Abstract
Background
This study describes a strategy to select and isolate spontaneous riboflavin-overproducing strains of Lactobacillus (Lb.) plantarum, Leuconostoc (Lc.) mesenteroides and Propionibacterium (P.) freudenreichii.
Results
The toxic riboflavin analogue roseoflavin was used to isolate natural riboflavin-overproducing variants of the food grade micro-organisms Lb. plantarum, Lc. mesenteroides and P. freudenreichii strains. The method was successfully employed for strains of all three species. The mutation(s) responsible for the observed overproduction of riboflavin were identified for isolates of two species.
Conclusion
Selection for spontaneous roseoflavin-resistant mutants was found to be a reliable method to obtain natural riboflavin-overproducing strains of a number of species commonly used in the food industry. This study presents a convenient method for deriving riboflavin-overproducing strains of bacterial starter cultures, which are currently used in the food industry, by a non-recombinant methodology. Use of such starter strains can be exploited to increase the vitamin content in certain food products.
Background
The term "nutraceutical" was first coined in 1989 as a contraction of the words "nutrition" and "pharmaceutical", and refers to a food compound that not only supplements the diet but also aids in the prevention and/or treatment of disease and/or disorder [1]. Similarly, functional foods contain at least one component, whether it be a nutrient or not, that affects a target function of the organism in a specific, positive way, thereby generating a physiological effect beyond its nutritional value [2]. Functional food ingredients include probiotics, prebiotics, vitamins and minerals, and can be found in such diverse products as fermented dairy products, sports drinks and chewing gum [3,4]. Interest in and acceptance of functional foods is gaining impetus, for a number of reasons that include changing consumer demands and social attitudes, scientific evidence of the health benefits of particular ingredients and commercially driven interests to add value to existing products. Consumer awareness on nutrition raises the demand for healthy food options, ideally delivered in a convenient form [5]. Functional foods have a significant and growing global market with recent estimates indicating up to a $50 billion annual share [4].
Since Metchnikoff first theorised that fermented milk products provided health benefits, including longer life expectancy, these products have been viewed as "healthy" by consumers [6]. In many modern societies fermented dairy products make up a substantial proportion of the total daily food consumption. Lactic acid bacteria (LAB) are an industrially important group of micro-organisms used all over the world for centuries in a large variety of food fermentations, such as those of meat and vegetables, but undoubtedly with the major application in the dairy industry. LAB have been shown to be ideal cell factories because the biosynthetic capacity and metabolic versatility of these bacteria is generally quite limited, their physiology is relatively simple, while their energy metabolism and biosynthesis processes are almost completely separated [7]. It has therefore been possible to exploit LAB for the production of many nutraceuticals [8]. The dairy propionibacteria, especially P. freudenreichii ssp. shermanii, are the main ripening flora in Swiss-type cheeses, where they play a critical role in the development of the characteristic flavour and "eye" production [9]. Propionibacteria are also used for the production of vitamin B12 [10] and there is increasing interest in their potential use as probiotics [11]. Considering the current widespread use of LAB and propionibacteria in the food industry, coupled with consumer demand for healthier foods, the potential to use these food grade microorganisms as "vitamin factories" was investigated.
Riboflavin is a water-soluble vitamin produced by plants and many micro-organisms. However, this biosynthetic capability is lacking in higher animals and they must therefore obtain this essential nutrient from their diet. Riboflavin is the precursor of the enzyme cofactors FMN and FAD, which are vital in many of the body's enzymatic functions for the transfer of electrons in oxidation-reduction reactions. Riboflavin deficiency is most commonly seen in developing countries [12], among the elderly [13], and in chronic alcoholics [14]. Clinical symptoms of riboflavin deficiency are rarely seen in developed countries but the subclinical stage of deficiency, characterised by a change in biochemical indices, is seen in a significant portion of the population of these nations as exemplified by Ireland [15]. Riboflavin deficiency mainly manifests itself clinically in the mucocutaneous surfaces of the mouth, through the occurrence of cracks at the corners, and inflammation of the lips and tongue [16], but deficiency is also associated with vision deterioration and growth failure. In recent years the vitamin has been found to be effective in the treatment of migraine [17], malaria [18] and Parkinson's disease [19]. Riboflavin is commonly obtained in the diet from meat, eggs, fortified cereals and green leafy vegetables, in addition to dairy products, which contribute most significantly to riboflavin intake [20].
Riboflavin has been traditionally synthesised for food and feed fortification by chemical means but in more recent years biotechnological processes employing various bacteria, yeast and fungi have been found to be commercially competitive and are replacing chemical synthesis [21]. One of these biotechnological processes employs Bacillus (B.) subtilis as the riboflavin cell factory and much work has been carried out in characterising the vitamin's biosynthetic pathway in this organism (For a review see [22]). For B. subtilis and Lactococcus (L.) lactis it has been shown that mutants that are isolated on the basis of their resistance to the toxic riboflavin analogue roseoflavin also exhibit a riboflavin-overproduction phenotype [23,24]. Recently, it was demonstrated that fermented dairy products produced either with a roseoflavin-resistant strain of P. freudenreichii or L. lactis, was able to improve growth and riboflavin status of riboflavin-depleted animals [25,26].
The current study reports on the isolation of roseoflavin-resistant mutants of various strains used in the food industry and analysis of their resulting riboflavin-overproducing phenotype. In the case of Lb. plantarum and Lc. mesenteroides the mutations responsible for riboflavin overproduction were identified and the possible effects of these mutations on transcription of the rib operon are discussed. In the case of P. freudenreichii the riboflavin-overproducing strains were examined in comparison to a control strain in a yoghurt production model.
Results
Isolation of roseoflavin resistant Leuconostoc mesenteroides
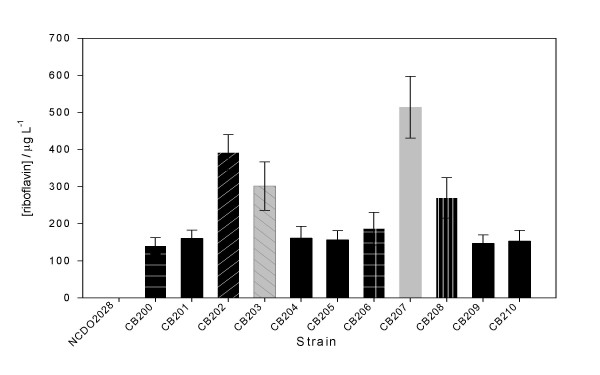
It has previously been shown in B. subtilis and L. lactis that spontaneous resistance to the toxic riboflavin analogue, roseoflavin, frequently coincides with a riboflavin-overproducing phenotype [24,27]. In order to determine whether this method was also applicable to Lc. mesenteroides, a wildtype strain of this species was plated at mid exponential growth phase on CDM containing 100 mg/L roseoflavin. In this way a total eleven mutants were isolated. These isolates were grown in CDM and as the cells entered the stationary phase the cell free supernatant was analysed for riboflavin content. All eleven roseoflavin-resistant mutants were found to be riboflavin overproducers (Fig. 1), leading to accumulation of the vitamin into the medium although at clearly different levels ranging from 120 to 500 μg/L.
Figure 1.
Riboflavin produced by roseoflavin-resistant Lc. mesenteroides NCDO2028 mutants. The riboflavin concentration was determined in the cell free supernatant as the cells entered stationary phase. The various patterns indicate different mutations.
Identification of mutations resulting in riboflavin overproduction
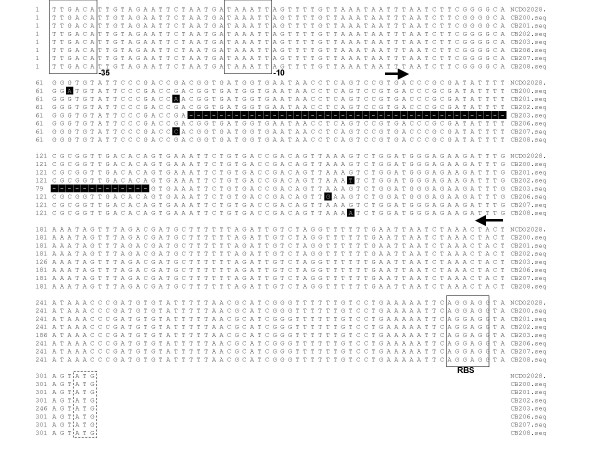
B. subtilis roseoflavin-resistant mutants have been shown to carry mutations either in the regulatory region of the rib operon or in ribC, encoding the flavokinase/FAD synthetase responsible for the conversion of riboflavin to its active derivatives FMN and FAD [28-30]. Roseoflavin resistance in L. lactis seems to be due to mutations that reside exclusively in the regulatory region upstream of the rib operon [24]. It has been found that this region acts as a riboswitch causing premature transcription termination of the rib operon in the presence of FMN [31]. The RFN element is a highly conserved domain found within this 5'-untranslated region consisting of 5 stem-loop structures [32]. A predicted RFN element was identified upstream of the Lc. mesenteroides rib operon using RFAM (indicated in Fig. 2). To investigate whether mutations were present in the homologous DNA regions in the roseoflavin-resistant Lc. mesenteroides mutants chromosomal DNA was isolated from each mutant after which the relevant regions were amplified by PCR and subjected to sequence analysis. No mutations were identified in the ribC gene, whereas in each of the isolated mutants the rib leader region [Genbank: DQ645591] was shown to contain a mutation (Fig. 2). In only one case (CB203) the mutation was represented by a deletion, while the other ten strains were shown to possess point mutations at one of four different locations within the rib leader region. At two of these locations different mutations occurred in different mutants (For example, the rib leader region of mutant CB201 contains a G to A substitution at position 77, while strain CB207 contains a G to C substitution at the corresponding position).
Figure 2.
Alignment of the rib operon regulatory region of Lc. mesenteroides NCDO2028 and seven different mutants isolated. The mutations are shaded. The predicted -10 and -35 recognition sequences and ribosomal binding site are boxed. The ribG start codon is boxed with dashed lines. The RFN element predicted by RFAM is indicated by the arrows below the sequence.
Isolation of riboflavin-overproducing Lb. plantarum and identification of mutations resulting in riboflavin overproduction
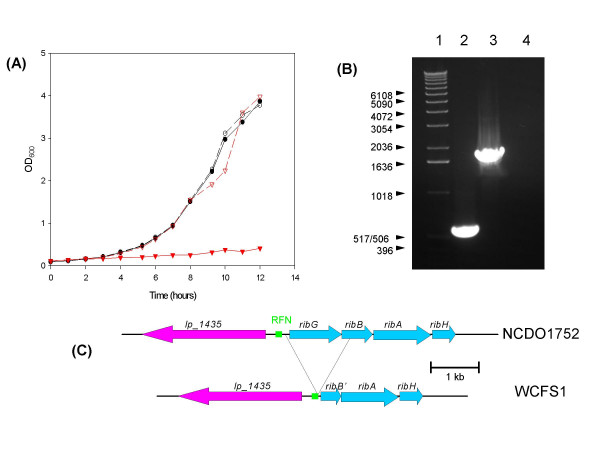
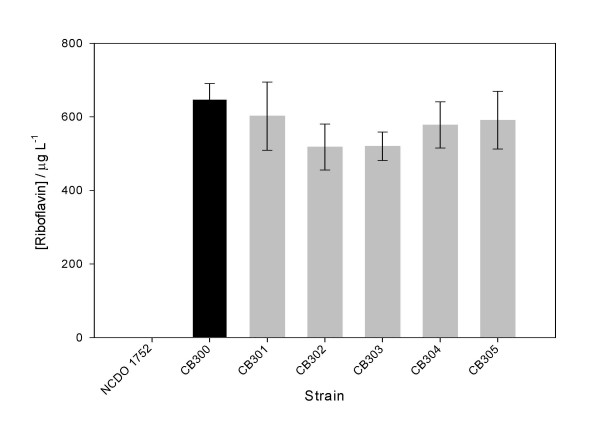
Bioinformatic analysis had previously shown that the sequenced strain of Lb. plantarum WCFS1 contains an incomplete rib operon, in which the entire ribG and part of ribB are absent from the genome [24,33]. As expected, this strain is unable to grow in the absence of riboflavin (Fig. 3(A)), in contrast to Lb. plantarum NCDO1752, which is capable of growth in the absence of the vitamin, thus indicating that this strain encodes a complete and functional riboflavin biosynthetic capability. The latter strain was exposed to 100 mg/L roseoflavin resulting in the isolation of six resistant mutants, which were also shown to be riboflavin overproducers (Fig. 4). In order to analyse the possible mutations in these mutants it was necessary to obtain the sequence of the region upstream of ribG, which is not present in the sequenced strain. PCR and sequence analysis of this region from NCDO1752 suggests that the WCFS1 genome harbours a deletion of approximately 1350 base pairs corresponding to the entire ribG gene, and part of ribB (Figs. 3(B) and 3(C)). The sequence of the regulatory region in NCDO1752 was determined [Genbank: DQ645592], allowing the identification of the putative RFN element using RFAM. This region was then amplified by PCR from the riboflavin-overproducing mutants and their sequence was determined. Only two types of mutations were found (Fig. 5). One strain (CB300) contained a G to T substitution, while the remaining strains (CB301 to 305) contained a 9 base pair deletion within the RFN element.
Figure 3.
(A) Growth of Lb. plantarum NCDO 1752 (black lines with circles) and Lb. plantarum WCFS1 (red lines with triangles) in CDM (solid lines, closed symbols) or CDM + 5 μM riboflavin (dashed lines, open symbols) (B) Amplification of the region from lp_1345 to the C terminal of ribB from Lb. plantarum strains chromosomal DNA. Lane 1: Molecular weight marker; Lane 2: Lb. plantarum WCFS1; Lane 3: Lb. plantarum NCDO 1752; Lane 4: negative control (H2O). The size of the marker bands are indicated to the left of the picture. (C) Schematic diagram of the deleted region in Lb. plantarum WCFS1 in comparison to Lb. plantarum NCDO 1752.
Figure 4.
Riboflavin produced by roseoflavin-resistant Lb. plantarum NCDO1752 mutants. The riboflavin concentration was determined in the cell free supernatant after the cells entered stationary phase. The two shades represent the two mutants isolated.
Figure 5.
Alignment of the rib operon regulatory region of Lb. plantarum NCDO1752 and the two mutants isolated. The mutations are shaded. The predicted -10 and -35 recognition sequences and ribosomal binding site are boxed. The ribG start codon is boxed with dashed lines. The RFN element predicted by RFAM is indicated by the arrows below the sequence.
Isolation of roseoflavin-resistant Propionibacterium freudenreichii
Two different strains of P. freudenreichii were selected in order to investigate whether the roseoflavin-resistance strategy for the isolation of riboflavin overproducers could also be applied to a representative species of a high-GC gram-positive bacterium. The method was indeed successful as shown by the isolation of twelve riboflavin over-producing mutants of this species, each varying in the amount of riboflavin produced (Fig. 6). Since the genome sequence of P. freudenreichii is not publicly available and the amplification of the rib leader region of this species using degenerate primers based on the rib leader sequence of P. acnes was unsuccessful, it was not possible to determine the nature of the mutation(s) resulting in the altered phenotype.
Figure 6.
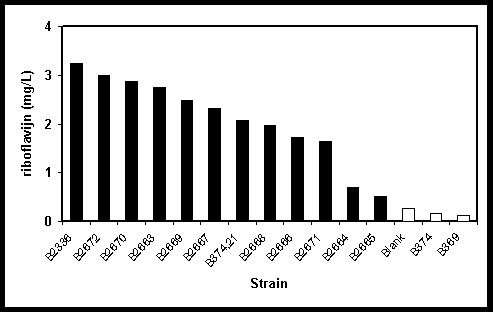
Riboflavin produced by roseoflavin-resistant P. freudenreichii B374 mutants. The riboflavin concentration was determined in the cell free supernatant after the cells entered stationary phase. The solid bars represent the levels produced by the roseoflavin-resistant strains, while the open bars represent the wildtype strain concentrations.
Stability study of riboflavin overproducing phenotype in P. freudenreichii
In order for any of the strains isolated in this study to have potential industrial usefulness it is necessary that the riboflavin-overproducing phenotype is stably maintained. To determine if this is the case, P. freudenreichii NIZO B374 and two of its riboflavin-overproducing derivatives were subcultured for sixty generations in the absence of the riboflavin analogue roseoflavin and extracellular riboflavin was determined throughout. It was found that the riboflavin-overproducing phenotype is stably maintained in the absence of the selection pressure for at least 60 generations (Fig. 7).
Figure 7.
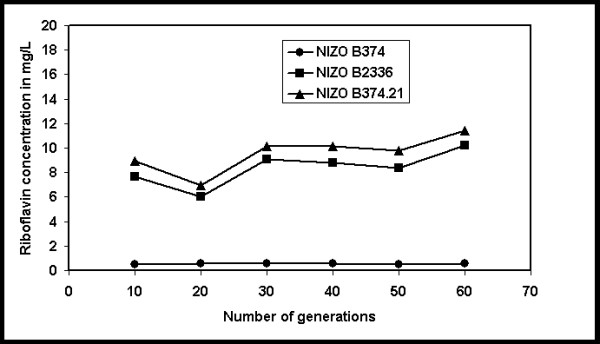
Riboflavin produced by P. freudenreichii B374 and its riboflavin overproducing derivatives B2336 and B374.21 after successive generations in roseoflavin free broth.
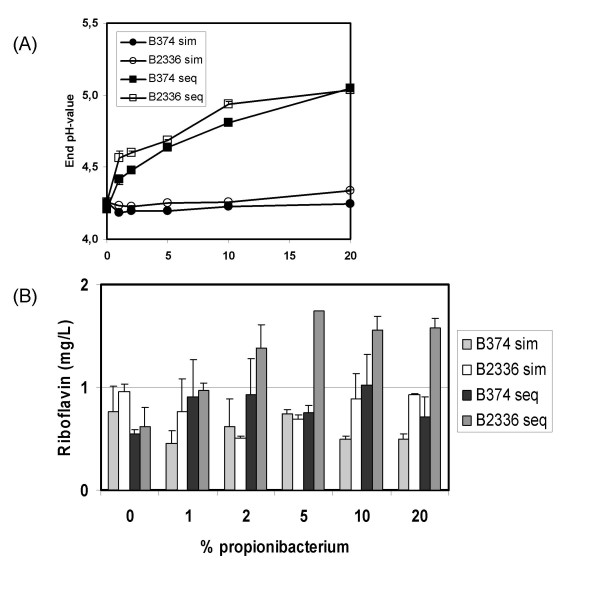
Yoghurt study using P. freudenreichii B374 and its riboflavin-overproducing derivative B2336
In order to determine if the cultures isolated in this study could have industrial potential, a yoghurt production trial was set up, which compared the use of P. freudenreichii NIZO B374 and its riboflavin-overproducing derivative P. freudenreichii NIZO B2336. To optimize the effect of propionibacteria in a yoghurt fermentation, we compared two different fermentation processes. The effect of co-inoculation of the propionibacteria with the yoghurt starter culture was compared with pre-fermentation by the propionibacteria prior to the addition of the yoghurt starter culture. The viable counts of propionibacteria, streptococci and lactobacilli were unaffected by the presence of the wildtype or riboflavin-overproducing propionibacterial strain. Also sequential inoculation or co-inoculation was did not affect viable counts of the different strains in the starter culture (data not shown). The final pH after fermentation is an important parameter in the yoghurt production process. Figure 8(A) shows the final pH value of the yoghurt at various inoculum levels using both the wildtype and riboflavin-overproducing strains in both sequential and simultaneous inoculation. Both strains have the same effect on acidification of the yoghurt. However, a marked difference is visible between the types of inoculation. Preculturing with propionibacteria counteracts acidification, while no effect was observed when the propionibacteria were added simultaneously with the starter culture. Figure 8(B) shows the final riboflavin concentration of the yoghurt at various inoculum levels of P. freudenreichii NIZO B374 and its riboflavin-overproducing derivative P. freudenreichii NIZO B2336. Co-inoculation of the propionibacteria with the starter culture has little impact on final riboflavin levels. However, addition of P. freudenreichii NIZO B2336 in the sequential fermentation process shows a doubling of the concentration of riboflavin in the fermented end-product.
Figure 8.
(A) Final pH of yoghurt with various inoculum levels of P. freudenreichii B374 or B2336 either added 24 hours beforehand or simultaneously with the yoghurt starter culture. (B) Final riboflavin levels in yoghurt with various inoculum levels of P. freudenreichii B374 or B2336 either added sequentially or simultaneously with the yoghurt starter culture.
Discussion
The work presented in this study demonstrates the broad applicability of the strategy to isolate strains with enhanced vitamin B2 production characteristics using sub-inhibitory levels of roseoflavin. The resultant strains are made through a non-recombinant method and may therefore be readily applicable in an industrial setting. Various biotechnological processes have been developed for industrial scale riboflavin biosynthesis using the ascomycetes Ashbya gossypii, different yeasts and the bacterium B. subtilis [21]. The latter organism has been successfully employed at a commercial scale to produce riboflavin for feed and food fortification purposes. However, since this production procedure involved a genetically recombinant organism regulatory approval was required where substantial equivalence of the product to non-recombinant riboflavin had to be established, while no DNA from the production strain was allowed to be present in the final product [34]. The purpose of the present study was not to develop strains which would challenge these already available processes, but rather to look at the potential to isolate strains with an improved riboflavin production phenotype that could replace the riboflavin-consuming parent strains in traditional food fermentation processes thus improving the bioavailability of riboflavin.
The Lc. mesenteroides strain, NCDO 2028 used in this study was originally isolated from beetroot silage, and was chosen as a representative of a species, which has many varied industrial applications in the dairy industry and various plant and vegetable fermentation processes [35-37]. Lb. plantarum, used in a large number of fermentation processes was chosen as another candidate. This organism is found in many diverse environments, owing to its metabolic flexibility [38]. In general, descriptions of the strain's nutritional requirements indicate that riboflavin is not required for growth, indicative of a functional riboflavin biosynthetic pathway [39]. Lb. plantarum NCDO 1752, a pickled cabbage isolate, is capable of growth in the absence of the vitamin illustrating its ability to synthesise the vitamin. However, in the course of this work it was revealed that the sequenced strain of Lb. plantarum, WCFS1 does not contain a functional rib operon and consequently is unable to grow in the absence of exogenous riboflavin [40]. Two strains of P. freudenreichii (both isolated from cheese), one subspecies freudenreichii (NIZO B374) and one subspecies shermanii (NIZO B369) were selected from the NIZO culture collection and the resistance strategy was applied also to this strain. All isolated mutants from these two subspecies were shown to be riboflavin overproducers. The successful application of the roseoflavin resistance strategy in three diverse bacterial species shows that it is a readily employable system in an industrial setting to isolate starter strains that produce and secrete vitamin B2. This is particularly appealing when one considers that some yoghurt cultures have been shown to actually decrease the concentration of riboflavin in some products due to their consumption of the vitamin [41]. In a simultaneously performed collaborative study it was demonstrated that a fermented dairy product produced with P. freudenreichii B2336 was able to improve growth and riboflavin status of riboflavin-depleted animals [25]. This proves that the riboflavin produced in yoghurt fermentation is available as a nutrient.
Although not all spontaneous or induced mutations causing riboflavin overproduction in B. subtilis have been analysed in detail, the characterised roseoflavin-induced mutations in this organism have been located either in ribC, the bifunctional flavokinase/FAD synthetase, which converts riboflavin to FMN and FAD [28,30], or in the regulatory region upstream of the rib operon [29]. This knowledge facilitated the identification of the mutations present in the roseoflavin-resistant LAB strains. No mutations were identified in the ribC homologue of any of the roseoflavin-resistant strains, but instead all characterised mutants were found to contain mutations in the regulatory leader region upstream of the rib operon. In both species different point mutations were identified, which were shown to differentially affect the level of riboflavin overproduction. Additionally, for both LAB species spontaneous roseoflavin-resistant mutants were isolated containing various deletions in the regulatory region of the rib operon. It is expected that such mutations are extremely stable. Furthermore, in P. freudenreichii it was shown that after sixty generations in non-selective media the riboflavin-overproducing phenotype does not revert to the wildtype phenotype. In analogy to what is known for the regulation of riboflavin biosynthesis in B. subtilis and L. lactis [24,31,42] it is assumed that also in the species used in this study regulation of the rib operon would be mediated by a termination-antitermination mechanism resulting from two different folding options of the RFN element upstream of the operon in response to riboflavin or FMN. RFN elements have been identified in the rib operon leader region of Lc. mesenteroides (Fig. 2) and Lb. plantarum (Fig. 5) using RFAM [43]. It is likely from the position of the mutations that they affect the stability of the terminator structure making it less energetically favourable for it to form, thus allowing continued transcription of the rib operon.
In order to study the potential usefulness of such strains in an industrial setting a pilot yoghurt trial was set up to compare addition of P. freudenreichii NIZO B374 or its riboflavin-overproducing derivative B2336 either 24 hours prior to the addition of the starter culture or simultaneously with the starter culture. Sequential addition of strain NIZO B2336 to the yoghurt was found to double the concentration of riboflavin in the final product in comparison to yoghurt containing the non-producing wildtype propionibacterium. Furthermore, the mutant showed no differences in comparison to the wildtype strain regarding final cell numbers of the starter culture or final pH of the product. This illustrates that there is a clear benefit of using such riboflavin-overproducing strains in fermentations as it increases the vitamin content of the final product, thus making it more appealing to consumers. Sequential inoculation of the propionibacteria was also found to counteract acidification resulting in a milder product, which could be used as another positive selling point.
The basic concepts of nutrition are changing. The traditional idea of an 'adequate diet', which provides enough nutrients to ensure the individual's survival and meet metabolic needs as well as satisfying hunger is now obsolete. More and more emphasis is being placed on the need for foods to promote health, improve well being and reduce the risk of illness through the adoption of the concept of an "optimum diet" [2]. Selection of strains that have been subjected to uncontrolled genetic alterations has been used in the dairy industry to improve certain intrinsic characteristics of the fermented end product. For example, a spontaneous IS element mediated deletion of the lacZ gene altered lactose metabolism resulting in a decreased fermentation of the sugar. Yoghurt made using this strain is not affected by further acidification [44]. Strains modified by induced mutations are considered non-GMO and are acceptable for deliberate release in the European Union [45]. Such strategies could have important implications for food fermentations as industry constantly strives to increase the marketability of their products to more health conscious consumers in an increasingly competitive market.
Methods
Bacterial strains, media and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Lc. mesenteroides and Lb. plantarum were routinely grown in MRS medium [46] or in chemically defined medium (CDM) (adapted by removal of folic acid, riboflavin and nucleotides) [47,48]. P. freudenreichii strains were routinely cultured in Lactate Broth (LB) containing tryptone (5 g/L), yeast extract (10 g/L), Na-acetate.3H2O (8 g/L) and Na-lactate molasses 60% (25 g/L) at 30°C. The pH of the medium was adjusted to pH 6.5 with 1N KOH before heat treatment for 15 min. at 121°C. The chemically defined medium (CDM) described by Jan et al. [49] was modified for the selection of roseoflavin-resistant mutants. The following ingredients were omitted: p-aminobenzoic acid, folic acid, riboflavin, L-phenylalanine, L-tryptophane, and L-tyrosine. Enumeration of propionibacteria was done on lactate broth agar plates.
Table 1.
Strains and plasmids used in this study
| Strains and plasmids | Details | Reference |
| Strains | ||
| Lc. mesenteroides NCDO2028 | Beetroot silage isolate | UCC culture collection |
| Lc. mesenteroides CB200 to CB210 | NCDO2028 derivatives that are resistant to roseoflavin | This study |
| Lb. plantarum WCFS1 | Sequenced strain | [33] |
| Lb. plantarum NCDO1752 | Pickled cabbage isolate | UCC culture collection |
| Lb. plantarum CB300 to CB305 | NCDO1752 derivatives that are resistant to roseoflavin | This study |
| P. freudenreichii NIZO B374 | CBS115915, formerly subsp. shermanii, now subsp. freudenreichii; cheese isolate | NIZO culture collection |
| P. freudenreichii NIZO B2336 | Roseoflavin resistant derivative of strain NIZO B374 | This study |
| P. freudenreichii NIZO B374.21 | Roseoflavin resistant derivative of strain NIZO B374 | This study |
| P. freudenreichii NIZO B369 | CBS115914; From Eidg. Milchw. Versuchsanstalt. Liebefeld | NIZO culture collection |
| P. freudenreichii NIZO B2663 to NIZO B2672 | Roseoflavin resistant derivatives of strain NIZO B369 | This study |
Isolation of roseoflavin-resistant mutants and sequence analysis of roseoflavin-resistant mutants
Spontaneous roseoflavin-resistant mutants of Lb. plantarum and Lc. mesenteroides were isolated by plating mid logarithmic phase cells on CDM containing 100 mg l-1 roseoflavin. Isolation of chromosomal DNA from Lc. mesenteroides and Lb. plantarum was performed as described by Leenhouts et al. [50,51]. In order to identify the mutations that were the probable cause of roseoflavin resistance and riboflavin overproduction the regulatory region upstream of ribG was amplified by PCR, purified using the JETquick PCR purification kit (Genomed, Löhne, Germany) and subjected to sequence analysis (MWG Biotech AG, Ebersberg, Germany). Spontaneous roseoflavin resistant mutants of P. freudenreichii strains were isolated by plating cultures on adapted chemically defined medium (see above) on solid medium made of the same chemically defined medium supplemented with 1% agar and 10 mg/L roseoflavin. After prolonged incubation at 30°C, roseoflavin-resistant colonies were isolated from the plates and further cultured on liquid chemically defined medium supplemented with 10 mg/L roseoflavin. Fully grown cultures were subsequently diluted in fresh medium in which the roseoflavin concentration was stepwise increased from 50 mg/L, then to 100 mg/L and finally to 200 mg/L. From the final culture with 200 mg/L roseoflavin, single colonies were isolated, and analysed for riboflavin production.
Bioinformatics
In Lc. mesenteroides and Lb. plantarum potential RFN elements were identified using RFAM, which is a collection of multiple sequence alignments and covariance models representing non-coding RNA families [43,52].
Quantitative analysis of riboflavin in culture medium
Extracellular riboflavin concentrations were measured by reverse phase HPLC. A Ultrasphere RP 4.6 mm × 25 cm column (Beckman Coulter, Fullerton, CA) was used and riboflavin was eluted with a linear gradient of acetonitrile from 3.6% to 30% at pH 3.2. Fluorescent detection was used and the excitation and emission wavelengths were 440 and 520 nm, respectively. Commercially obtained riboflavin and FMN were used as references and to obtain a standard curve for quantitative purposes (Sigma, Steinheim, Germany).
Stability study of riboflavin overproducing phenotype in P. freudenreichii
P. freudenreichii B374 and its riboflavin-overproducing derivatives were subcultured in lactate broth lacking roseoflavin for 60 generations and extracellular riboflavin levels were measured every 10 generations to determine the stability of the riboflavin-overproducing phenotype.
Yoghurt study using P. freudenreichii NIZO B374 and its riboflavin-overproducing derivative P. freudenreichii NIZO B2336
Two different methods were applied to produce yoghurt with propionibacteria. First the pasteurized milk was fermented by simultaneous addition of a traditional yoghurt starter culture NIZO S737 (inoculation level of 0.2%) and Propionibacterium freudenreichii NIZO B374 (wildtype) or P. freudenreichii NIZO B2336 (riboflavin-overproducing derivative of strain NIZO B374). The inoculated yoghurt milk was subsequently incubated for 16 hours at 30°C. P. freudenreichii NIZO B374 or NIZO B2336 were added at various inoculum levels (0, 1, 2, 5, 10 or 20%). Secondly, the pasteurized milk was pre-fermented for 24 hours at 30°C with the propionibacteria (inoculum levels of 0, 1, 2, 5, 10 or 20%), before addition of the yoghurt starter culture. The second phase of fermentation was again 16 hours at 30°C. At the end of fermentation the pH and viable counts were determined and the riboflavin content of the yoghurt was measured.
Nucleotide sequence accession numbers
The nucleotide sequence data of the 5' leader regions upstream of the riboflavin biosynthesis operons in Lc. mesenteroides NCDO 2028 and Lb. plantarum NCDO 1752 reported in this paper have been submitted to the Genbank database under accession numbers DQ645591 and DQ645592 respectively.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
CB performed all work and analyses on Lc. mesenteroides and Lb. plantarum including the isolation of the mutants, bioinformatic analysis and identification of mutations. CB also drafted the manuscript. GR performed all the work with P. freudenreichii including isolation of the mutants, the stability assay and the yoghurt trial. DvS and ES participated in the design of the study and the coordination and supervision of the work. DvS and ES also helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was funded by the European Union project QLK1-CT-2000-01376 (Nutra Cells). We wish to thank Jeroen Hugenholtz as overall coordinator of this project. We also would like to thank Anne Wiersma for technical assistance.
Contributor Information
Catherine M Burgess, Email: kaye.burgess@teagasc.ie.
Eddy J Smid, Email: Eddy.Smid@nizo.nl.
Ger Rutten, Email: Eddy.Smid@nizo.nl.
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
References
- Kalra EK. Nutraceutical-Definition and Introduction. AAPS Pharm Sci. 2003;5:Article 25. doi: 10.1208/ps050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou A. Food Safety and Functional Foods in the European Union: Obesity as a Paradigmatic Example for Novel Food Development. Nutr Rev. 2004;62:169–181. doi: 10.1301/nr.2004.jul.S169-S181. [DOI] [PubMed] [Google Scholar]
- Sanders ME. Overview on functional foods: emphasis on probiotic bacteria. Int Dairy J. 1998;8:341–347. doi: 10.1016/S0958-6946(98)00056-9. [DOI] [Google Scholar]
- Stanton C, Ross RP, Fitzgerald GF, Sinderen DV. Fermented functional foods based on probiotics and their biogenic metabolites. Current Opinion in Biotechnology. 2005;16:198–203. doi: 10.1016/j.copbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Stanton C, Gardiner G, Meehan H, Collins K, Fitzgerald GF, Lynch PB, Ross RP. Market potential for probiotics. Am J Clin Nutr. 2001;73(suppl):476S–483S. doi: 10.1093/ajcn/73.2.476s. [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. The prolongation of life. New York, NY, G. Putnam and Sons; 1908. [Google Scholar]
- Kleerebezem M, Hugenholtz J. Metabolic pathway engineering in lactic acid bacteria. Curr Opin Biotech. 2003;14:232–237. doi: 10.1016/S0958-1669(03)00033-8. [DOI] [PubMed] [Google Scholar]
- Hugenholtz J, Smid EJ. Nutraceutical production with food-grade organisms. Curr Opin Biotech. 2002;13:497–507. doi: 10.1016/S0958-1669(02)00367-1. [DOI] [PubMed] [Google Scholar]
- Thierry A, Maillard MB, Herve C, Richoux R, Lortal S. Varied volatile compounds are produced by Propionibacterium freudenreichii in Emmental cheese. Food Chem. 2004;87:439–446. doi: 10.1016/j.foodchem.2003.12.018. [DOI] [Google Scholar]
- Gardner N, Champagne CP. Production of Propionibacterium shermanii biomass and vitamin B12 on spent media. J Appl Microbiol. 2005;99:1236–1245. doi: 10.1111/j.1365-2672.2005.02696.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Adams MC. In vitro assessment of the gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int J Food Microbiol. 2004;91:253–260. doi: 10.1016/j.ijfoodmicro.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Blanck HM, Bowman BA, Serdula MK, Khan LK, Kohn W, Woodruff BA. Angular stomatitis and riboflavin status among adolescent Bhutanese refugees living in southeastern Nepal. Am J Clin Nutr. 2002;76:430–435. doi: 10.1093/ajcn/76.2.430. [DOI] [PubMed] [Google Scholar]
- McKinley MC, McNulty H, McPartlin J, Strain JJ, Scott JM. Effect of riboflavin supplementation on plasma homocysteine in elderly people with low riboflavin status. Eur J Clin Nutr. 2002;56:850–856. doi: 10.1038/sj.ejcn.1601402. [DOI] [PubMed] [Google Scholar]
- Langohr HD, Petruch F, Schroth G. Vitamin B 1, B 2 and B 6 deficiency in neurological disorders. J Neurol. 1981;225:95–108. doi: 10.1007/BF00313323. [DOI] [PubMed] [Google Scholar]
- O'Brien MM, Kiely M, Harrington KE, Robson PJ, Strain JJ, Flynn A. The North/South Ireland Food Consumption Survey: vitamin intakes in 18-64-year-old adults. Public Health Nutr. 2001;4:1069–1079. doi: 10.1079/PHN2001188. [DOI] [PubMed] [Google Scholar]
- Baku TK, Dickerson JWT. Vitamins in Human Health and Disease. Wallinford, Oxon, CAB International; 1996. [Google Scholar]
- Boehnke C, Reuter U, Flach U, Schuh-Hofer S, Einhaupl KM, Arnold G. High-dose riboflavin treatment is efficacious in migraine prophylaxis: an open study in a tertiary care centre. Eur J Neurol. 2004;11:475–477. doi: 10.1111/j.1468-1331.2004.00813.x. [DOI] [PubMed] [Google Scholar]
- Akompong T, Ghori N, Haldar K. In vitro activity of riboflavin against the human malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2000;44:88–96. doi: 10.1128/AAC.44.11.3107-3111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra CG, Junqueira VB. High doses of riboflavin and the elimination of dietary red meat promote the recovery of some motor functions in Parkinson's disease patients. Braz J Med Biol Res. 2003;36:1409–1417. doi: 10.1590/S0100-879X2003001000019. [DOI] [PubMed] [Google Scholar]
- Cooperman JM, Lopez R. Riboflavin. In: Machlin LJ, editor. Handbook of Vitamins. Second. New York, Marcel Dekker Inc.; 1991. [Google Scholar]
- Stahmann KP, Revuelta JL, Seulberger H. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol. 2000;53:509–516. doi: 10.1007/s002530051649. [DOI] [PubMed] [Google Scholar]
- Perkins JB, Pero J. Vitamin Biosynthesis. In: Sonenshein A, Hoch J and Losick R, editor. Bacillus subtilis and Its Closest Relatives: from Genes to Cells. Washington D.C., ASM Press; 2002. pp. 271–286. [Google Scholar]
- Kukanova AI, Zhdanov VG, Stepanov AI. Bacillus subtilis mutants resistant to roseoflavin. Genetika. 1982;18:319–321. [PubMed] [Google Scholar]
- Burgess C, O' Connell-Motherway M, Sybesma W, Hugenholtz J, van Sinderen D. Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods? Appl Environ Microbiol. 2004;70:5769–5777. doi: 10.1128/AEM.70.10.5769-5777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JG, Rutten G, Bruinenberg P, Sesma F, Savoy de Giori G, Smid EJ. A novel dairy product fermented with Propionibacterium freudenreichii improves the riboflavin status of deficient rats. Nutrition. 2006;22:645–651. doi: 10.1016/j.nut.2006.01.002. [DOI] [PubMed] [Google Scholar]
- LeBlanc JG, Burgess C, Sesma F, Savoy de Giori G, van Sinderen D. Ingestion of milk fermented by genetically modified Lactococcus lactis improves the riboflavin status of deficient rats. J Dairy Sci. 2005;88:3435–3442. doi: 10.3168/jds.s0022-0302(05)73027-7. [DOI] [PubMed] [Google Scholar]
- Perkins JB, Pero J, Sloma A. Riboflavin overproducing strains of bacteria. 1991. European patent EP0405370.
- Kreneva RA, Perumov DA. Genetic mapping of regulatory mutations of Bacillus subtilis riboflavin operon. Mol Gen Genet. 1990;222:467–469. doi: 10.1007/BF00633858. [DOI] [PubMed] [Google Scholar]
- Kil YV, Mironov VN, Gorishin IY, Kreneva RA, Perumov DA. Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. Mol Gen Genet. 1992;233:483–486. doi: 10.1007/BF00265448. [DOI] [PubMed] [Google Scholar]
- Coquard D, Huecas M, Ott M, van Dijl JM, van Loon AP, Hohmann HP. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol Gen Genet. 1997;254:81–84. doi: 10.1007/s004380050393. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/S0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretzel W, Schurter W, Ludwig B, Kupfer E, Doswald S, Pfister M, van Loon AP. Commercial riboflavin production by recombinant Bacillus subtilis: down-stream processing and comparison of the composition of riboflavin produced by fermentation or chemical synthesis. J Ind Microbiol Biotech. 1999;22:19–26. doi: 10.1038/sj.jim.2900604. [DOI] [Google Scholar]
- Coulin P, Farah Z, Assanvo J, Spillmann H, Puhan Z. Characterisation of the microflora of attieke, a fermented cassava product, during traditional small-scale preparation. Int J Food Microbiol. 2006;106:131–136. doi: 10.1016/j.ijfoodmicro.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Plengvidhya V, Breidt F, Fleming HP. Use of RAPD-PCR as a method to follow the progress of starter cultures in sauerkraut fermentation. Int J Food Microbiol. 2004;93:287–296. doi: 10.1016/j.ijfoodmicro.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/S0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- Nicoloff H, Arsene-Ploetze F, Malandain C, Kleerebezem M, Bringel F. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J Bacteriol. 2004;186:6059–6069. doi: 10.1128/JB.186.18.6059-6069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes WP. The genus Lactobacillus. In: Wood BJB and Holzapfel WH, editor. The Genera of Lactic Acid Bacteria. Vol. 2. Glasgow, Blackie Academic and Professional; 1995. pp. 19–54. [Google Scholar]
- Teusink B, van Enckevort FHJ, Francke C, Wiersma A, Wegkamp A, Smid EJ, Siezen R. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: Comparing predictions of nutrient requirements with those from growth experiments. Appl Environ Microbiol. 2005;71:7253–7262. doi: 10.1128/AEM.71.11.7253-7262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmadfa I, Heinzle C, Majchrzak D, Foissy H. Influence of a probiotic yoghurt on the status of vitamins B(1), B(2) and B(6) in the healthy adult human. Ann Nutr Metab. 2001;45:13–18. doi: 10.1159/000046700. [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/S0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet B, Delley M. Spontaneous deletion formation within the beta-galactosidase gene of Lactobacillus bulgaricus. J Bacteriol. 1990;172:5670–5676. doi: 10.1128/jb.172.10.5670-5676.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Union Council Directive on the deliberate release into the environment of genetically modified organisms. 2001/18/EC. 2001. (European Union Council).
- de Man JC, Rogosa M, Sharp ME. A medium for the cultivation of Lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- Otto R, ten Brink B, Veldkamp H, Konings WN. The relation between growth rate and electrochemical gradient in Streptococcus cremoris. FEMS Microbiol Lett. 1983;16:69–74. doi: 10.1111/j.1574-6968.1983.tb00261.x. [DOI] [Google Scholar]
- Poolman B, Konings WN. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan G, Leverrier P, Pichereau V, Boyavall P. Changes in Protein Synthesis and Morphology during Acid Adaptation of Propionibacterium freudenreichii. Appl Environ Microbiol. 2001;67:2029–2036. doi: 10.1128/AEM.67.5.2029-2036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts KJ, Kok J, Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts KJ, Kok J, Venema G. Replacement recombination in Lactococcus lactis. J Bacteriol. 1991;173:4794–4798. doi: 10.1128/jb.173.15.4794-4798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RFAM (RNA family database) http://www.sanger.ac.uk/Software/Rfam/ [DOI] [PMC free article] [PubMed]