Abstract
The serine/threonine protein kinase Sgk1 (serum- and glucocorticoid-inducible kinase 1) is characterized by a short half-life and has been implicated in the control of a large variety of functions in different subcellular compartments and tissues. Here, we analysed the influence of the N-terminus of Sgk1 on protein turnover and subcellular localization. Using green fluorescent protein-tagged Sgk1 deletion variants, we identified amino acids 17–32 to function as an anchor for the OMM (outer mitochondrial membrane). Subcellular fractionation of mouse tissue revealed a predominant localization of Sgk1 to the mitochondrial fraction. A cytosolic orientation of the kinase at the OMM was determined by in vitro import of Sgk1 and protease protection assays. Pulse–chase experiments showed that half-life and subcellular localization of Sgk1 are inseparable and determined by identical amino acids. Our results provide evidence that Sgk1 is primarily localized to the OMM and shed new light on the role of Sgk1 in the control of cellular function.
Keywords: half-life, localization, mitochondria, outer mitochondrial membrane, serum- and glucocorticoid-inducible kinase 1 (Sgk1), topology
Abbreviations: BiP, immunoglobulin heavy-chain binding protein; DAPI, 4′,6-diamidino-2-phenylindole; DIABLO, direct IAP (inhibitor of apoptosis protein) binding protein with low pI; DMEM, Dulbecco's modified Eagle's medium; ENaC, epithelial sodium channel; FOXO3a, forkhead box O3a; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; Grp78, 78 kDa glucose-regulated protein; HEK-293 cells, human embryonic kidney cells; Hsp60, heat-shock protein 60; Nedd4-2, neural precursor cell expressed, developmentally down-regulated 4-2; NP40, Nonidet P40; OMM, outer mitochondrial membrane; Sgk1, serum- and glucocorticoid-inducible kinase 1; Smac, second mitochondria-derived activator of caspase
INTRODUCTION
The serum- and glucocorticoid-inducible serine/threonine kinase Sgk1 (serum- and glucocorticoid-inducible kinase 1) consists of 431 amino acids and belongs to a family of structurally related enzymes termed AGC-type kinases (protein kinase A/protein kinase G/protein kinase C-family kinases). Among these, Sgk1 shares approx. 50% sequence similarity at the amino acid level with protein kinase B/Akt, protein kinase C, ribosomal protein S6 kinase and cAMP-dependent protein kinase [1]. Sgk1 has been described to regulate a variety of cellular processes. In vitro assays suggested that Sgk1 might control signalling pathways and cell survival by phosphorylation of glycogen synthase kinase 3, B-raf kinase and forkhead family member FOXO3a (forkhead box O3a) [2–4]. In addition, the action of Sgk1 has been implicated in the control of a variety of plasma membrane transport proteins, including sodium channels ENaC (epithelial sodium channel) and SCN5A (sodium channel, voltage-gated, type V, α-subunit), chloride channel ClC2, the epithelial calcium channel TRPV5 (transient receptor potential cation channel, subfamily V, member 5), glucose transporter SGLT1 (sodium-coupled glucose transporter 1), the potassium channels Kv1.3, ROMK (renal outer medullary potassium channel) and KCNE1 (potassium voltage-gated channel, Isk-related family, member 1), and glutamate transporter EAAT1 (glial excitatory amino acid transporter 1) [5–14]. In the kidney, Sgk1 contributes to sodium homoeostasis by mediating the aldosterone-induced up-regulation of ENaC in the plasma membrane of distal tubule cells. Accordingly, Sgk1 knockout mice display an impaired sodium retention during dietary sodium restriction [15].
Sgk1 was first identified as an immediate early gene expressed in a rat mammary tumour cell line following stimulation with serum or glucocorticoids [1,16]. Depending on the specific tissues or cell lines examined, Sgk1 was shown to be induced by diverse factors, such as changes in osmolarity [17], aldosterone [18] and various growth factors, including fibroblast growth factor, platelet-derived growth factor [19], follicle-stimulating hormone [20] and transforming growth factor β [21]. Activation of the kinase requires at least two phosphorylation events in the activation loop and the C-terminal hydrophobic motif [22,23] and is further limited by a short half-life, resulting in low levels of protein expression in many tissues and cell lines. In line with these observations, Sgk1 is rapidly ubiquitinated and degraded by the proteasome [24]. Interestingly, although lysine residues at the N-terminus are not subject to ubiquitination, a variant of Sgk1 lacking the N-terminal 60 amino acids has a much longer half-life [24]. Another level of regulation was attributed to the subcellular localization of Sgk1. Various studies suggested that Sgk1 is translocated from the cytoplasm to the nucleus in a stimulus- and cell-cycle-dependent manner via binding to importin-α [25–27]. In the nucleus, Sgk1 is thought to phosphorylate specific targets such as FOXO3a [4]. In contrast, two recent reports showed an association of Sgk1 with the plasma membrane in the rat kidney and in different cell lines, and that the N-terminus of the protein plays a major role in membrane targeting [24,28].
In order to gain further insight into the regulation and the range of cellular functions of Sgk1, the goals of the present study were to examine the intracellular localization of Sgk1 and to determine the region responsible for localization and degradation.
EXPERIMENTAL
Molecular cloning
Sgk1–GFP (green fluorescent protein) fusion protein variants and variants with a C-terminal FLAG tag were generated by PCR using a Pwo polymerase (Roche Biochemicals, Mannheim, Germany). Primers were extended with restriction sites and fragments were cloned in frame into pEGFP-N3 (BD Clontech, Palo Alto, CA, U.S.A.), or in case of the FLAG-tagged variants into pcDNA3.1(+) (Invitrogen). The 5′ primers contained a Kozak sequence surrounding the ATG start codon. For variants starting with amino acids 17, 33 and 60, we made use of the internal ATGs coding for the respective methionine residues. In variants starting at position 48, Tyr47 was replaced with methionine to obtain a start codon. All constructs were confirmed by DNA sequencing.
Cell culture and transfections
HEK-293 cells (human embryonic kidney cells) were maintained in DMEM (Dulbecco's modified Eagle's medium; Biochrom, Berlin, Germany), supplemented with 5% (v/v) fetal calf serum (Biochrom) and 2 mM L-glutamine. For pulse–chase experiments and immunoblot analysis, 2×106 cells were resuspended in 500 μl of serum-free medium and, after addition of 20 μg of the respective plasmid-DNA, electroporated at 170 V and 960 μF in 4 mm gap electroporation cuvettes (Bio-Rad, Munich, Germany) using a Bio-Rad Gene Pulser II. Transfected cells were plated on a 10 cm diameter dish. For confocal microscopy, 50000 cells were grown on 12 mm coverslips in 24-wells and transfected with 100 ng of Mito-DsRedII (BD Clontech) and 200 ng of GFP or the indicated Sgk1–GFP fusion protein variants using the FuGENE™ transfection reagent (Roche Biochemicals) according to the manufacturer's instructions.
Confocal laser scanning microscopy
HEK-293 cells were washed with PBS 48 h after transfection, and fixed with 4% (w/v) paraformaldehyde, and nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) before mounting. Microscopy was performed with a Zeiss LSM510 inverted laser scanning microscope (Carl Zeiss, Jena, Germany) using the following configuration: DAPI: λexc=364 nm, band-pass filter λem=385–470 nm; GFP: λexc=488 nm, band-pass filter λem=505–550 nm; DsRed2 (Discosoma red fluorescent protein 2): λexc=543 nm, low-pass filter λem=560 nm. Images were processed with the Zeiss LSM image browser software.
Pulse–chase experiments and immunoprecipitation
Transfected HEK-293 cells were incubated for 45 min at 37 °C in methionine- and cysteine-free DMEM, and then pulse-labelled for 10 min with 200 μCi of [35S]methionine/cysteine labelling mixture (Hartmann, Braunschweig, Germany). Incorporation of label was stopped by the addition of complete medium supplemented with 1.5 mM non-radioactive methionine and 0.5 mM non-radioactive cysteine to a final volume of 5 ml. After 0 min, 15 min, 60 min and 4 h, cells from 1 ml of suspension were pelleted and lysed in 50 mM Tris (pH 7.5), 5 mM MgCl2, 0.5% NP40 (Nonidet P40), 1 mM PMSF and 1.5 μg/ml aprotinin for 30 min at 4 °C. Lysates were depleted of nuclei and cell debris by centrifugation for 10 min at 20800 g in a microcentrifuge and precleared with normal rabbit serum. GFP fusion proteins were recovered by immunoprecipitation with anti-GFP rabbit serum (a gift from Ralf Schülein, Leibniz-Institute for Molecular Pharmacology, Berlin, Germany) and Protein A–Sepharose (Sigma, Steinheim, Germany), followed by five washes in 50 mM Tris/HCl (pH 7.5), 5 mM EDTA, 150 mM NaCl and 0.5% NP40. Sepharose beads were boiled in reducing sample buffer [50 mM Tris, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol, 100 mM dithiothreitol, 0.1% (w/v) Bromophenol Blue] and subjected to PAGE. Gels were dried and signals were obtained with 2,5-diphenyloxazole fluorography.
Subcellular fractionation
Fractions were prepared from livers and kidneys of Sgk1 knockout mice and their wild-type littermates [15]. Organs were homogenized in TES buffer (10 mM Tris, 250 mM sucrose and 1 mM EDTA, pH 7.4) in a Dounce homogenizer by 12 strokes with a tight-fitting pestle. After centrifugation at 500 g for 10 min, crude mitochondria were pelleted from postnuclear supernatant at 10000 g for 20 min. The pellet was resuspended in TES, layered on a discontinuous sucrose gradient consisting of 1 and 1.5 M sucrose and centrifuged at 80000 g for 1 h. Highly purified mitochondria were harvested at the 1 M/1.5 M interphase and washed twice in TES. Cytosolic and microsomal fractions were prepared by ultracentrifugation of the postmitochondrial supernatant for 1 h at 48000 rev./min in a TLA 110 rotor (Beckman). The pellet served as microsomal fraction. For the purification of nuclei and plasma membranes from the 500 g pellet, the pellet was resuspended in 120 mM Tricine, 150 mM KCl and 30 mM MgCl2 (pH 7.5) supplemented with 1 mM dithiothreitol and 0.1% Triton X-100, and nuclei and plasma membrane sheets were pelleted by centrifugation through a 1.75 M sucrose cushion in a Beckman SW-32Ti rotor for 2 h at 38000 g. In some experiments, mice were injected with 100 μg/kg body weight dexamethasone (Sigma) 4 h before the preparation.
In vitro protein import into mitochondria
Coupled in vitro transcriptions and translations were performed in the presence of [35S]methionine according to the manufacturer's instructions using the Promega TnT kit (Promega, Mannheim, Germany). Reticulocyte lysates containing the radiolabelled Sgk1 variants were cleared of low-molecular-mass components by passage through Sephadex G-50 and added for the indicated periods of time at room temperature (22 °C) to purified C57BL/6 kidney mitochondria in import buffer consisting of 250 mM sucrose, 80 mM KCl, 10 mM Mg(OAc)2, 2.5 mM EDTA, 2 mM KH2PO4, 2.5 mM ATP, 5 mM NADH, 2.5 mM disodium succinate, 50 mM Hepes (pH 7.3) and 0.2 mg/ml BSA. Mitochondria were washed twice in ice-cold import buffer and subjected to PAGE. For the analysis of membrane association and orientation, in vitro-translated proteins were added to 1 ml of mouse kidney postnuclear supernatant from C57BL/6 mice supplemented with 2.5 mM ATP, 5 mM NADH and 2.5 mM disodium succinate, where import in mitochondria occurred for 20 min at 30 °C. Mitochondria were pelleted at 10000 g for 15 min, and were washed once with TES.
Membrane association, sodium carbonate extraction and protease protection assays
For protease protection assays, mitochondria were incubated with 200 μg/ml proteinase K (Roth, Karlsruhe, Germany) for 15 min at room temperature with or without 0.5% Triton X-100. The reaction was stopped by addition of 5 mM PMSF.
Mitochondria were sonicated and then separated into soluble and membrane fractions by ultracentrifugation for 1 h at 85000 rev./min in a Beckman TLA 100.3 rotor. For sodium carbonate extractions, a membrane pellet was resuspended in 100 mM Na2CO3, incubated for 1 h at 4 °C under agitation and again pelleted for 1 h at 300000 g. Samples were separated by PAGE and radiolabelled proteins were visualized by fluorography.
Immunoblots
Transfer of proteins on to nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany) or on to PVDF membranes for anti-Sgk1 and anti-FLAG M2 blots (Immobilon P; Millipore) was conducted using a tank blot chamber (Bio-Rad). After blocking in 5% (w/v) non-fat dry milk in PBS containing 0.05% Tween 20 (PBS-T), antibodies were applied in PBS-T as follows: anti-Sgk1 (Sigma) 1:50000, anti-Sgk1 (Cell Signaling Technology, Danvers, MA, U.S.A.) 1:2000, anti-Sgk1 [28] 1:5000, anti-Sgk1 [29] 1:2000, anti-Sgk1 (the present study, see below) 1:10000, anti-FLAG M2 (Sigma) 1:10000, anti-Hsp60 (heat-shock protein 60; Stressgen, San Diego, CA, U.S.A.) 1:4000, anti-cytochrome c (Santa Cruz Biotechnology, Heidelberg, Germany) 1:5000, anti-Smac (second mitochondria-derived activator of caspase)/DIABLO [direct IAP (inhibitor of apoptosis protein) binding protein with low pI] (ProSci, Poway, CA, U.S.A.) 1:1000, anti-BiP (immunoglobulin heavy-chain binding protein)/Grp78 (78 kDa glucose-regulated protein) (Stressgen) 1:500, anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Chemicon, Temecula, CA, U.S.A.) 1:2500000, anti-Na/K-ATPase α-subunit (Upstate Biotechnology) 1:10000, and anti-histone H1 (Santa Cruz Biotechnology) 1:1000. For detection of Sgk1 with a sheep anti-Sgk1 antibody [29], membranes were blocked with Rotiblock reagent (Carl Roth, Karlsruhe, Germany) instead of dry milk. Membranes were washed, incubated at room temperature for 2 h with peroxidase-conjugated secondary antibodies (Vector Laboratories), washed again and subjected to chemiluminescence detection.
We produced an anti-Sgk1 antibody by immunization of a rabbit five times with 500 μg of Sgk1 amino acids 290–431 fused to an N-terminal His tag. The protein was expressed in Escherichia coli and purified using Ni-NTA (Ni2+-nitrilotriacetate) spin columns (Qiagen, Hilden, Germany). IgGs were purified from the serum using Protein A–Sepharose.
RESULTS
A signal peptide between amino acids 17 and 32 targets Sgk1 to mitochondria
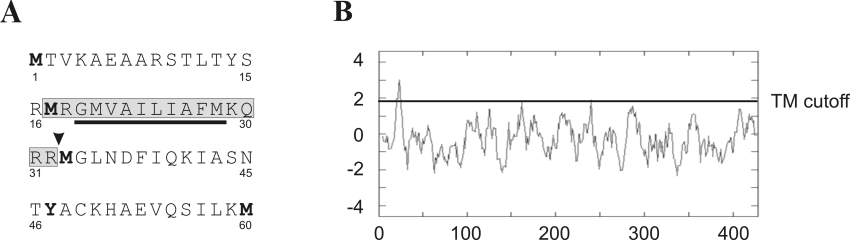
Previous studies indicated that Sgk1 is localized to the cytosol, the nucleus and the plasma membrane in response to different external stimuli [24–28]. We used the PSORTII prediction algorithm [30] to calculate the probability of Sgk1 localization to different cellular compartments. In this analysis, Sgk1 has a 43.5% probability to be a cytosolic protein and a 34.8% probability to localize to mitochondria. According to TargetP V1.0 [31] and MitoProt II 1.0a4 [32], two programs that predict mitochondrial localization, Sgk1 is likely to be imported into mitochondria, and a predicted mitochondrial signal peptide might be cleaved at amino acid position 33 (Figure 1A). Analysis using the Kyte-Doolittle plot suggested that the N-terminus of Sgk1 might contain a transmembrane segment (Figure 1B).
Figure 1. Amino acid sequence and hydropathy of the Sgk1 N-terminus.
(A) The N-terminal 60 amino acids of murine Sgk1. Residues are shown in the one-letter code. Starting amino acids for the constructs used in the present study are in boldface; in some constructs, Tyr47 was replaced by methionine. Grey box highlights a predicted mitochondrial targeting sequence, and arrowhead indicates a potential cleavage site for the mitochondrial processing peptidase at amino acid position 33. The black bar indicates a potential transmembrane segment as predicted by the Kyte-Doolittle analysis. (B) Kyte-Doolittle hydropathy plot of Sgk1. The N-terminus of Sgk1 contains a potential transmembrane (TM) segment.
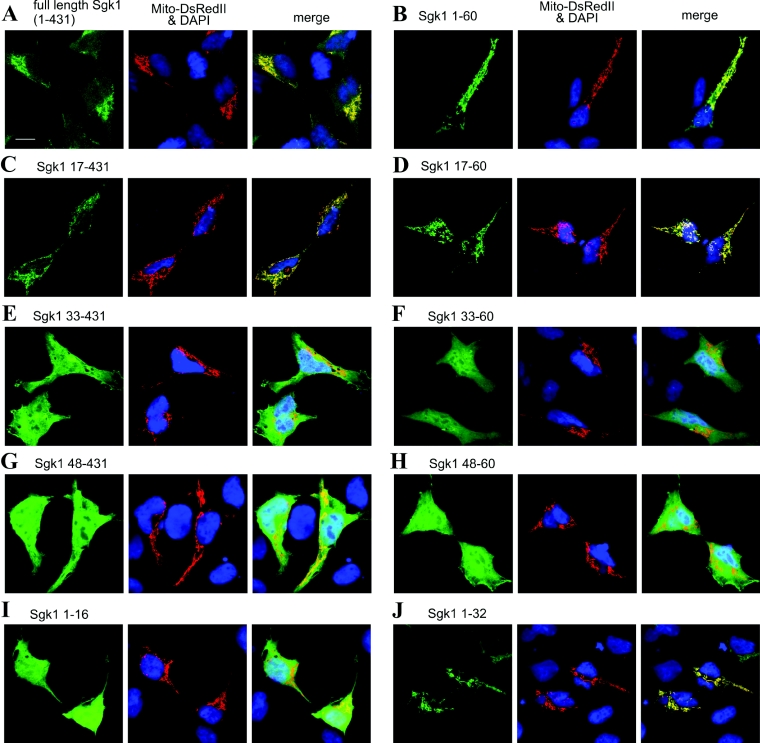
To examine the subcellular localization of Sgk1 experimentally, we transfected HEK-293 cells with a series of Sgk1 variants that harboured a C-terminally fused GFP for analysis with confocal laser scanning microscopy. We focused on the N-terminus of the kinase, because the first 60 amino acids were previously proposed to be responsible for plasma membrane association [24] and the mitochondrial targeting signal was predicted to reside in this region. When we transiently transfected HEK-293 cells with full-length Sgk1–GFP, we obtained a reticulo-tubular staining pattern that is characteristic for mitochondria in this cell line. To corroborate mitochondrial localization of Sgk1–GFP, we co-transfected mito-DsRedII as mitochondrial marker and stained nuclei with DAPI. Our results demonstrate that GFP- and DsRedII-fluorescence signals were overlapping (Figure 2A). To narrow down the region responsible for organelle targeting, we next generated and analysed N-terminal deletion variants starting with amino acids 17, 33 and 48 (Figures 2C, 2E and 2G). Our analysis shows that amino acids 1–16 are dispensable for protein targeting, as an Sgk1 variant containing amino acids 17–431 is still localized to mitochondria (Figure 2C). In contrast, Sgk1-(33–431) (Figure 2E) and Sgk1-(48–431) (Figure 2G) show a fluorescence pattern indistinguishable from that of GFP alone (results not shown). To investigate if the C-terminal regions of Sgk1 contribute to protein targeting, we fused the same deletion variants but ending at amino acid position 60 of Sgk1 to GFP. Confocal analyses of HEK-293 cells transfected with the variants Sgk1-(1–60)–GFP (Figure 2B), Sgk1-(17–60)–GFP (Figure 2D), Sgk1-(33–60)–GFP (Figure 2F) and Sgk1-(48–60)–GFP (Figure 2H) revealed no difference between these and the long variants, arguing against an influence of the C-terminal parts of Sgk1 on protein targeting. Additionally, we analysed the variants Sgk1-(1–16)–GFP and Sgk1-(1–32)–GFP. Sgk1-(1–16)–GFP was distributed throughout the cell (Figure 2I), whereas Sgk1-(1–32)–GFP localized to mitochondria (Figure 2J). Taken together, our results demonstrate that the region between amino acids 17 and 32 is necessary and sufficient for mitochondrial localization of Sgk1. These findings are in good accordance with the amino acid composition of Sgk1 in this region (Figure 1). Here, negatively charged residues are rare and arginine is over-represented, as is the case in most mitochondrial targeting sequences [31]. In a few cells, Sgk1–GFP signals were not restricted to mitochondria. However, these cells showed a strong overexpression of the transgene. In no instance were any Sgk1–GFP variants enriched in the nucleus even when cells were serum-stimulated or exposed to heat shock (results not shown). Moreover, none of the fusion protein variants was detectable at the plasma membrane.
Figure 2. Amino acids 17–33 target Sgk1 to mitochondria in HEK-293 cells.
HEK-293 cells were transfected with the mitochondrial marker cytochrome c oxidase subunit VIII coupled with DsRedII and either full-length Sgk1–GFP (A) or Sgk1–GFP deletion variants (B–J). Cells were grown on coverslips and formaldehyde-fixed, and nuclei were stained with DAPI. Left panels show GFP fluorescence signals (green), middle panels show DAPI (blue) and mitochondrial staining (red) and right panels show merged images. Yellow colour indicates co-localization.
Sgk1 is localized to mitochondria in vivo
To confirm a mitochondrial localization of Sgk1 in vivo, we performed subcellular fractionation experiments from mouse tissue. We first tested five commercial and non-commercial antibodies for specificity of Sgk1 binding in Western-blot analyses. Although all antibodies readily detected overexpressed Sgk1 in cell lines, we were not able to detect Sgk1 expression in any mouse tissue, including kidney and liver (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/399/bj3990069add.htm). We therefore increased levels of Sgk1 protein expression by administration of dexamethasone. Specificity of the Sgk1 signal was verified by direct comparison with corresponding tissue from knockout animals. We fractionated, livers from Sgk1 knockout mice and their wild-type littermates into cytosol, nuclei and plasma membranes, microsomes and mitochondria by differential centrifugation, 4 h after intraperitoneal application of dexamethasone, and processed them for immunoblot analyses. The purity of the fractions was examined with antibodies directed against GAPDH, histone H1, Na/K-ATPase, BiP/Grp78, cytochrome c and the α-subunit of the Na/K-ATPase (Figure 3). We obtained robust Sgk1 signals from the mitochondrial fraction and weak signals from the cytosolic and microsomal lanes (Figure 3). The signals originated from Sgk1, as they were absent from the knockout tissue fractions.
Figure 3. Sgk1 localizes to mitochondria in vivo.
The distribution of the Sgk1 protein in subcellular fractions from liver tissue was analysed 4 h after intraperitoneal administration of 100 μg/kg body weight dexamethasone. To be able to identify Sgk1 signals, Sgk1 knockouts (−/−) and their wild-type littermates were analysed in parallel. Signals were obtained predominantly from the mitochondrial fraction and only weakly from the cytosolic and microsomal fractions. Lower panels show immunoblots with antibodies against the indicated organelle markers to demonstrate purity of the fractions. Protein (20 μg/lane) was loaded for detection of Sgk1 and 10 μg of protein was loaded for detection of organelle markers. For the detection of Sgk1, we employed the antibody obtained from Sigma. PM, plasma membrane.
Sgk1 is localized to the cytosolic surface of the OMM (outer mitochondrial membrane)
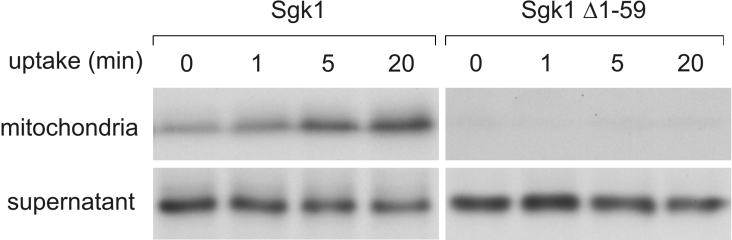
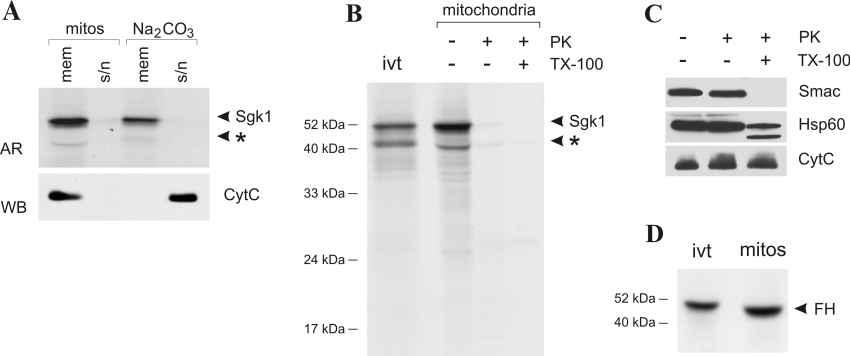
Mitochondrial proteins can be localized to the inner or OMM, the intermembrane space or the matrix. To overcome the low endogenous Sgk1 protein expression levels, we investigated the localization and orientation of in vitro-translated radiolabelled Sgk1 after import into intact mouse kidney mitochondria. Figure 4 shows that full-length Sgk1 is imported by purified mitochondria in a time-dependent manner. In contrast, the Δ1–59 variant, lacking the mitochondrial targeting sequence, completely remained in the supernatant after pelleting the organelles. When mitochondria were disrupted and separated into a soluble and a membrane fraction after protein import, Sgk1 was detectable only in the membrane fraction even after treatment with 100 mM sodium carbonate (Figure 5A, upper panel). Under these conditions, peripheral membrane proteins are released to the supernatant, as is the case for cytochrome c, which is attached to the inner mitochondrial membrane mainly by electrostatical force (Figure 5A, lower panel). These experiments indicate that Sgk1 is an integral mitochondrial membrane protein. Based on the observation that Sgk1 can be phosphorylated by cytosolic kinases, we speculated that Sgk1 might be localized to the OMM, with the kinase domain and the C-terminus facing the cytosol. To test this assumption, imported Sgk1 was subjected to protease protection assays. As a control for correct protein import, we used the in vitro-translated and radiolabelled mitochondrial matrix enzyme fumarate hydratase (Figure 5D). Proteinase K treatment of mitochondria led to complete digestion of Sgk1. To demonstrate that mitochondria remained intact during protease treatment, we performed Western-blot analyses with antibodies directed against the intermembrane-space protein Smac/DIABLO and the matrix protein Hsp60. Figure 5(C) shows that both proteins were protected unless Triton X-100 was added. Cytochrome c is extremely resistant to digestion by proteinase K [33] and served as loading control (Figure 5C, lowest panel). The degradation of Sgk1 suggests a largely cytoplasmic orientation at the OMM. From these studies, we conclude that Sgk1 is targeted and anchored via amino acids 16–33 to the OMM and faces the cytosol with the C-terminus containing the catalytic domain.
Figure 4. Sgk1 associates with purified mitochondria in a time-dependent manner.
Radiolabelled in vitro-translated full-length Sgk1 and Sgk1 Δ1–59 were exposed to highly purified mitochondria from mouse kidneys for the indicated times. Mitochondria were washed, lysed and separated by PAGE. Full-length Sgk1 is imported in a time-dependent manner, whereas the truncated variant is not. Lower panels show aliquots from the supernatants of the import reaction. Signals were visualized by fluorography.
Figure 5. Sgk1 associates with the OMM.
(A) Membrane association of imported radiolabelled Sgk1 was assessed by preparation of mitochondrial (mitos) soluble (s/n) and membrane (mem) fractions. Radiolabelled proteins were visualized by autoradiography (AR). The peripheral membrane protein cytochrome c (CytC) was released by Na2CO3 treatment and visualized by Western blot (WB, lower panel). Star symbol: this signal sometimes occurred after in vitro translation and is most likely a degradation product (also in B). (B) After import of in vitro-translated radiolabelled Sgk1, mitochondria were treated with 200 μg/ml proteinase K (PK) in the presence or absence of Triton X-100 or left untreated. Sgk1 was subject to complete digestion, indicating a localization to the cytoplasmic face of the OMM. ivt; an aliquot of the in vitro translation product. (C) Aliquots of the protease protection assay were subjected to Western-blot analyses with antibodies directed against the matrix protein Hsp60, and the intermembrane space proteins Smac/DIABLO and cytochrome c (CytC). CytC is resistant to proteinase K treatment under the conditions used, even when the membranes are permeabilized by Triton X-100 [33], and therefore served as loading control. Hsp60 and Smac/DIABLO remain intact in the absence of Triton X-100, demonstrating the integrity of the organelles. (D) The capability of the preparation to import proteins correctly is shown by the import of in vitro-translated fumarate hydratase (FH) in parallel. Imported fumarate hydratase migrates slightly faster than the in vitro translation product (ivt) because of the removal of the signal peptide by the processing peptidase after import into the matrix.
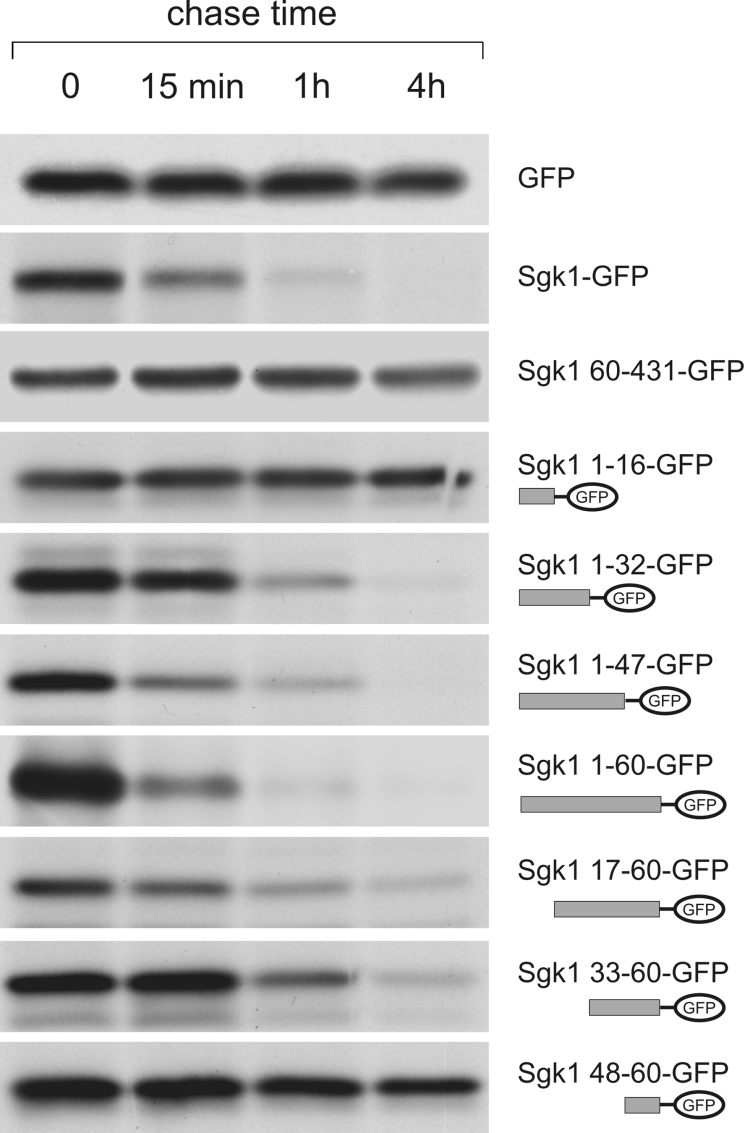
Half-life of Sgk1 is determined by amino acids 17–47
It has been documented previously [23,24] that Sgk1 protein levels are very low even after overexpression in cell lines because of rapid ubiquitin-mediated degradation by the proteasome [23,24]. The short life span is conferred by the first 60 amino acids of Sgk1, but is not due to ubiquitination of lysine residues within this region [24]. To more precisely determine the region responsible for proteasomal degradation, we performed pulse–chase experiments using Sgk1–GFP deletion variants. After a 10 min pulse with [35S]methionine/cysteine, transfected HEK-293 cells were immediately lysed or chased for 15 min, 60 min and 4 h. Sgk1 fusion proteins were recovered by immunoprecipitation with an anti-GFP serum and subjected to autoradiography (Figure 6). GFP alone does not show any apparent degradation within a 4 h chase, nor does Sgk1-(60–431), as previously described [24]. In contrast, full-length Sgk1 and Sgk1-(1–60) are rapidly degraded. Fine mapping of the regions responsible for the fast turnover showed that amino acids 17–32 and 33–47 are sufficient for targeting Sgk1 for proteasomal degradation (Figure 6). Therefore the short half-life of Sgk1 and mitochondrial targeting are conveyed by the identical peptide sequence.
Figure 6. Rapid turnover of Sgk1 is determined by amino acids 17–47.
Sgk1–GFP fusion protein variants or GFP alone were transfected into HEK-293 cells and subjected to pulse–chase analysis. After a pulse of 10 min, proteins were chased for 0 min, 15 min, 1 h or 4 h and the respective fusion proteins were recovered by immunoprecipitation with an anti-GFP antiserum. All variants containing amino acids 17–47 were rapidly degraded.
DISCUSSION
Here, we demonstrate that Sgk1 is anchored via the N-terminus to the OMM, that mitochondrial targeting is conferred by amino acids 17–32, that the C-terminus containing the kinase domain faces the cytosol, and that mitochondrial targeting is inseparably linked to the rapid degradation of Sgk1 by the proteasome. Using an in vitro import assay, confocal microscopy of Sgk1–GFP deletion variants in HEK-293 cells and biochemical fractionation of murine liver tissue, we provide three independent experimental lines of evidence that Sgk1 is predominantly localized to mitochondria and resides at the OMM. Mitochondrial localization is mediated by amino acids 17–32, which resemble a bona fide mitochondrial targeting signal. However, this region is not a classical mitochondrial presequence, since Sgk1 is not imported into the mitochondrial matrix and is not cleaved by the mitochondrial processing peptidase. Several OMM proteins with cytoplasmic orientation are targeted by a non-cleavable N-terminal signal anchor consisting of a hydrophobic transmembrane domain and a net positive charge at the corresponding C-terminal flanking segment [34,35]. Similarly, the N-terminal region of Sgk1 is hydrophobic and is followed by three positively charged residues (Figure 1). Although the carbonate extraction data support that Sgk1 is an integral component of the OMM, a stretch of ten hydrophobic amino acids as indicated by the Kyte-Doolittle plot is generally considered to be too short for a membrane-spanning segment. However, even if not integral, the experiments indicate that this segment at least confers an unusually strong peripheral membrane association. The cytoplasmic orientation of Sgk1 meets the requirement to be accessible for activation by cytosolic kinases [22,23].
Our results provide strong evidence that Sgk1 is primarily localized to the OMM, but do not exclude other sites of localization. Previous reports describe that Sgk1 is found in association with membranes [24,28]. Our results indicated that these membranes might have been of mitochondrial origin. Moreover, our results support the observation of Brickley et al. [24] that a variant of Sgk1 lacking the first 60 residues does not exhibit membrane association, is not ubiquitinated, and consequently, has a prolonged half-life. Finally, the N-terminus of Sgk1 does not contain a classical targeting signal, and targeting of the kinase to different membrane systems and compartments might depend on the specific cell lines or culture conditions used. For example, the Bcl-2 (B-cell lymphocytic-leukaemia proto-oncogene 2) protein can associate with both the OMM and microsomal membranes via a C-terminal hydrophobic domain [36].
At the plasma membrane, Sgk1 was proposed to interact with and phosphorylate the E3 ubiquitin ligase Nedd4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) and thereby control plasma membrane insertion of ENaC sodium transporters [5]. However, using surface plasmon resonance analysis, a direct interaction of Sgk1 with Nedd4-2 could not be confirmed [37]. Moreover, consistent with our results, a recent report described activation of ENaC by Sgk1 independent of Nedd4-2 [6].
Using confocal fluorescence microscopy, we also never observed significant localization of Sgk1 to the nucleus, not even after serum stimulation or heat shock, two stimuli that were reported to lead to nuclear translocation of Sgk1 [25–27]. Consistent with this, we demonstrate that endogenous Sgk1 is not present at detectable levels in the nuclear fraction of mouse liver tissue. However, we would like to point out that in these experiments, pretreatment of mice with steroid hormones was necessary to induce detectable levels of Sgk1 protein expression. In mammary epithelial and tumour cells, steroid hormone treatment was shown to lead to a translocation of Sgk1 from the nucleus to a perinuclear compartment that contains mitochondria [25,27]. Moreover, different cell lines and tissues are likely to differ in their capacity to import Sgk1 into the nucleus.
We here show that amino acids 17–32 are essential for organelle targeting and determine the half-life of Sgk1 protein. This makes it likely that degradation of Sgk1 is facilitated rapidly upon arrival at its mitochondrial destination. A corollary of this finding is that a large number of studies using an active variant of Sgk1, lacking the N-terminal 60 residues, need to be critically revisited. This long-lived variant, which has often been used in kinase assays for the detection or validation of kinase targets, is not directed to its physiological subcellular localization and is therefore prone to interact with and phosphorylate non-physiological protein targets. Moreover, our results indicate that the use of Sgk1 variants with N-terminal tags should be avoided, because these are likely to alter the subcellular localization. In this context, it is interesting to note that Helms et al. [38] reported that a Δ1–60 variant showed less biological activity than full-length Sgk1 on ENaC function.
The influence of Sgk1 on a broad spectrum of cellular functions and its activation by several different stimuli indicate that Sgk1 plays a central role in cell signalling. Our results provide evidence that Sgk1 acts at the OMM. This finding may lay the basis for a further analysis of the function of Sgk1. The low protein expression levels of Sgk1, however, render a thorough biochemical examination of this molecule difficult. Detailed analysis of the molecules involved in the degradation of Sgk1 will clearly provide more insight into the biology of this kinase.
Online data
Acknowledgments
We thank Burkhard Wiesner from the confocal imaging unit of the Institute for Molecular Pharmacology (Berlin, Germany) for help with laser scanning microscopy, Ralf Schülein for providing anti-GFP rabbit serum, and Cecilia Canessa (Yale University School of Medicine, New Haven, CT, U.S.A.) and James Murray (University of Dundee, Dundee, Scotland, U.K.) for providing anti-Sgk1 antibodies. We also thank Volker Haucke (Free University of Berlin, Berlin, Germany) for discussions.
References
- 1.Webster M. K., Goya L., Ge Y., Maiyar A. C., Firestone G. L. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol. Cell. Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakoda H., Gotoh Y., Katagiri H., Kurokawa M., Ono H., Onishi Y., Anai M., Ogihara T., Fujishiro M., Fukushima Y., et al. Differing roles of Akt and serum- and glucocorticoid-regulated kinase in glucose metabolism, DNA synthesis, and oncogenic activity. J. Biol. Chem. 2003;278:25802–25807. doi: 10.1074/jbc.M301127200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B. H., Tang E. D., Zhu T., Greenberg M. E., Vojtek A. B., Guan K. L. Serum- and glucocorticoid-inducible kinase SGK phosphorylates and negatively regulates B-Raf. J. Biol. Chem. 2001;276:31620–31626. doi: 10.1074/jbc.M102808200. [DOI] [PubMed] [Google Scholar]
- 4.Brunet A., Park J., Tran H., Hu L. S., Hemmings B. A., Greenberg M. E. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol. Cell. Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Munster C., Chraibi A., Pratt J. H., Horisberger, et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diakov A., Korbmacher C. A novel pathway of epithelial sodium channel activation involves a serum- and glucocorticoid-inducible kinase consensus motif in the C terminus of the channel's α-subunit. J. Biol. Chem. 2004;279:38134–38142. doi: 10.1074/jbc.M403260200. [DOI] [PubMed] [Google Scholar]
- 7.Boehmer C., Wilhelm V., Palmada M., Wallisch S., Henke G., Brinkmeier H., Cohen P., Pieske B., Lang F. Serum and glucocorticoid inducible kinases in the regulation of the cardiac sodium channel SCN5A. Cardiovasc. Res. 2003;57:1079–1084. doi: 10.1016/s0008-6363(02)00837-4. [DOI] [PubMed] [Google Scholar]
- 8.Palmada M., Dieter M., Boehmer C., Waldegger S., Lang F. Serum and glucocorticoid inducible kinases functionally regulate ClC-2 channels. Biochem. Biophys. Res. Commun. 2004;321:1001–1006. doi: 10.1016/j.bbrc.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 9.Embark H., Setiawan I., Poppendieck S., Van De Graaf S., Boehmer C., Palmada M., Wieder T., Gerstberger R., Cohen P., Yun C., et al. Regulation of the epithelial Ca2+ channel TRPV5 by the NHE regulating factor NHERF2 and the serum and glucocorticoid inducible kinase isoforms SGK1 and SGK3 expressed in Xenopus oocytes. Cell. Physiol. Biochem. 2004;14:203–212. doi: 10.1159/000080329. [DOI] [PubMed] [Google Scholar]
- 10.Dieter M., Palmada M., Rajamanickam J., Aydin A., Busjahn A., Boehmer C., Luft F. C., Lang F. Regulation of glucose transporter SGLT1 by ubiquitin ligase Nedd4-2 and kinases SGK1, SGK3, and PKB. Obes. Res. 2004;12:862–870. doi: 10.1038/oby.2004.104. [DOI] [PubMed] [Google Scholar]
- 11.Henke G., Maier G., Wallisch S., Boehmer C., Lang F. Regulation of the voltage gated K+ channel Kv1.3 by the ubiquitin ligase Nedd4-2 and the serum and glucocorticoid inducible kinase SGK1. J. Cell. Physiol. 2004;199:194–199. doi: 10.1002/jcp.10430. [DOI] [PubMed] [Google Scholar]
- 12.Yun C. C., Palmada M., Embark H. M., Fedorenko O., Feng Y., Henke G., Setiawan I., Boehmer C., Weinman E. J., Sandrasagra S., et al. The serum and glucocorticoid-inducible kinase SGK1 and the Na+/H+ exchange regulating factor NHERF2 synergize to stimulate the renal outer medullary K+ channel ROMK1. J. Am. Soc. Nephrol. 2002;13:2823–2830. doi: 10.1097/01.asn.0000035085.54451.81. [DOI] [PubMed] [Google Scholar]
- 13.Embark H. M., Bohmer C., Vallon V., Luft F., Lang F. Regulation of KCNE1-dependent K+ current by the serum and glucocorticoid-inducible kinase (SGK) isoforms. Pflügers Arch. 2003;445:601–606. doi: 10.1007/s00424-002-0982-y. [DOI] [PubMed] [Google Scholar]
- 14.Boehmer C., Henke G., Schniepp R., Palmada M., Rothstein J. D., Broer S., Lang F. Regulation of the glutamate transporter EAAT1 by the ubiquitin ligase Nedd4-2 and the serum and glucocorticoid-inducible kinase isoforms SGK1/3 and protein kinase B. J. Neurochem. 2003;86:1181–1188. doi: 10.1046/j.1471-4159.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 15.Wulff P., Vallon V., Huang D. Y., Volkl H., Yu F., Richter K., Jansen M., Schlunz M., Klingel K., Loffing J., et al. Impaired renal Na+ retention in the sgk1-knockout mouse. J. Clin. Invest. 2002;110:1263–1268. doi: 10.1172/JCI15696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster M. K., Goya L., Firestone G. L. Immediate-early transcriptional regulation and rapid mRNA turnover of a putative serine/threonine protein kinase. J. Biol. Chem. 1993;268:11482–11485. [PubMed] [Google Scholar]
- 17.Waldegger S., Barth P., Raber G., Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4440–4445. doi: 10.1073/pnas.94.9.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naray-Fejes-Toth A., Canessa C., Cleaveland E. S., Aldrich G., Fejes-Toth G. Sgk is an aldosterone-induced kinase in the renal collecting duct: effects on epithelial Na+ channels. J. Biol. Chem. 1999;274:16973–16978. doi: 10.1074/jbc.274.24.16973. [DOI] [PubMed] [Google Scholar]
- 19.Mizuno H., Nishida E. The ERK MAP kinase pathway mediates induction of SGK (serum- and glucocorticoid-inducible kinase) by growth factors. Genes Cells. 2001;6:261–268. doi: 10.1046/j.1365-2443.2001.00418.x. [DOI] [PubMed] [Google Scholar]
- 20.Alliston T. N., Maiyar A. C., Buse P., Firestone G. L., Richards J. S. Follicle stimulating hormone-regulated expression of serum/glucocorticoid-inducible kinase in rat ovarian granulosa cells: a functional role for the Sp1 family in promoter activity. Mol. Endocrinol. 1997;11:1934–1949. doi: 10.1210/mend.11.13.0033. [DOI] [PubMed] [Google Scholar]
- 21.Waldegger S., Klingel K., Barth P., Sauter M., Rfer M. L., Kandolf R., Lang F. h-sgk serine-threonine protein kinase gene as transcriptional target of transforming growth factor β in human intestine. Gastroenterology. 1999;116:1081–1088. doi: 10.1016/s0016-5085(99)70011-9. [DOI] [PubMed] [Google Scholar]
- 22.Park J., Leong M. L., Buse P., Maiyar A. C., Firestone G. L., Hemmings B. A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T., Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 24.Brickley D. R., Mikosz C. A., Hagan C. R., Conzen S. D. Ubiquitin modification of serum and glucocorticoid-induced protein kinase-1 (SGK-1) J. Biol. Chem. 2002;277:43064–43070. doi: 10.1074/jbc.M207604200. [DOI] [PubMed] [Google Scholar]
- 25.Buse P., Tran S. H., Luther E., Phu P. T., Aponte G. W., Firestone G. L. Cell cycle and hormonal control of nuclear-cytoplasmic localization of the serum- and glucocorticoid-inducible protein kinase, Sgk, in mammary tumor cells: a novel convergence point of anti-proliferative and proliferative cell signaling pathways. J. Biol. Chem. 1999;274:7253–7263. doi: 10.1074/jbc.274.11.7253. [DOI] [PubMed] [Google Scholar]
- 26.Maiyar A. C., Leong M. L., Firestone G. L. Importin-α mediates the regulated nuclear targeting of serum- and glucocorticoid-inducible protein kinase (Sgk) by recognition of a nuclear localization signal in the kinase central domain. Mol. Biol. Cell. 2003;14:1221–1239. doi: 10.1091/mbc.E02-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leong M. L. L., Maiyar A. C., Kim B., O'Keeffe B. A., Firestone G. L. Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. J. Biol. Chem. 2003;278:5871–5882. doi: 10.1074/jbc.M211649200. [DOI] [PubMed] [Google Scholar]
- 28.Alvarez de la Rosa D., Coric T., Todorovic N., Shao D., Wang T., Canessa C. M. Distribution and regulation of expression of serum- and glucocorticoid-induced kinase-1 in the rat kidney. J. Physiol. 2003;551:455–466. doi: 10.1113/jphysiol.2003.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray J. T., Campbell D. G., Morrice N., Auld G. C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 31.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 32.Claros M. G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 33.Uren R. T., Dewson G., Bonzon C., Lithgow T., Newmeyer D. D., Kluck R. M. Mitochondrial release of pro-apoptotic proteins: electrostatic interactions can hold cytochrome c but not Smac/DIABLO to mitochondrial membranes. J. Biol. Chem. 2005;280:2266–2274. doi: 10.1074/jbc.M411106200. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H., Maeda M., Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 2002;115:1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- 35.Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janiak F., Leber B., Andrews D. W. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 37.Henry P. C., Kanelis V., O'Brien M. C., Kim B., Gautschi I., Forman-Kay J., Schild L., Rotin D. Affinity and specificity of interactions between Nedd4 isoforms and the epithelial Na+ channel. J. Biol. Chem. 2003;278:20019–20028. doi: 10.1074/jbc.M211153200. [DOI] [PubMed] [Google Scholar]
- 38.Helms M. N., Fejes-Toth G., Naray-Fejes-Toth A. Hormone-regulated transepithelial Na+ transport in mammalian CCD cells requires SGK1 expression. Am. J. Physiol. Renal Physiol. 2002;284:F480–F487. doi: 10.1152/ajprenal.00299.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.