Z., Shoelson S. E., Cantley L. C., Band H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J. Biol. Chem. 1997;272:33140–33144. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- 42.Lupher M. L., Jr, Rao N., Lill N. L., Andoniou C. E., Miyake S., Clark E. A., Druker B., Band H. Cbl-mediated negative regulation of the Syk tyrosine kinase. A critical role for Cbl phosphotyrosine-binding domain binding to Syk phosphotyrosine 323. J. Biol. Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- 43.Andoniou C. E., Lill N. L., Thien C. B., Lupher M. L., Jr, Ota S., Bowild-typeell D. D., Scaife R. M., Langdon W. Y., Band H. The Cbl proto-oncogene product negatively regulates the Src-family tyrosine kinase Fyn by enhancing its degradation. Mol. Cell. Biol. 2000;20:851–867. doi: 10.1128/mcb.20.3.851-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Kerkhof P., Alves dos Santos C. M., Sachse M., Klumperman J., Bu G., Strous G. J. Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell. 2001;12:2556–2566. doi: 10.1091/mbc.12.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thömmes K., Lennartsson J., Carlberg M., Rönnstrand L. Identification of Tyr-703 and Tyr-936 as the primary association sites for Grb2 and Grb7 in the c-Kit/stem cell factor receptor. Biochem. J. 1999;341:211–216. [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J., Hubbard S. R. Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 2005;280:18943–18949. doi: 10.1074/jbc.M414157200. [DOI] [PubMed] [Google Scholar]