Abstract
WD (tryptophan-aspartic acid dipeptide)-repeat proteins play a central role in signal transduction cascades by co-ordinating the interaction of key signalling molecules. We identified a novel propeller-FYVE [domain identified in Fab1p, YOTB, Vac1p and EEA1 (early endosome antigen 1)] protein, ProF, which is expressed in various cell lines and tissues and consists of seven WD-repeats and a FYVE domain. WD-repeat proteins offer a platform for protein–protein interactions by folding into a seven-bladed propeller-like structure, while the FYVE domain binds to phosphatidylinositol 3-phosphate present mainly on intracellular membranes. The ProF protein partially co-localizes with EEA1 on vesicular structures and binds to the protein kinases Akt and PKCζ/λ (protein kinase Cζ/λ) via its WD-repeat propeller. ProF interacts more strongly with the kinases after hormonal stimulation. Endogenously expressed ProF and the two kinases interact in brain and in the preadipocyte cell line 3T3-L1, suggesting a role in secretory vesicular processes. In summary, we describe a new binding partner for kinases, located on vesicular structures in specialized cells, which may play a role for the spatial organization of signalling cascades.
Keywords: Akt, PC12 cell, FYVE domain, 3T3-L1 adipocyte, vesicle, WD-repeat
Abbreviations: BLAST, basic local alignment search tool; Bcr, break point cluster region; DMEM, Dulbecco's modified Eagle's medium; EEA1, early endosome antigen 1; FCS, fetal calf serum; FYVE, domain identified in Fab1p, YOTB, Vac1p and EEA1; FENS-1, FYVE domain protein localized to endosomes 1; FOXO, forkhead box O; GFP, green fluorescent protein; GLUT4, glucose transporter type 4; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; HEK-293T, HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40); IGF-1, insulin-like growth factor 1; m/p-Akt1, myristoylated and palmitoylated Akt1; mTOR, mammalian target of rapamycin; NGF, nerve growth factor; PDK1, phosphoinositide-dependent kinase 1; PH domain, pleckstrin homology domain; PI3K, phosphoinositide 3-kinase; PI3P, phosphatidylinositol 3-phosphate; PIP3, phosphatidylinositol 3,4,5-trisphosphate; PKC, protein kinase C; ProF, propeller-FYVE protein; RACK, receptor of activated C-kinase; RT, reverse transcriptase; SARA, Smad anchor for receptor activation; siRNA, small interfering RNA; SMART, simple modular architecture research tool; SNARE, soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor; TfR, transferrin receptor; TGF, transforming growth factor; TPEN, N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine; TRITC, tetramethylrhodamine β-isothiocyanate; VAMP2, vesicle-associated membrane protein 2; WD, tryptophan-aspartic acid dipeptide
INTRODUCTION
WD (tryptophan-aspartic acid dipeptide)-repeat proteins belong to a large structurally conserved protein family. Members of this family are characterized by the presence of several partially conserved sequence repeats of 40–60 amino acids. These repeats typically end with a WD at the C-terminus [1]. The exact number of WD-repeats in a protein can vary, but a cluster of seven WD-repeats is the predominant form. The crystal structure of the β-subunit of the GTP-binding protein transducin has shown a highly symmetrical propeller-like structure of seven folded WD-repeats [2] and all WD-repeat proteins are speculated to fold into circularized β-propellers. WD domains have no intrinsic catalytic activity, but they serve as a platform for protein–protein interactions. The family of WD-repeat proteins has been implicated in a wide range of cellular functions as signal transduction, RNA synthesis and processing, chromatin and cytoskeletal assembly, vesicular trafficking, cell-cycle control, and apoptosis (for a review, see [3]). Well-characterized seven-bladed WD-repeat proteins involved in signalling are the RACKs (receptors of activated C kinase). RACK proteins bind activated PKC (protein kinase C) isoforms, thereby enabling the interaction with their substrates (summarized in [4]).
Most WD-repeats are part of multidomain proteins. These proteins contain in addition to WD-repeats zinc-binding motifs, actin-binding motifs, leucine zippers, SH3 (Src homology 3) domains binding to proline-rich sequences, BROMO (derived from brahma by analogy to the chromodomain) domains as found in chromatin-associated proteins, or the catalytic domains of kinases (for a review, see [3]). The associated protein domains can determine the subcellular localization of the propeller by binding to specific subcellular compartments and can bring together interaction partners at this location.
The FYVE domain, which was originally identified in Fab1p, YOTB, Vac1p and EEA1 (early endosome antigen 1), represents one such compartment-specific targeting domain. FYVE domains are conserved from yeast to human [5]. More than 30 FYVE domain-containing proteins are known in humans. The domain enables binding to PI3P (phosphatidylinositol 3-phosphate), which is mainly constitutively produced on endosomal membranes by PI3K (phosphoinositide 3-kinase). Most FYVE domain-containing proteins serve as regulators of endocytotic membrane trafficking, whereby a few proteins have a role in signalling or cytoskeleton remodelling [5]. Many FYVE proteins contain additional domains such as coiled-coil domains in EEA1, ankyrin repeats in SARA (Smad anchor for receptor activation), or WD-repeats in FENS-1 (FYVE domain protein localized to endosomes 1). While the function of FENS-1 is unknown [6,7], SARA presents mediators of the TGF-β (transforming growth factor-β) signalling cascade to the TGF-β receptor for phosphorylation, and EEA1 plays an important role in vesicle fusion during endocytosis by controlling endosome docking (summarized in [5]).
We identified a WD-repeat propeller and FYVE domain-containing protein, ProF (propeller-FYVE protein), as interaction partner of the serine/threonine kinase Akt. Akt regulates multiple biological processes, including inhibition of apoptosis, cell-cycle promotion and insulin signalling (summarized in [8]). We have previously described a cross-talk between Akt- and Raf-mediated signal transduction [9–11]. Akt has three isoforms, Akt1, Akt2, and Akt3, each with overlapping but distinct cellular functions. Akt1 plays an important role in regulation of growth and apoptosis, whereas Akt2 functions primarily as a regulator of glucose metabolism (summarized in [12]). Akt is activated by various growth factors and hormones such as IGF-1 (insulin-like growth factor 1) and insulin. Activation occurs at the plasma membrane after PI3K-dependent generation of PIP3 (phosphatidylinositol 3,4,5-trisphosphate). Thereby Akt is recruited to the plasma membrane via its PH (pleckstrin homology) domain and is activated by phosphorylation at Thr308 by PDK1 (phosphoinositide-dependent kinase 1) followed by phosphorylation at Ser473 by the mTOR (mammalian target of rapamycin)–rictor (rapamycin-insensitive companion of mTOR) complex [13]. In addition to Akt, also atypical PKC isoforms, PKCζ/λ, were found here to bind to the WD-FYVE protein. PDK1 also phosphorylates Thr410/Thr403 in the activation loop of PKCζ/λ in an insulin-dependent mechanism (summarized in [14]). PKCζ shows 86% overall amino acid sequence identity with PKCλ, but only 45–55% with other PKC isoforms. PKCζ/λ is involved in many signal transduction pathways, including cell polarity and NF-κB (nuclear factor κB) signalling [14]. The binding of Akt and PKCζ/λ to ProF suggests a role of ProF in cellular systems in which both kinases are involved, such as insulin signalling [15].
We describe here a new WD-repeat FYVE protein, ProF, which is expressed in the mouse pre-adipocyte cell line 3T3-L1 and the rat neuroendocrine cell line PC12 as well as in other tissues and cell lines. The FYVE domain of ProF targets the protein to vesicles. The WD-repeat propeller binds to the two kinases Akt and PKCζ/λ in a stimulation-dependent manner. Our results suggest that ProF functions as a vesicle-located binding partner of the two kinases with a putative role in signal transduction.
MATERIALS AND METHODS
Materials
Cell-culture media, supplements and Novex® 8–16% and 10–20% Tris/glycine gradient gels were purchased from Invitrogen. TPEN [N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine], NGF (nerve growth factor) and IGF-1 were obtained from Calbiochem, and wortmannin and TRI Reagent were from Sigma. Restriction enzymes were purchased from New England Biolabs, polymerases from Promega, the Access RT (reverse transcriptase)–PCR system was from Promega and the QuikChange™ Site-Directed Mutagenesis kit from Stratagene. Unlabelled ATP for in vitro kinase assay was obtained from Roche. Oligonucleotides were purchased from Microsynth (Balgach, Switzerland). The transfection reagents Lipofectamine™ and Fugene-6 were purchased from Invitrogen and Roche respectively. Protease inhibitors were from Roche, Protein G–Sepharose beads and Redivue [γ-32P]ATP were obtained from Amersham Biosciences. Mouse monoclonal antibody 9E10 against Myc and rat monoclonal 3F10 against HA (haemagglutinin) were obtained from Roche; rabbit polyclonal A14 against Myc, rabbit polyclonal Y-11 against HA, mouse monoclonal DF1513 against CD71/TfR (transferrin receptor), rabbit polyclonal to PKC isoforms α/β, δ, ϵ, η and ζ/λ and a goat polyclonal against PKCζ/λ were obtained from Santa Cruz Biotechnology; mouse monoclonal 262K against HA, rabbit polyclonal against Akt, and rabbit polyclonal against phospho-Akt-Ser473 were from Cell Signaling Technology; mouse monoclonal GD11 against c-Src and PKC substrate peptide were from Upstate Biotechnology; mouse monoclonal clone 14 against EEA1 was from BD Transduction Laboratories; the anti-rabbit IgG–phycoerythrin conjugate was from Sigma. Fluorescein (FITC)-, rhodamine [TRITC (tetramethylrhodamine β-isothiocyanate)]- and Cy™5-conjugated anti-donkey antibodies against mouse, rabbit or sheep were purchased from Jackson Biologicals.
Cell culture
The cell lines HEK-293 (human embryonic kidney cells) (A.T.C.C. number CRL-1573) and HEK-293T [HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40)] (A.T.C.C. number CRL-11268), the human epithelial cell line HeLa (A.T.C.C. number CCL-2), the simian kidney cell line COS-7 (A.T.C.C. number CRL-1651), the mouse neuroblastoma cell line Neuro2A (CCL-131), the rat phaeochromocytoma cell line PC12 (CRL-1721), the mouse Schwann cell line Sw10 (CRL-2766), the mouse cerebellum cell line C8-D30 (CRL-2534) and the mouse fibroblastic cell line 3T3-L1 (A.T.C.C. number CL-173) were grown in DMEM (Dulbecco's modified Eagle's media) containing 10% (v/v) FCS (fetal calf serum; Seratec). Penicillin and streptomycin were added to cultures of 3T3-L1 cells. PC12 cells were seeded (3×105/cm2) on collagen-coated glass bottom culture dishes (MatTek, Ashland, MA, U.S.A.) in DMEM with 10% FCS and grown overnight. Then the cells were washed twice with PBS, and were grown for 3 days in DMEM with 0.1% FCS and 40 ng/ml NGF.
Yeast two-hybrid screen
A yeast two-hybrid screen was performed as described in [16] using full-length Akt1 as bait. A B-cell-specific cDNA library was obtained from S.J. Elledge (Baylor College of Medicine, Houston, TX, U.S.A.). One of two cDNAs identified in the yeast two-hybrid screen was subcloned into pBluescriptKS (Stratagene).
Computational analysis of ProF protein
Analysis of secondary structure elements was performed using SMART (simple modular architecture research tool; [17]) and a WD motif program (http://bmerc.bu.edu/projects/wdrepeat/). The three-dimensional model of ProFΔFYVE was generated by 3D-PSSM program [18] with the WD-repeat protein Tup1 (yeast dTMP uptake 1) (PDB 1ERJ) as structural template [19]. The calculated fold-structure had a certainty of fit better than 95%.
Recombinant DNA procedures
A ProF fragment containing a double Myc-tag 5′ to the cDNA was generated by PCR and the amplified fragment was cloned into pCMV5 generating pCMV5-Myc-ProF.
The deletion mutant lacking the FYVE domain, ProFΔFYVE (pCMV5-Myc-ProFΔFYVE), was generated by introducing a BspMI restriction site between the FYVE domain and the last WD-repeat domain by site-directed mutagenesis using pCMV5-Myc-ProF as a template and the following primers: 5′-GGCCATCACAGATGAAGAACCTGCACCCACAGCCACCTTCC-3′ and the complementary reverse primer. Subsequently, the plasmid was digested with MunI and BspMI restriction enzymes to excise the FYVE domain. The plasmid was annealed with a short partially overlapping oligonucleotide linker (forward primer 5′-AATTGATCTCCTGTGGCGGTGATGGTGGGATTGTCGTCGGGAACATGGACGTGGAGGAACGTGCACCC-3′ and reverse primer 5′-CTGTGGGTGCACGTTCCTCCACGTCCATGTTCCAGACGACAATCCCACCATCACCGCCACAGGAGATC-3′) and re-ligated.
The ProFΔFYVE mutants lacking blades 1–3 and blades 4–7 of the WD-repeat propeller were generated by site-directed mutagenesis using pCMV5-Myc-ProFΔFYVE as the template generating pCMV5-Myc-ProF 1–3 (primers: 5′-GCAATTTGCCTGGCACTAGTCTGAGAGTGGGCAGC-3′ and reverse complementary oligonucleotide) and pCMV5-Myc-ProF 4–7 (primer: 5′-CGAACAAAAACTTATTTCTGAAGAAGATCTGCTATGCTCTGAGAGTGGGCAGCGCCTGGGAGG-3′). A translational GFP (green fluorescent protein)–ProF fusion was generated by cloning Myc–ProF into the BglII- and SalI-digested plasmid pEGFP-C2 (Clontech).
FUW–FLAG–ProF was cloned using cFUW (lentiviral vector cFUW [24]) linearized with XbaI and HpaI, the BglII–SmaI fragment of pCMV5-MycProf and an oligonucleotide encoding the FLAG tag (upper: CTAGACCACCATGGACTACAAAGACGATGACGATAAACTA; and lower: CTAGACCACCATGGACTACAAAGACGATGACGATAAACTA). FUW–GFP–ProF was cloned using cFUW linearized with XbaI and BamHI and the NheI–BamHI fragment of pEGFPc2ProF. The latter plasmid was derived by insertion of a EcoRI- and BamHI-digested PCR fragment obtained from pCMV5-Myc-ProF (upper: CCGGAATTCATGGCGGCGGAGATCCAGCCCAAGCCTCTGACC; and lower: CGCGGATCCTCATCAAGACACGACTGGGGTCATATCCC) into pEGFPc2 linearized with EcoRI and BamHI. HA–Bcr (break point cluster region) wild-type was cloned in pCDNA3 (Invitrogen). The sequence of all plasmids was verified by DNA sequencing.
HA-tagged Akt1 wild-type [20], HA–Akt2 wild-type [21] and HA–m/p-Akt1 (myristoylated and palmitoylated Akt1) [22] have been described previously. c-Src wild-type cloned in pUSE plasmid was purchased from Upstate Biotechnology. HA–PKCζ encoding plasmid was obtained from the Fraunhofer Institute, Stuttgart, Germany.
RNA extraction and RT–PCR
RNA was extracted using TRI Reagent and RT–PCR was performed with 1 μg of total RNA. Primers for human and murine ProF were: forward (nt 658–678): GATCACTCTGTCATCATGTGG; reverse (nt 894–915): CTTACTGTCCCACATTTGCTTG. Primers for murine β-actin as internal control were designed as described in [23]; primers for human β-actin were: forward (nt 607–629): ACGGCCGAGCGGGAAATCGTGCG, reverse (nt 987–1009): ACTTGCGCTCAGGAGGAGCAATG. First-strand cDNA was synthesized at 48 °C for 45 min, and amplified by 2 min at 94 °C and 22 cycles (for β-actin) or 30 cycles (for ProF) for 30 s at 94 °C, 1 min at 68 °C, and 2 min at 68 °C.
ProF antiserum
An anti-ProF rabbit polyclonal antiserum was raised in rabbits against a peptide corresponding to the 15 amino acids at the C-terminus of murine and human ProF. The animal was boosted five times and the serum was recovered. Some of the antibody was thereafter affinity-purified to increase the specificity of binding (Eurogentech, Belgium). The same peptide was used for competition.
Viral transduction and generation of stably transduced 3T3-L1 fibroblasts
For siRNA (small interfering RNA) down-regulation, the sequence used was nt 1154–1172 of ProF open reading frame, targeted against exon 11 (5′-GAACTGACAAGGTAATTAA-3′). A 64-nt oligonucleotide, containing the target both in sense and antisense orientation, was cloned into pSUPER and then together with the H1 promoter into the lentiviral vector FUGW [24]. HEK-293T cells (4×105) were transfected with 0.6 μg of the lentiviral expression construct, 0.6 μg of HCMV-G and 0.6 μg of pCMVΔR8.3 helper virus plasmids using Lipofectamine™ 2000 (Invitrogen), followed by medium change after 24 h. The supernatant was used to infect 8×104 cells of low passage 3T3-L1 cells, yielding a pool of siProF-expressing cells.
The identical procedure was performed for the lentiviral transduction of 3T3-L1 and PC12 cells with FUW–FLAG–ProF and FUW–GFP–ProF.
Immunofluorescence
COS-7 or HeLa cells seeded on glass coverslips were transiently transfected with Fugene-6 or Lipofectamine™. For inhibitor studies, cells were treated with 250 μM TPEN or 100 nM wortmannin (for 30 min). Cells were fixed with 3% (w/v) paraformaldehyde and permeabilized in PBS containing 0.25% Triton X-100. 3T3-L1 cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in PBS. Afterwards, the cells were incubated with appropriate primary and secondary antibodies for 1 h each at 20 °C and mounted in Mowiol (Hoechst Pharmaceuticals). Secondary antibodies were used at 1:100 dilution. The cells were examined by sequential excitation at 488 nm (FITC) and 568 nm (TRITC) using a confocal laser microscope (Leica SP2) and a ×40 1.25 oil immersion objective (Leica). The images were processed using IMARIS (Bitplane). The time-lapse study of PC12 cells was performed at 37 °C and 5% CO2. Pictures were captured every 5 s.
Immunoprecipitation and Western-blot analysis
HEK-293 and HEK-293T cells were transfected with expression vectors encoding the indicated proteins and processed as described in [9]. IGF-1 stimulation was performed with 100 ng/ml IGF-1 for 15 min. Murine tissues were extracted in a glycerol buffer containing 150 mM NaCl, 20 mM Tris/HCl (pH 7.5), 0.2% Nonidet P40, 10% (v/v) glycerol and 1 mM EDTA using a tight-fitting tissue blender. Complete homogenization was achieved after 6–10 strokes, 10 s each. Lysates were solubilized by shaking for 30 min at 4 °C.
Lysates of HEK-293T cells were immunoprecipitated with 1 μg of the appropriate antibody for at least 2 h at 4 °C, followed by additional 45 min with 10 μl of Protein G–Sepharose. Lysates and the resulting immunoprecipitates were subjected to SDS/PAGE and immunoblot analysis as described in [9].
Mouse brain lysates (∼1.25×106 cells/ml of lysate) were subjected to immunoprecipitation with 1 μg of affinity-purified antibody and incubated overnight at 4 °C with or without a 100-fold molar excess of the competing peptide. The endogenous ProF protein was detected as 44 kDa form in direct lysates and precipitates.
In 3T3-L1 cells, immunoprecipitation of endogenous ProF was performed with anti-ProF antiserum, covalently coupled with Protein G–Sepharose in order to avoid overlapping of predominant signal of the IgG antibody heavy chain. In brief, 10 μl of Protein G–Sepharose (Amersham) was incubated with 2 μg of anti-ProF antiserum in 500 μl of Washing-Binding Buffer (50 mM sodium borate, pH 8.2; all buffers were from Pierce) for 30 min at room temperature (20 °C), afterwards washed twice with this buffer and incubated for 1 h at 20 °C in 260 μl of Cross-linking Buffer (0.2 M triethanolamine, pH 8.2) with 850 μl of freshly added dimethylpimelimidate. Thereafter, Protein G–Sepharose was incubated for 1 h at room temperature and washed twice with Cross-linking Buffer, incubated for 10 min with Blocking Buffer (0.1 M ethanolamine, pH 8.2) at room temperature, washed twice with Blocking Buffer, washed three times with Elution Buffer (Immunopure IgG elution buffer; Pierce), and then equilibrated with Washing-Binding Buffer. Lysate of 3T3-L1 cells was added to beads and incubated overnight at 4 °C.
RESULTS
Sequence and putative structure of the WD-repeat FYVE protein ProF
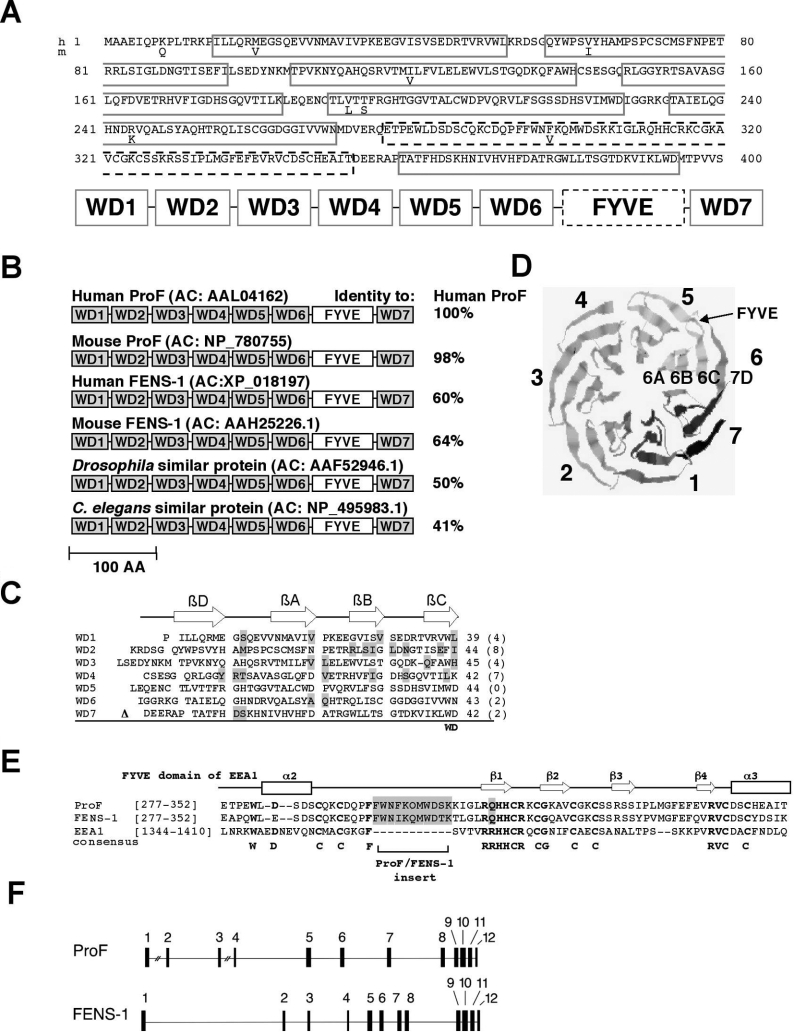
We performed a yeast two-hybrid screen using a human B-cell-specific embryonic cDNA library and full-length Akt1 as bait [16]. One of the identified clones encoded a novel protein of 400 amino acids and a predicted molecular mass of 44 kDa (GenBank® Entrez Protein database accession number AAL04162) (Figure 1A). Preliminary structural analysis using the SMART program [17] revealed one FYVE domain and five WD-repeats (WD1, WD3 and WD5–7 in Figure 1A). Further inspection using the specific WD motif search program BMERC (http://bmerc.bu.edu/projects/wdrepeat/) identified two additional WD-repeats (WD2 and WD4, Figure 1A). We designated the WD-repeat propeller-FYVE protein as ProF.
Figure 1. Characterization of ProF.
(A) Deduced amino acid sequences of human (h) ProF [GenBank® Entrez Protein database accession number (AC): AAL04162] and the murine (m) homologue (AC: NP_780755), differing in eight indicated amino acids, are shown. The protein contains seven WD-repeats (WD 1–7), indicated as grey boxes, and one FYVE domain, indicated as black dashed box. (B) The domain structure of ProF is shown, with the FYVE domain and seven WD domains (WD1–7) indicated. Molecules structurally related to human ProF are found in human (human FENS-1; AC: XP_018197), mouse (mouse ProF; AC: NP_780755; mouse FENS-1; AC: AAH25226.1), Drosophila (AC: AAF52946.1) and C. elegans (AC: NP_495983.1). Their amino acid sequence identity with human ProF is given in percentage. Scale bar indicates 100 amino acids. (C) The amino acid sequences of the WD-repeats 1 to 7 of ProF are listed. A typical WD-repeat unit contains four β-strands, βD, βA, βB and βC (arrows), and consists of approx. 40 amino acids. Mismatches to a postulated WD-repeat consensus sequence are grey-shaded and their numbers are listed in parentheses. The FYVE domain is located between WD6 and WD7 and is indicated by Δ. (D) A model of the three-dimensional structure of ProF without the FYVE domain was generated by the 3D-PSSM program with the yeast protein Tup1 as a template. The resulting β-propeller consists of seven blades formed by the WD-repeats (1–7), whereby the D-strand of each WD-repeat is part of the previous blade. The FYVE domain is located between the β-strands 6C and 7D (arrow). (E) The amino acid sequences of the FYVE domains of ProF, FENS-1 and EEA1 are aligned and compared with the predicted secondary structure of EEA1 containing four β-strands and two α-helices, α2 and α3 (top). Highly conserved sequences are RRHHCR, WXXD, CG, RVC and the eight cysteine residues (boldface letters). The FYVE domains of ProF and FENS-1 have a similar insert of 11 amino acids and a R to Q mutation in the conserved RRHHCR region (grey-shaded). (F) The genomic organization of ProF, found on chromosome 13 (13q13.3–13q14.3), and FENS-1, found on chromosome 2 (2q36), is schematically shown. The genomic structure of ProF and FENS-1 is characterized by 12 exons represented by black boxes. In the case of ProF, the gap between exons 1 and 2 has a length of 75 kb, and the gap between exons 3 and 4 is 28 kb long. The complete open reading frame of ProF has a length of 180 kb, and the complete open reading frame of FENS-1 has a length of 65 kb. In both cases, the FYVE domain is encoded by exons 9 and 10.
Database mining using the BLAST (basic local alignment search tool) program [25] revealed a murine homologue of unknown function (GenBank® Entrez Protein database accession number: NP_780755) with only eight divergent amino acids (Figure 1A). Figure 1(B) shows alignments of some proteins with identical domain structure. Homologues of ProF were encountered in Drosophila melanogaster and Caenorhabditis elegans, indicating that the protein may have a conserved function in animals. No homologues of the protein were found in other eukaryotes or prokaryotes. The FYVE domain of human ProF, indicated as a white box in Figure 1(B), is approx. 80% identical with the other FYVE domains listed. The WD-repeats show a higher divergence. The individual WD-repeats of the same protein (WD1–7) are only 5–26% identical with each other, while the corresponding WD-repeats among the various species are approx. 39–55% identical, indicating that the repeating elements of the WD motif exhibit only little sequence, but high structural, conservation.
The individual WD-repeats of human ProF are shown in Figure 1(C) with the aberrations compared with the consensus sequence [1] highlighted in grey and their number indicated in parentheses. The WD-repeats WD2 and WD4 show the highest diversity from the WD consensus motif, consistent with our finding that those WD-repeats were not recognized by the SMART program. Analysis of the secondary structure suggested that all seven repeats are able to form four antiparallel β-strands, A to D, indicated as arrows on the top of Figure 1(C). Seven WD-repeats can fold into a highly symmetrical β-propeller [2], whereby one propeller blade contains the last three strands of a repeat unit followed by the first strand of the next repeat unit. Therefore we used a protein structure prediction approach to find out whether ProF was able to fold into a seven-blade-containing propeller (Figure 1D). As template we used the WD-repeat-containing protein Tup1, a co-repressor of transcription in yeast [19]. Indeed, the amino acid sequence of the protein without the FYVE domain could be modelled into a seven-blade-containing propeller (Figure 1D) arranged around a central axis with a certainty of fit better than 95%.
The FYVE domain of ProF is located between WD-repeats 6 and 7 and comprises two zinc ions co-ordinated by eight conserved cysteine residues to form zinc fingers [5]. To compare the FYVE domain of ProF with that of other proteins, we aligned the amino acid sequences of the FYVE domains of ProF, the human ProF-homologue FENS-1, and EEA1 in Figure 1(E). The predicted secondary structure of the EEA1-FYVE domain is shown at the top. It contains four β-strands and two α-helices, as indicated by arrows and boxes (Figure 1E). The FYVE domains of ProF and FENS-1 are highly conserved, with 80% amino acid sequence identity, but both show only limited identities with the FYVE domain of EEA1, 27–28% respectively. ProF and FENS-1 share a change of an R to Q, R-Q-H-H-C-R in the conserved R-(R/K)-H-H-C-R motif and an 11-amino-acid insert between α2 and β1 (Figure 1E, highlighted in grey). These two characteristics are different from all other FYVE domain proteins [6]. They may account for a higher affinity towards PI3P and may make the binding of a second protein obsolete, which is usually important for membrane targeting [26].
Furthermore, the genomic organization of ProF and FENS-1 is shown (Figure 1F). ProF is encoded on chromosome 13 and was identified in the Human Genome database by using the BLAST program. The gene consists of 12 exons spread over more than 180 kb (Figure 1F, top). The ProF-homologue FENS-1 is encoded on chromosome 2 and the complete open reading frame of FENS-1 is 65 kb long including all introns (Figure 1F, bottom). ProF and FENS-1 show a similar chromosomal arrangement of 12 exons of very similar size. The two distinct genes may be the result of a gene duplication that did not occur in invertebrates (C. elegans and Drosophila, see Figure 1B).
Analysis of expression of ProF
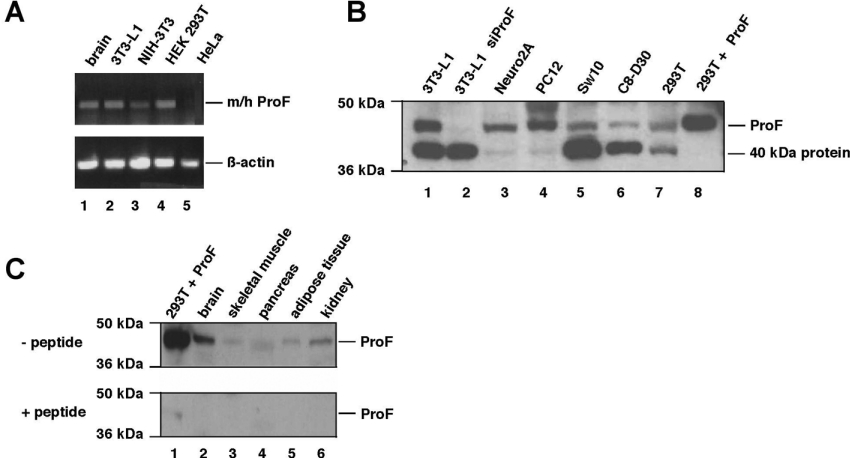
To detect mRNA expression of endogenous ProF, we performed RT–PCR using primers that recognize both the murine as well as the human ProF (m/h ProF). Endogenous ProF mRNA was detectable in murine brain extract, 3T3-L1 preadipocytes, NIH 3T3 cells, and in HEK-293T cells, but undetectable in HeLa cells (Figure 2A).
Figure 2. Expression of ProF.
(A) mRNA expression levels in different cells were analysed by RT–PCR from total RNA with primers specific for murine and human (m/h) ProF as indicated (top). β-Actin expression was used as control (bottom). (B) ProF protein expression levels in different cell lines were analysed by Western blotting with the peptide antibody raised against the 15 C-terminal amino acids of ProF. 3T3-L1 siProF (lane 2) shows down-regulation of the ProF protein in 3T3-L1 cells using an siRNA targeted against ProF. 293T+ProF (lane 8) indicates a lysate of HEK-293T cells overexpressing untagged ProF. In the case of 293T+ProF, only 1/20 of the protein content of lanes 1–7 was loaded to avoid excess signal strength by detection. (C) ProF protein expression levels in different cell lines were analysed by Western blotting of murine organs using the peptide antibody used in (B). 293T+ProF (lane 1) indicates a lysate of HEK-293T cells overexpressing untagged ProF. Antibody specificity was verified by immunoblotting with a 100-fold excess of competing peptide (+peptide, lower part).
For detection of ProF protein expression, we raised an antiserum against a peptide corresponding to the 15 C-terminal amino acids of mouse and human ProF. The antibody recognized a band at 44 kDa, corresponding to ProF as demonstrated with overexpressed untagged ProF (Figure 2B, lane 8) and the down-regulation of the band by an siRNA targeted against ProF (Figure 2B, lane 2). We tested various cell lines and found relatively high expression levels of ProF protein in 3T3-L1 (Figure 2B, lane 1) and PC12 cells (Figure 2B, lane 4). PC12, a rat neuroendocrine cell line, is a model system for neurons [27], whereas 3T3-L1 cells, a mouse preadipocyte cell line, is used as an in vitro model of adipogenesis and to study insulin pathways and obesity [28]. In a number of cell lines, an additional band was detected at 40 kDa (Figure 2B, lanes 1, 2 and 5–7), which was not related to ProF. Furthermore, immunoblots of different mouse tissues revealed the presence of ProF in various organs, with the highest expression levels detected in brain (Figure 2C). The specificity of the band was verified by peptide competition.
Subcellular localization of ProF
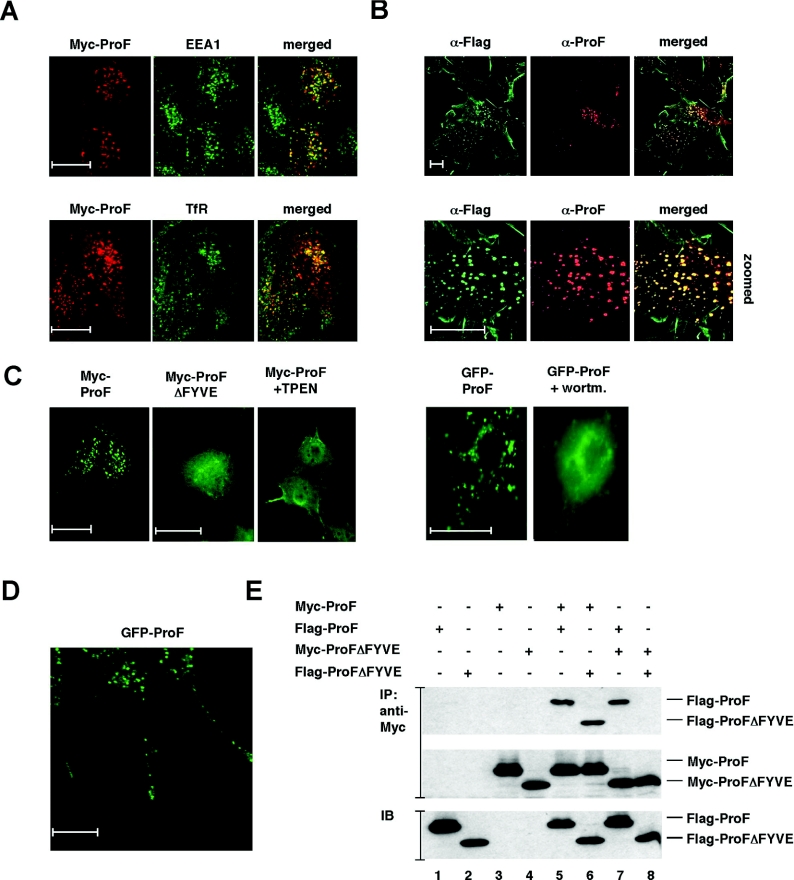
Because FYVE domains bind to PI3P present on vesicles, we investigated the subcellular localization of ProF. For that purpose, we transiently transfected HeLa cells with a Myc-tagged ProF construct and performed confocal immunofluorescence microscopy. Myc–ProF partially co-localized with EEA1, a marker for early endosomes, and to a lesser extent with the TfR, a more general marker for the recycling endocytic compartment (Figure 3A), and mannose-6-phosphate receptor, a marker for late endosomes and lysosomes (results not shown). These results indicate that a subpopulation of ProF is located at the early endosome compartment, where most of the PI3P- and FYVE domain-containing proteins reside [26]. The vesicular localization of ProF was also found in 3T3-L1 cells stably transduced with a FLAG-tagged construct of ProF (Figure 3B). Vesicular ProF was recognized by an antibody against the FLAG tag as well as by the peptide antibody against ProF (Figure 3B).
Figure 3. Subcellular localization of ProF.
(A) HeLa cells were transiently transfected with Myc–ProF and subjected to confocal immunofluorescence microscopy with antibodies against the Myc epitope, EEA1 and TfR. Co-localization of Myc-tagged ProF (red) with endogenous EEA1 (green) on early endosomes and with endogenous TfR (green) on recycling endosomes is visualized in yellow in the merged pictures. Scale bars indicate 20 μm. (B) Immunofluorescence microscopy analysis of undifferentiated 3T3-L1 cells stably transduced with a lentiviral vector encoding FLAG-tagged ProF with antibodies against ProF and the FLAG tag. Scale bars indicate 20 μm. (C) Immunofluorescence microscopy analysis of COS-7 cells transiently transfected with plasmid DNA encoding Myc–ProF, Myc–ProFΔFYVE and GFP–ProF. Cells were treated for 30 min with 250 μM TPEN or 100 nM wortmannin. Scale bars indicate 20 μm. (D) Immunofluorescence microscopy analysis of differentiated PC12 cells (day 3), stably transduced with a lentiviral vector encoding GFP-tagged ProF. Scale bar indicates 20 μm. (E) Oligomerization: lysates prepared from HEK-293T cells transiently transfected with the indicated constructs were subjected to immunoprecipitation (IP) with an antibody against the Myc epitope tag. Dimerization capabilities of ProF and ProFΔFYVE were assessed by immunoblotting (IB) with an antibody against the FLAG epitope and by SDS/PAGE (upper panel). The two lower panels show controls for protein expression levels of Myc- and FLAG-tagged proteins.
To analyse the role of the FYVE domain, we compared the localization of Myc–ProF and Myc–ProFΔFYVE, a mutant of ProF lacking the FYVE domain. Indeed, the vesicular localization is dependent on the FYVE domain (Figure 3C, left panel). Myc–ProFΔFYVE is distributed in the cytoplasm and the punctuated staining is strongly reduced in comparison with the full-length protein. Furthermore, the vesicular localization of Myc–ProF was lost in the presence of the Zn2+-chelator TPEN, which destroys zinc finger configurations. Additionally, the vesicular staining of a GFP–ProF fusion protein was abolished in the presence of wortmannin (Figure 3C, right panel). The PI3K inhibitor wortmannin prevents the formation of PI3P, which is required for the binding of FYVE domains to vesicles. These findings indicate that the FYVE domain targets the protein to PI3P-containing vesicles.
To test whether ProF also binds to vesicles in cell types with specialized vesicular trafficking systems, we stably expressed FLAG-tagged ProF in PC12 cells, which were later differentiated with 40 ng/ml NGF. Indeed, we could observe the vesicular localization of ProF in differentiated PC12 cells by confocal immunofluorescence microscopy (Figure 3D). Furthermore, time-lapse studies were performed and revealed that ProF is partially localized in a highly dynamic subpopulation of vesicles (see Supplementary Movie 1 at http://www.BiochemJ.org/bj/399/bj3990009add.htm). These results indicate that ProF rapidly moves along the neurites in both directions.
Oligomerization of ProF
The ability to dimerize is a property of a subset of FYVE domain proteins and important for their affinity to endosomal membranes [26]. Dimerization of EEA1 proteins is assumed to enhance the avidity to PI3P. To demonstrate the interaction of ProF with itself, we performed co-immunoprecipitation assays with Myc- and FLAG-tagged ProF. Indeed, the interaction was clearly detectable (Figure 3E). Since the dimerization motif of EEA1 is localized in the FYVE domain [7], we tested the interaction of the ProF mutant ProFΔFYVE. The interaction also takes place when only one of the interaction partners contains a FYVE domain (Figure 3E). This suggests that sequences outside of the FYVE domain contribute to the interaction. This had consequences for our effort to design dominant-negative mutants of ProF. Putative dominant-negative ProF mutants deleted for their FYVE domains can still heterodimerize with wild-type ProF and can therefore behave similarly to the wild-type protein.
Interaction of ProF with Akt and PKCζ/λ
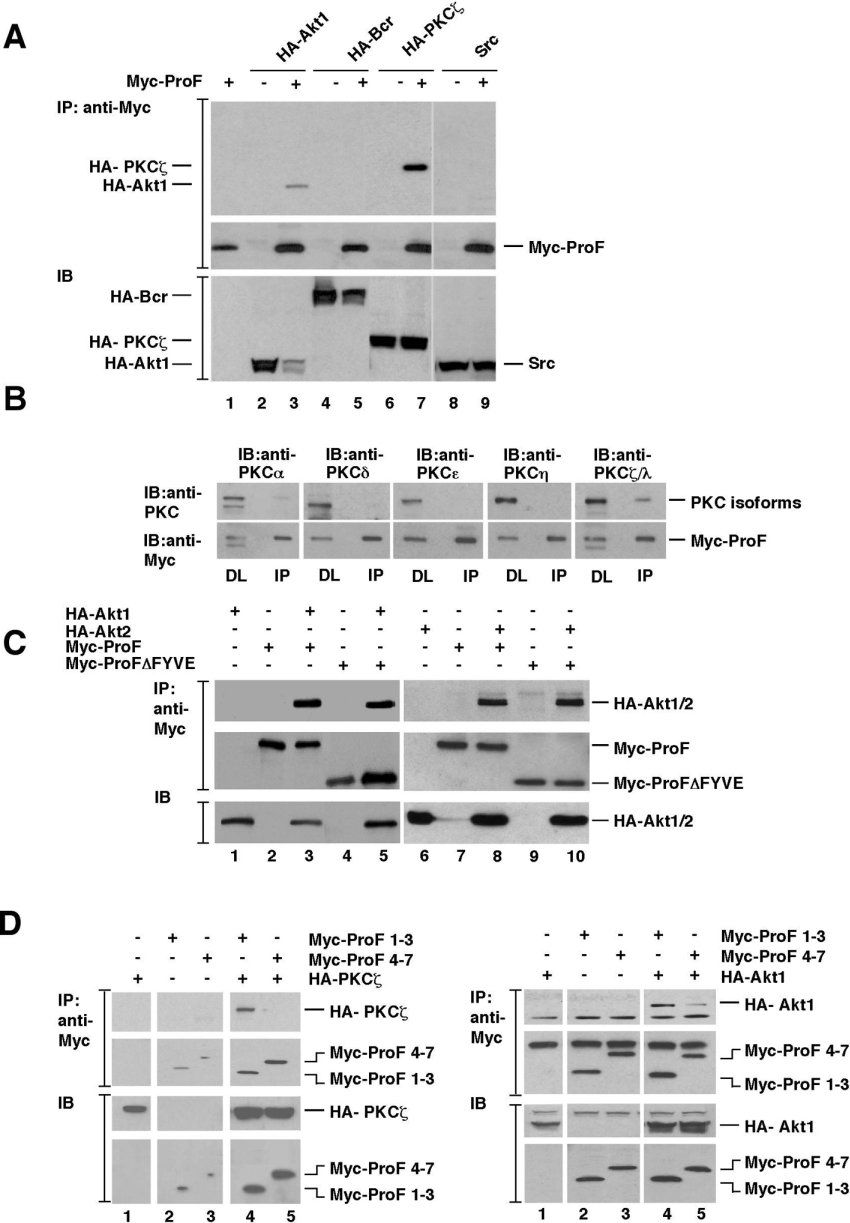
Since ProF was identified as an interaction partner of Akt1, we wanted to confirm this interaction by co-immunoprecipitation assays. Furthermore, we wanted to test the binding to other kinases which are known to interact with WD-repeat proteins, such as members of the PKC and the Src kinase families. Additionally, the multidomain protein Bcr was tested, since it contains a PH domain and various PH domain-containing proteins are known to bind to WD-repeat proteins [4]. We tested the kinases HA–Akt1, HA–PKCζ and HA–Bcr as well as untagged Src by using transiently transfected HEK-293T cells and antibodies against the HA tag or against Src (Figure 4A). Myc–ProF co-immunoprecipitated HA–Akt1 and HA–PKCζ (Figure 4A, lanes 3 and 7), but not HA–Bcr or untagged Src.
Figure 4. Interaction of ProF with overexpressed Akt and PKCζ.
(A) HEK-293T cells were transiently transfected with HA–Akt1, HA–Bcr, HA–PKCζ and Src in the presence or absence of Myc–ProF. Interaction of ProF with HA–Akt1, HA–Bcr, HA–PKCζ and Src was analysed by immunoprecipitation (IP) with an antibody to Myc tag followed by immunoblotting (IB) with antibodies against Src, HA and Myc epitopes (IP, top). Direct lysates are shown as expression controls (IB, bottom). (B) HEK-293T cells were transiently transfected with Myc–ProF. Interaction of Myc–ProF with various PKC isoforms was analysed by IP with an antibody against Myc followed by IB with antibodies against PKC isoforms and Myc-tagged ProF (IP, right). Direct lysates (DL) are shown as expression controls (DL, left). Samples were loaded on one gel; separating lines were included for clarity. (C) HEK-293T cells were transiently transfected with Myc–ProF or Myc–ProFΔFYVE in the presence of HA–Akt1 (left) or HA–Akt2 (right). Interaction of Myc–ProF or Myc–ProFΔFYVE with HA–Akt1 and HA–Akt2 was analysed by IP with an antibody to Myc tag followed by IB with antibodies against HA (top) and Myc epitopes (middle lane). Direct lysates are shown as expression controls (IB, bottom). (D) For mapping of the interaction sites of PKCζ and Akt on ProF, HEK-293T cells were transiently transfected with the Myc–ProF constructs Myc–ProF 1–3 (a mutant lacking the FYVE domain and the blades 4–7) and Myc–ProF 4–7 (a mutant lacking the FYVE domain and the blades 1–3) together with HA–PKCζ (left) and HA–Akt1 (right). The interaction of ProF deletion mutants with PKCζ and Akt was analysed by IP with an antibody to the Myc tag, followed by IB with antibodies against HA and Myc epitopes. The two upper panels show the interaction (IP), the two lower ones show direct lysates as expression controls (IB). Samples were loaded on one gel; separating lines were included for clarity.
To further investigate the binding specificity of ProF for certain kinases, we transiently transfected HEK-293T cells with constructs expressing Myc–ProF and immunoprecipitated ProF with an antibody against Myc. Overexpressed ProF co-precipitated endogenous atypical PKC isoforms PKCζ/λ in HEK-293T cells, but not the novel PKC isoforms PKCδ, PKCϵ and PKCη and bound only very weakly to the classical PKC isoform PKCα (Figure 4B). These results suggest a specificity of ProF for particular kinases and even isoforms within a kinase family.
To further characterize the binding properties of ProF, we transiently transfected HEK-293T cells with constructs expressing two isoforms of Akt, HA–Akt1 and HA–Akt2, together with Myc–ProF and Myc–ProFΔFYVE. Myc–Prof immunoprecipitated both Akt isoforms equally well (Figure 4C). Furthermore, the deletion of the FYVE domain did not affect the binding of ProF to Akt (Figure 4C, lanes 5 and 10), indicating that the WD-repeat propeller mediates the interaction of ProF with the kinases.
To delineate the interaction sites of Akt and PKCζ with the ProF protein, we constructed mutants of ProF. Deletion of single amino acids, larger fragments, or individual blades did not show reduced binding (results not shown). Therefore we generated mutants embracing larger sections of ProF. One mutant comprised the blades 1–3 (Myc–ProF 1–3, a mutant lacking the FYVE domain and the blades 4–7), the other one blades 4–7 (Myc–ProF 4–7, a mutant lacking the FYVE domain and the blades 1–3). In both cases, the mutants did not harbour the FYVE domain, which did not affect the binding of ProF to the kinases. The ProF mutants were tested by co-transfection with HA–Akt and HA–PKCζ (Figure 4D). Akt was co-precipitated by both mutants, but bound more strongly to blades 1–3, whereby PKCζ exclusively bound to blades 1–3. Akt truncation mutants containing either the PH domain or the kinase and regulatory domains were still able to bind Myc–ProF (results not shown). These results suggest that several binding sites contribute to the interaction between the kinases and ProF. Nevertheless, they indicate a specific binding of Akt1, Akt2 and PKCζ to ProF involving mainly or exclusively blades 1–3.
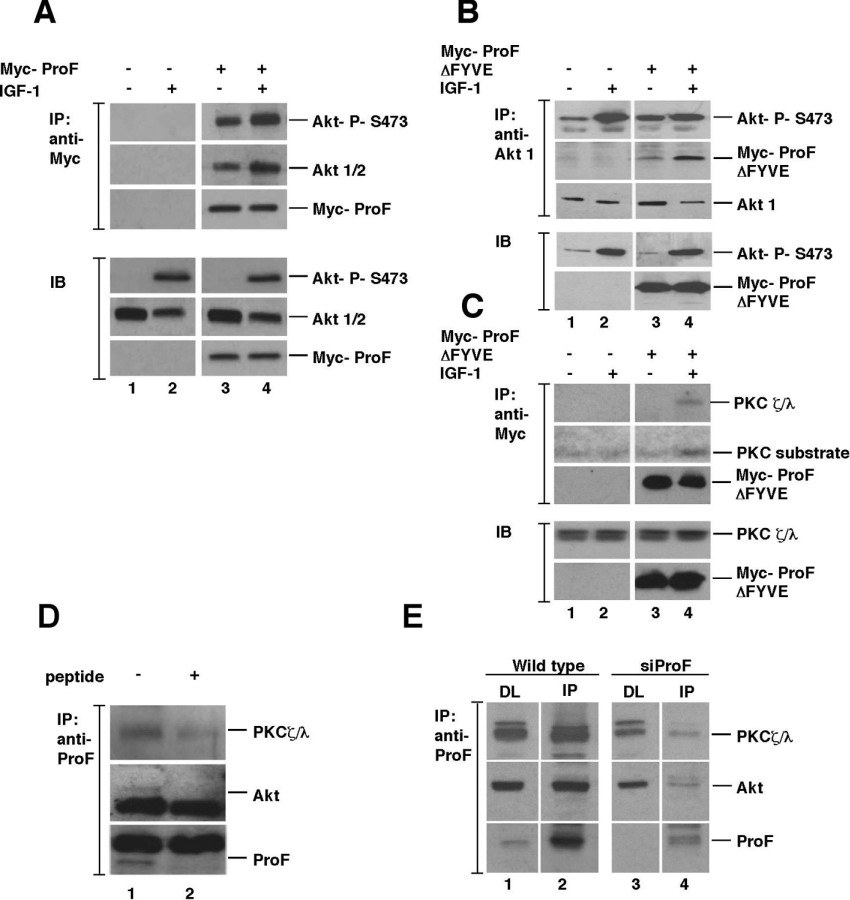
Hormonal stimulation of cells often regulates the binding of kinases to WD-repeat proteins [4]. In order to test this, HEK-293T cells were stimulated by IGF-1. Immunoprecipitates of overexpressed Myc–ProF from stimulated cells indeed contained both more total Akt and more phosphorylated Akt in comparison with non-stimulated cells (Figure 5A). Even in unstimulated cells, which contained only a low amount of activated Akt, phosphorylated Akt was preferentially recruited to ProF, indicating a very efficient selection of activated Akt in unstimulated cells. In summary, these results suggest a stronger interaction of ProF with the kinase Akt after hormonal stimulation. Furthermore, they confirm the interaction of ProF with Akt in a semi-endogenous system.
Figure 5. Semi-endogenous and endogenous interactions of ProF with Akt and PKCζ/λ.
(A) HEK-293T cells transiently expressing Myc–ProF were stimulated with 100 ng/ml IGF-1 and subjected to immunoprecipitation (IP) with an antibody towards the Myc epitope. Association of Myc–ProF with endogenous Akt1/2 was detected by an anti-Akt antibody and phosphorylation of Akt by an antibody to phosphorylated Ser473 (P-S473), indicating increased co-IP of Akt after stimulation by immunoblotting (IB) analysis. Expression and IP of Myc–ProF were verified by IB of direct lysates and IP with an antibody against the Myc epitope. Samples were loaded on one gel; separating lines were included for clarity. (B) HEK-293 cells transiently expressing Myc–ProFΔFYVE were stimulated with 100 ng/ml IGF-1 and subjected to IP with an antibody to Akt1. Association of Myc–ProFΔFYVE with endogenous Akt1 was detected by an anti-Myc antibody and phosphorylation of Akt by an antibody to P-S473 in an IB analysis. Expression of Myc–ProFΔFYVE were verified by IB of direct lysates with an antibody against the Myc epitope. Samples were loaded on one gel; separating lines were included for clarity. (C) HEK-293 cells transiently expressing Myc–ProFΔFYVE were stimulated with 100 ng/ml IGF-1 and subjected to IP with an antibody to Myc. PKCζ/λ co-precipitates with ProF and the interaction is increased upon IGF-1 stimulation. The kinase activity of the co-precipitated PKCζ/λ was tested with a PKC substrate peptide. Expression and IP of Myc–ProFΔFYVE were verified by IB of direct lysates and IP with an antibody against the Myc epitope. Samples were loaded on one gel; separating lines were included for clarity. (D) Mouse brain extract was treated with anti-ProF antibody in the absence (−) or presence (+) of an excess of ProF peptide to compete for the antibody followed by IB. Lane 2 shows competition of the ProF protein by addition of an excess of competing peptide. (E) Undifferentiated 3T3-L1 cells, which were untransduced (lanes 1 and 2) or transduced with an siRNA targeted against ProF to down-regulate ProF expression (lanes 3 and 4), were subjected to IP in lanes 2 and 4. IP was performed with the peptide antibody against ProF, which was cross-linked to Sepharose beads by dimethylpimelimidate. Association of endogenous ProF with endogenous kinases was detected in untransduced control cells (lane 2) and in cells with siRNA-mediated down-regulation of ProF (lane 4). Lane 1 shows the direct lysates of control cells, and lane 3 shows the direct lysates of siProF-transduced cells. Lysates were loaded on one gel; separating lines were included for clarity.
To further investigate the interaction of ProF with the endogenous kinases after hormonal stimulation of the cell, we overexpressed Myc–ProFΔFYVE in HEK-293 cells, which were stimulated by IGF-1. Indeed, we found that upon IGF-1 stimulation, binding of the ProF mutant to Akt1 (Figure 5B) and PKCζ/λ (Figure 5C) was increased. We found that the interaction of the WD-repeat propeller of ProF alone with the kinases was stronger after stimulation than the interaction of the full-length protein. This could be due to higher availability of the Myc–ProFΔFYVE mutant as binding partner for the cytoplasmatic kinases, when compared with the wild-type protein, which is mainly located on vesicles (Figure 3C). In summary, this experiment demonstrated that the stimulation-dependent binding capacity of ProF is mediated by the WD-repeat propeller.
Interaction of ProF with endogenous kinases
To investigate whether endogenous ProF also interacted with endogenous Akt and PKCζ/λ, we analysed mouse brain, where all three proteins are expressed. By using brain lysate for immunoprecipitation of ProF, we were able to co-precipitate Akt and PKCζ/λ (Figure 5D). The specificity of the immunoprecipitation was controlled by competition of the antibody with an excess of the peptide used as antigen (Figure 5D, lane 2).
Furthermore, the interaction of endogenous ProF with endogenous kinases was also demonstrated in 3T3-L1 cells (Figure 5E, lane 2), where we found comparatively high expression levels of ProF (Figure 2B). To control for the specificity of the interaction, we performed the immunoprecipitation in 3T3-L1 cells, transduced with a lentiviral vector expressing siRNA targeted against ProF (Figure 5E, lane 4). These results verified the interaction of ProF with Akt and PKCζ/λ with endogenous proteins in brain tissue and preadipocytes.
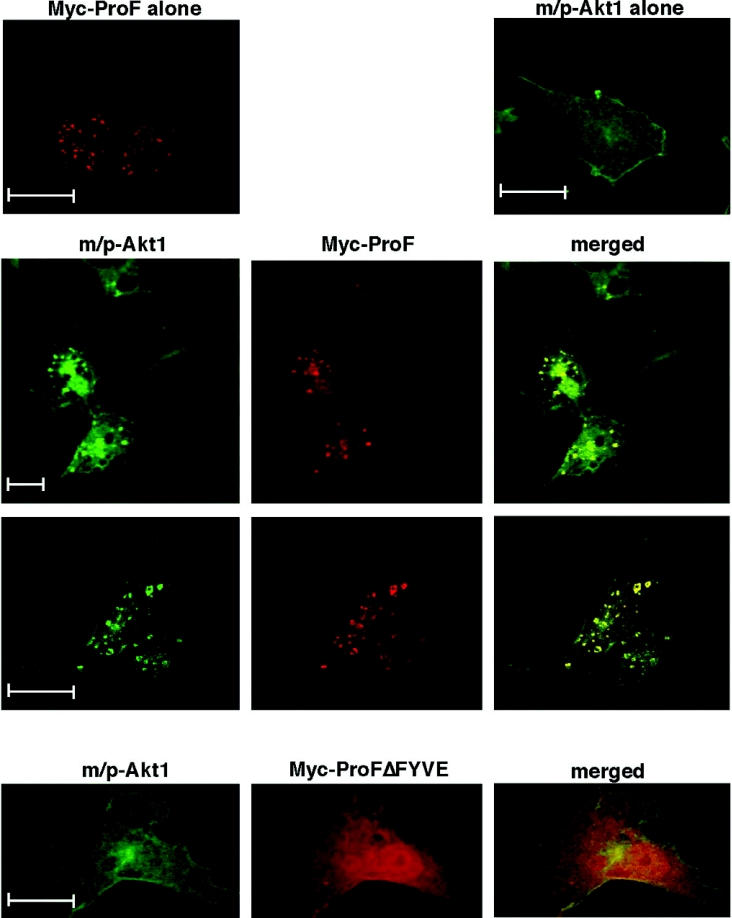
Interaction of ProF with membrane-targeted Akt alters the subcellular localization of Akt
In order to clarify whether the subcellular localization of ProF influenced its interaction with Akt, we performed confocal immunofluorescence microscopy. We used COS-7 cells because their large cytoplasm and small nuclei are favourable for immunofluorescence studies. Presence of Myc–ProF in COS-7 cells led to the formation of large vesicles (Figure 6, top left). This has also been observed previously with other FYVE domain-containing proteins, such as SARA [29]. Next, a membrane-targeted Akt (m/p-Akt1) mutant was expressed together with Myc–ProF. We found that m/p-Akt1 was pulled into the vesicular structures on which the co-expressed Myc–ProF was located, suggesting an interaction strong enough to overcome the anchoring of m/p-Akt1 to the cell-surface membrane (Figure 6, middle lane). Deletion of the FYVE domain led to a loss of m/p-Akt1 dragging to the vesicular structures (Figure 6, bottom lane). This experiment further substantiated our finding that the FYVE domain is essential for targeting ProF to internal vesicles, but it also showed that the binding of ProF to Akt is strong enough to alter the subcellular localization of m/p-Akt1.
Figure 6. Interaction of ProF with membrane-targeted Akt.
COS-7 cells were transiently transfected with Myc–ProF alone (top, left), HA–m/p-Akt1 alone (top, right) and together with Myc–ProF (two middle lanes) or Myc–ProFΔFYVE (bottom lane). Confocal microscopy analysis with HA- or Myc-specific antibodies revealed areas of co-localization as visualized in yellow on the merged pictures (two middle lanes). In all cases, scale bars indicate 20 μm.
Thus this experiment suggests that ProF can target the kinases to vesicles. Furthermore, it brings together the capacity of ProF as an interaction partner of kinases with its property as a vesicular protein.
DISCUSSION
We have identified a WD-repeat protein, ProF, which contains a FYVE domain and interacts with the kinases Akt and PKCζ/λ. The WD-repeats of ProF form a putative propeller structure that binds Akt and PKCζ/λ, preferentially after hormonal stimulation. Interestingly, several WD-repeat proteins are known to favour binding of activated kinases such as RACK1 and RACK2. RACK1 selectively binds activated PKC isoforms on ribosomes (for a review, see [30]) and also eIF6 (eukaryotic translation initiation factor 6), which is phosphorylated by PKC, leading to increased translation. RACK2 recruits activated PKCϵ to Golgi membranes, where it might be involved in control of secretion and vesicular trafficking [31]. These examples show that WD-repeat proteins can bring together various kinases and their substrates to integrate signalling pathways.
The WD-repeats of ProF are linked to a FYVE domain, which is responsible for the localization to intracellular vesicles. Overexpressed Myc- or FLAG-tagged ProF was observed on vesicular structures and was found to partially co-localize with EEA1 or the TfR. The vesicular localization was dependent on the presence of the FYVE domain and was lost in cells that had been treated with wortmannin to deplete PIP3 and PI3P or with TPEN to chelate Zn2+, which is the co-ordinating ion of the FYVE domain. A vesicular localization of ProF has been shown in various cell types such as HeLa, COS-7, 3T3-L1 preadipocytes and PC12 cells. Although we could not specifically detect endogenous ProF by confocal immunofluorescence microscopy, cell fractionation revealed that endogenous ProF was present in the low-density microsomal fraction (results not shown) which contains endosomes and other intracellular membranes [32]. Additionally, we showed in differentiated PC12 cells the localization of GFP–ProF fusion protein on vesicular structures, which either randomly move within the cell body or migrate in both directions through neurites.
These results suggest that ProF plays a role in association with vesicles. ProF could be involved in numerous vesicular processes in brain and pancreas, where its expression levels are comparatively high, as well as in many other tissues. For example, in the pancreas, it could be involved in insulin release from pancreatic β-cells upon elevation of glucose levels [33], or zymogen granule release from pancreatic acinal cells upon stimulation [34]. In neurons, ProF could play a role in synaptic vesicle exocytosis [35]. Additionally, ProF as a vesicular protein could play a role in numerous intracellular trafficking events such as vesicle transport from endosomes to lysosomes or from endosomes to the trans-Golgi network (summarized in [36]) or in vesicle-related signal transduction events.
ProF is a WD-repeat and FYVE domain-containing protein. Database mining using the SMART program revealed that apart from ProF there are two other proteins in humans which contain both WD-repeats and a FYVE domain, FENS-1 and WDF3. The protein FENS-1 shows a relatively high amino acid sequence identity of 60% with ProF and an identical domain organization. Furthermore, both proteins, ProF and FENS-1, contain a unique and highly conserved FYVE domain. FYVE domains bind exclusively and specifically to PI3P [26], but only a small subset of FYVE-containing proteins can translocate to endosome membranes without the need for additional interaction domains (summarized in [7]). Interestingly, the FYVE domain of FENS-1 has been demonstrated to display the highest binding capacity to PI3P among all of the FYVE domains investigated so far [6,7]. Similar to FENS-1, the FYVE domain of ProF was found to be sufficient for membrane localization of the protein, indicating that ProF also has a high PI3P-binding capacity. FENS-1 has previously been called WDF1 for WD40- and FYVE domain-containing protein 1, whereas ProF has been called WDF2, without assignment of any further citation. The third WD-repeat FYVE protein, which exists in humans, was designated WDF3. It contains a BEACH domain [beige and CHS (Chediak–Higashi syndrome) domain], five WD-repeats and a FYVE domain, and has also been named Alfy (autophagy-linked FYVE domain) [37]. WDF3 is conserved in animals, plants and yeast. It has a domain organization that is different from that of ProF or FENS-1. Furthermore, the overall identity of the deduced amino acid sequence of the WD-repeats and the FYVE domain of WDF3 with those of human ProF or its homologues from other species is only 21%.
It is difficult to conclude anything on the function of ProF from the known properties of WD-repeat propellers or FYVE domains. Based on the fact that ProF interacts with Akt and PKCζ/λ, we speculate that ProF could play a role in cellular functions in which both kinases are involved. Co-operation of Akt and PKCζ/λ has been described in two cellular systems: firstly, ceramides can activate PKCζ/λ for phosphorylation and negative regulation of Akt activity [38]; secondly, both kinases, Akt and PKCζ/λ, have been shown to regulate insulin-dependent glucose uptake in adipocytes (reviewed in [39]).
As a binding partner of Akt and PKCζ/λ, ProF could directly influence the activation and phosphorylation of the kinases after hormonal stimulation. However, because overexpression and knockdown of ProF in 3T3-L1 cells did not affect Akt phosphorylation (results not shown), we speculate that ProF might rather serve as an adaptor to bring together the kinases and their substrates to play a role in signalling events.
Therefore, to understand the function of ProF as a binding partner of Akt and PKCζ/λ, it would be important to determine vesicular substrates of ProF-binding kinases. ProF could bind the activated kinases on vesicles to bring them to the vicinity of their substrates for facilitated phosphorylation. A PKCζ substrate on vesicles in PC12 and 3T3-L1 cells could be the VAMP2 (vesicle-associated membrane protein 2) [40]. VAMP2 is a v-SNARE [vesicular SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor)] protein, which interacts with t-SNARE (target membrane SNARE) protein to promote vesicle fusion. VAMP2 mediates the stimulation-dependent plasma membrane translocation of GLUT4 (glucose transporter type 4) in 3T3-L1 adipocytes (summarized in [41]) and synaptic vesicle exocytosis [35]. The kinase Akt has numerous substrates (summarized in [8]). An example of a vesicular Akt substrate in 3T3-L1 cells is AS160 (Akt substrate of 160 kDa), a protein that participates in the translocation of GLUT4-containing vesicles to the plasma membrane [42] and is found on GLUT4-containing vesicles in unstimulated cells [43].
Akt also plays a key role in the process of adipocyte differentiation in 3T3-L1 cells [44] and neuronal differentiation in PC12 cells [45]. Several Akt substrates involved in these processes have emerged recently. One indirect target of Akt implicated in 3T3-L1 adipocyte differentiation is the serine/threonine kinase mTOR. Akt activates mTOR via direct phosphorylation and inhibition of the tumour suppressor protein TSC2 (tuberous sclerosis protein 2), which otherwise inhibits mTOR activity (summarized in [46]). mTOR is found predominantly on internal vesicles of mammalian cells and is highly up-regulated during adipocyte differentiation [47]. Other targets of Akt are the FOXO (forkhead box O) transcription factors. Phosphorylation of FOXO1, the most abundant FOXO isoform in 3T3-L1 adipocytes, by Akt plays a role in the regulation of adipocyte differentiation [48], whereas NGF-mediated phosphorylation of FOXO3 by Akt might be involved in neuronal differentiation and survival in PC12 cells [49]. Currently, we are investigating the potential role of ProF as an adaptor protein for the phosphorylation of Akt and PKCζ substrates.
In summary, we demonstrated that ProF is targeted to vesicular membranes by the FYVE domain and serves via its WD-repeat propeller as a binding platform for the kinases Akt and PKCζ/λ. The binding of the kinases occurs preferentially after hormonal stimulation, suggesting a role of ProF in signalling and raising the question about substrates. ProF may act as an adaptor protein in a variety of tissues, possibly in secretory pathways. ProF could exert its function by binding to additional signalling proteins and to their effectors and substrates.
Multimedia adjunct
Acknowledgments
We thank: Brian A. Hemmings (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) and Franz-Josef Johannes (Fraunhofer Institute for Interfacial Engineering, Stuttgart, Germany) for providing the Akt and the PKCζ vectors respectively; Jonathan S. Bogan (Section of Endocrinology, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, U.S.A.) for expert advice, discussions and help; Ralf Kretschmar (Institute of Medical Virology, Zurich, Switzerland) for initial contributions to the study; the Electronmicroscopical Laboratory at the University of Zurich for providing the facilities for confocal microscopy and for expert help; Professor Dr Martin Altwegg (Institute of Medical Microbiology, Zurich, Switzerland; now: Bioanalytica AG, Luzern 6, Switzerland) for providing the real-time PCR facilities; Dr M. Schwemmle and Dr D. Mayer (both from Institute of Medical Virology, Zurich, Switzerland) for initial contributions to siRNA studies. The individual contributions of the authors are: A.S.B. made initial contributions in 1999; S.Z. contributed initially and continuously to this study; T.F.: Figures 1(A)–1(F), 2A, 4(B)–4(D), 5(A), 5(C) and 5(E); G.B. and E.H.: immunofluorescence except Figures 3(B) and 3(D); G.B.: Figure 4(A), mutants for Figure 4(D); E.H., Figure 3(E); M.S.: antibody reporter system; A.F.: Figures 2(C) and 5(D); J.H.: siRNA, PC12 cell culture, immunofluorescence of Figures 3(B) and 3(D); J.H. and M.B.: adipocyte culture, Figure 5(B); A.C.: Figure 2(B).
References
- 1.Neer E. J., Schmidt C. J., Nambudripad R., Smith T. F. The ancient regulatory-protein family of WD-repeat proteins. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 2.Sondek J., Bohm A., Lambright D. G., Hamm H. E., Sigler P. B. Crystal structure of a G-protein βγ dimer at 2.1 Å resolution. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 3.Li D., Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schechtman D., Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 5.Stenmark H., Aasland R., Driscoll P. C. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 6.Ridley S. H., Ktistakis N., Davidson K., Anderson K. E., Manifava M., Ellson C. D., Lipp P., Bootman M., Coadwell J., Nazarian A., et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 7.Blatner N. R., Stahelin R. V., Diraviyam K., Hawkins P. T., Hong W., Murray D., Cho W. The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]
- 8.Lawlor M. A., Alessi D. R. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- 9.Rommel C., Clarke B. A., Zimmermann S., Nunez L., Rossman R., Reid K., Moelling K., Yancopoulos G. D., Glass D. J. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann S., Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 11.Moelling K., Schad K., Bosse M., Zimmermann S., Schweneker M. Regulation of Raf-Akt cross-talk. J. Biol. Chem. 2002;277:31099–31106. doi: 10.1074/jbc.M111974200. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z. Z., Tschopp O., Baudry A., Dummler B., Hynx D., Hemmings B. A. Physiological functions of protein kinase B/Akt. Biochem. Soc. Trans. 2004;32:350–354. doi: 10.1042/bst0320350. [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 14.Hirai T., Chida K. Protein kinase C zeta (PKC zeta): activation mechanisms and cellular functions. J. Biochem. 2003;133:1–7. doi: 10.1093/jb/mvg017. [DOI] [PubMed] [Google Scholar]
- 15.Zick Y. Uncoupling insulin signalling by serine/threonine phosphorylation: a molecular basis for insulin resistance. Biochem. Soc. Trans. 2004;32:812–816. doi: 10.1042/BST0320812. [DOI] [PubMed] [Google Scholar]
- 16.Schneider S., Buchert M., Georgiev O., Catimel B., Halford M., Stacker S. A., Baechi T., Moelling K., Hovens C. M. Mutagenesis and selection of PDZ domains that bind new protein targets. Nat. Biotechnol. 1999;17:170–175. doi: 10.1038/6172. [DOI] [PubMed] [Google Scholar]
- 17.Schultz J., Milpetz F., Bork P., Ponting C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley L. A., MacCallum R. M., Sternberg M. J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 19.Sprague E. R., Redd M. J., Johnson A. D., Wolberger C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000;19:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 21.Meier R., Alessi D. R., Cron P., Andjelkovic M., Hemmings B. A. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J. Biol. Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 22.Andjelkovic M., Alessi D. R., Meier R., Fernandez A., Lamb N. J., Frech M., Cron P., Cohen P., Lucocq J. M., Hemmings B. A. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 23.Takamura T., Nohara E., Nagai Y., Kobayashi K. Stage-specific effects of a thiazolidinedione on proliferation, differentiation and PPARγ mRNA expression in 3T3-L1 adipocytes. Eur. J. Pharmacol. 2001;422:23–29. doi: 10.1016/s0014-2999(01)01053-6. [DOI] [PubMed] [Google Scholar]
- 24.Lois C., Hong E. J., Pease S., Brown E. J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Stenmark H., Aasland R. FYVE-finger proteins–effectors of an inositol lipid. J. Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- 27.Greene L. A., Tischler A. S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U.S.A. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green H., Kehinde O. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell. 1976;7:105–113. doi: 10.1016/0092-8674(76)90260-9. [DOI] [PubMed] [Google Scholar]
- 29.Itoh F., Divecha N., Brocks L., Oomen L., Janssen H., Calafat J., Itoh S., Dijke Pt P. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-β/Smad signalling. Genes Cells. 2002;7:321–331. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson J., Sengupta J., Frank J., Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csukai M., Chen C.-H., De Matteis M. A., Mochly-Rosen D. The coatomer protein β′-COP, a selective binding protein (RACK) for protein kinase C epsilon. J. Biol. Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- 32.Simpson I. A., Yver D. R., Hissin P. J., Wardzala L. J., Karnieli E., Salans L. B., Cushman S. W. Insulin-stimulated translocation of glucose transporters in the isolated rat adipose cells: characterization of subcellular fractions. Biochim. Biophys. Acta. 1983;763:393–407. doi: 10.1016/0167-4889(83)90101-5. [DOI] [PubMed] [Google Scholar]
- 33.Regazzi R., Wollheim C. B., Lang J., Theler J. M., Rossetto O., Montecucco C., Sadoul K., Weller U., Palmer M., Thorens B. VAMP-2 and cellubrevin are expressed in pancreatic beta-cells and are essential for Ca2+-but not for GTPγS-induced insulin secretion. EMBO J. 1995;14:2723–2730. doi: 10.1002/j.1460-2075.1995.tb07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C. C., Ng C. P., Lu L., Atlashkin V., Zhang W., Seet L. F., Hong W. A role of VAMP8/endobrevin in regulated exocytosis of pancreatic acinar cells. Dev. Cell. 2004;7:359–371. doi: 10.1016/j.devcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Burgoyne R. D., Morgan A. Secretory granule exocytosis. Physiol. Rev. 2003;83:581–632. doi: 10.1152/physrev.00031.2002. [DOI] [PubMed] [Google Scholar]
- 36.Hong W. SNAREs and traffic. Biochim. Biophys. Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- 37.Simonsen A., Birkeland H. C., Gillooly D. J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 38.Powell D. J., Hajduch E., Kular G., Hundal F. S. Ceramide disables 3-phosphoinositide binding to the pleckstrin homology domain of protein kinase B (PKB)/Akt by a PKC zeta-dependent mechanism. Mol. Cell. Biol. 2003;23:7794–7808. doi: 10.1128/MCB.23.21.7794-7808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saltiel A. R., Kahn C. R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature (London) 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 40.Braiman L., Alt A., Kuroki T., Ohba M., Bak A., Tennenbaum T., Sampson S. R. Activation of protein kinase C zeta induces serine phosphorylation of VAMP2 in the GLUT4 compartment and increases glucose transport in skeletal muscle. Mol. Cell. Biol. 2001;21:7852–7861. doi: 10.1128/MCB.21.22.7852-7861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grusovin J., Macaulay S. L. Snares for GLUT4 – mechanisms directing vesicular trafficking of GLUT4. Front. Biosci. 2003;8:d620–d641. doi: 10.2741/1052. [DOI] [PubMed] [Google Scholar]
- 42.Kane S., Sano H., Liu S. C. H., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. A method to identify serine kinase substrates – Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 43.Larance M., Ramm G., Stockli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- 44.Kohn A. D., Summers S. A., Birnbaum M. J., Roth R. A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J. Biol. Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y., Seger R., Suresh Babu C. V., Hwang S. Y., Yoo Y. S. A positive role of the PI3-K/Akt signaling pathway in PC12 cell differentiation. Mol. Cells. 2004;18:353–359. [PubMed] [Google Scholar]
- 46.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Withers D. J., Ouwens D. M., Nave B. T., van der Zon G. C., Alarcon C. M., Cardenas M. E., Heitman J., Maassen J. A., Shepherd P. R. Expression, enzyme activity, and subcellular localization of mammalian target of rapamycin in insulin-responsive cells. Biochem. Biophys. Res. Commun. 1997;241:704–709. doi: 10.1006/bbrc.1997.7878. [DOI] [PubMed] [Google Scholar]
- 48.Nakae J., Kitamura T., Kitamura Y., Biggs W. H., III, Arden K. C., Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 49.Wang X., Chen L., Maures T. J., Herrington J., Carter-Su C. SH2-B is a positive regulator of nerve growth factor-mediated activation of the Akt/Forkhead pathway in PC12 cells. J. Biol. Chem. 2004;279:133–141. doi: 10.1074/jbc.M310040200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.