Abstract
hBRCA1 and hBARD1 are tumor suppressor proteins that are involved as heterodimer via ubiquitinylation in many cellular processes, such as DNA repair. Loss of BRCA1 or BARD1 results in early embryonic lethality and chromosomal instability. The Arabidopsis genome carries a BRCA1 homologue, and we were able to identify a BARD1 homologue. AtBRCA1 and the putative AtBARD1 protein are able to interact with each other as indicated by in vitro and in planta experiments. We have identified T-DNA insertion mutants for both genes, which show no visible phenotype under standard growth conditions and are fully fertile. Thus, in contrast to animals, both genes have no indispensable role during development and meiosis in plants. The two single as well as the double mutant are to a similar extent sensitive to mitomycin C, indicating an epistatic interaction in DNA crosslink repair. We could further demonstrate that in Arabidopsis BARD1 plays a prominent role in the regulation of homologous DNA repair in somatic cells.
Keywords: Arabidopsis thaliana , BARD1, breast cancer genes, DNA repair, homologous recombination
Introduction
Germline mutations of the hBRCA1 (breast cancer susceptibility 1) gene are known to be responsible for about 50% of all inherited breast cancer cases (Miki et al, 1994). The human BRCA1 gene codes for an 1863 amino acids (aa) long nuclear protein with two functionally important motifs. The first motif is located at the N-terminus of the protein and codes for a RING-finger domain, consisting of 40–60 aa. Many RING finger containing proteins function as ubiquitin E3 ligase (Wu et al, 1996; Joazeiro and Weissman, 2000). The second motif is located at the C-terminus and encodes two repeats of approximately 80 aa. These repeats were designated as Breast cancer C-terminal repeats (BRCT; Callebaut and Mornon, 1997; Koonin et al, 1996). These BRCT domains are present in a large number of cell cycle checkpoint proteins ranging from bacteria to humans (Koonin et al, 1996; Callebaut and Mornon, 1997). Both RING and BRCT domains of hBRCA1 are well conserved and serve as common sites for missense mutations that predispose women to early-onset breast cancer (Ruffner et al, 2001; Rodriguez et al, 2004). Protein interaction studies using either the RING or BRCT domain of BRCA1 identified several interacting proteins (Jensen et al, 1998; Yarden and Brody, 2001). Interestingly, a protein found to interact with the N-terminal RING domain contained itself both a RING as well as two BRCT domains similar to BRCA1 (Wu et al, 1996). As further studies demonstrated that both proteins are able to form a heterodimer through their common N-terminal RING domain, this protein was designated BARD1, breast cancerassociatedRINGdomain (Meza et al, 1999; Joukov et al, 2001). This hBRCA1/hBARD1 heterodimer complex functions as an E3 ubiquitin ligase that catalyses the synthesis of polyubiquitin chains (reviewed by Baer and Ludwig, 2002).
DNA damage poses a continuous threat to genomic integrity in eukaryotic cells. A particularly lethal form of DNA damage is the DNA double-strand break (DSB). Cells have two major pathways for the repair of DSBs, homologous recombination (HR) and nonhomologous end joining (NHEJ) (reviewed by Puchta, 2005). Although NHEJ is a process in which the ends of a DSB might be modified, HR precisely restores the continuity of a broken DNA molecule using an intact and homologous DNA strand as template.
For a decade, multiple analyses have been performed to elucidate the biological role of BRCA1. Evidence for the involvement of BRCA1 in the repair of DSB originates from its association with hRAD51 (Scully et al, 1997a), and from the formation of foci at sites of DSBs after genotoxic stress (Scully et al, 1997b; Paull et al, 2000). Disruption of BRCA1 in mice results in embryonic lethality that is accompanied by growth retardation, apoptosis, cell cycle defects and genetic instability (Gowen et al, 2000). Taken together, these results demonstrate a very important role for BRCA1 in promoting HR and thus in maintaining genomic integrity.
In contrast to BRCA1 very few and partially indirect functional studies on BARD1 homologues were performed. Besides its function as E3 ubiquitin ligase in a complex with BRCA1, some studies indicated that the protein might also be involved in homologous DSB repair (Westermark et al, 2003; Stark et al, 2004). Recently, studies on a BARD1 homologue in Caenorhabditis elegans showed that depletion of the BARD1 protein resulted in germination defects and radiation sensitivity (Boulton et al, 2004).
Until 2003 orthologues of BRCA1 were only identified in other animal genomes, for example, C. elegans and Xenopus laevis (Joukov et al, 2001; Boulton et al, 2004). Surprisingly, Lafarge and Montane identified in 2003 a BRCA1 orthologue in the genome of the model plant Arabidopsis. Similar to its orthologue from humans, this protein has the characteristic RING and BRCT domains. Furthermore, it was shown that the transcription of the AtBRCA1 gene was strongly induced by γ-irradiation (Lafarge and Montane, 2003). However, the study did not address the biological function of the protein in plants.
We have now been able to identify a hBARD1 homologue in Arabidopsis and in the following we characterise the biological role of AtBARD1 and AtBRCA1 in plants.
Results
Identification of a hBARD1 homologue in A. thaliana
The characteristic feature of both hBRCA1 and hBARD1 is the presence of a conserved RING as well as two BRCT domains. Orthologues of hBRCA1 and hBARD1 in Mus musculus, C. elegans or X. laevis display a similar domain structure (Szabo et al, 1996; Joukov et al, 2001; Boulton et al, 2004). To identify putative BARD1 homologues in the Arabidopsis genome, a database search was carried out in TAIR-BLASTP using hBARD1 as template (NP000456). This search resulted in two significant hits: At4g21070, which had previously been classified as the hBRCA1 homologue of Arabidopsis (Lafarge and Montane, 2003), and At1g04020. The homology of AtBRCA1 to the hBARD1 protein is restricted to the previously mentioned conserved RING and BRCT domains. However, At1g04020 has additional homology to hBARD1 outside the RING and BRCT domains, in total 22% amino-acid identity and 38% similarity (Figure 1A and B). We therefore assumed that At1g04020 might be the BARD1 homologue of Arabidopsis.
Figure 1.
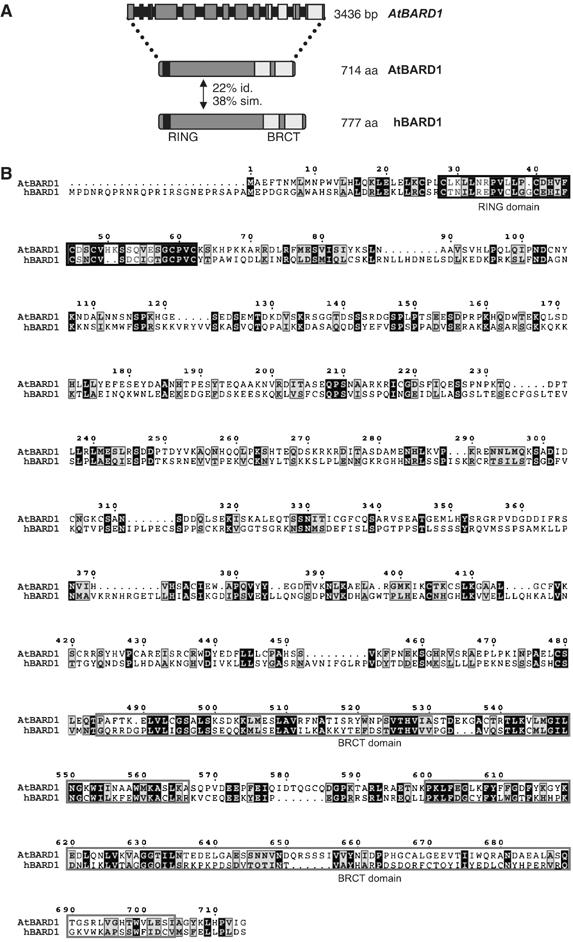
Gene structure of AtBARD1 and comparison of the AtBARD1 and hBARD1 proteins. (A) A schematic representation of the AtBARD1 intron–exon structure. Exons are represented by grey boxes, introns by black bars. In total, the AtBARD1 gene counts 13 exons, the gene has a length of 3436 bp encoding a protein of 714 aa. The AtBARD1 protein has a similar structure as the hBARD1 protein, also containing a conserved RING domain (black regions) and two BRCT domains (light grey regions). Both proteins have an identity of 22% and a similarity of 38%. (B) Protein sequence alignment of AtBARD1 against hBARD1. Identical amino acids are shaded black whereas similar amino acids are shown in grey. Conserved RING and BRCT domain structures are indicated by black and light grey frames, respectively.
Using mRNA from Arabidopsis flowers as template, the cDNA from At1g04020 could be amplified by polymerase chain reaction (PCR). By RACE-PCR with nested gene-specific primers, 5′ and 3′ ends were obtained. The ORF of full-length AtBARD1 has a total length of 2145 bp, contains 13 exons, and codes for a protein of 714 aa (Figure 1A). A cDNA clone (BX815982) from the GenBank confirmed this structure.
The ORF of AtBRCA1 was also determined by RACE-PCR. In line with cDNA clones from SALK (U24692, R24692 and AF515728), we identified the ORF of full-length AtBRCA1 consisting of 2826 bp, containing 14 exons and coding for a protein of 941 aa. This is in contrast to the original report of Lafarge and Montane (2003) who identified the ORF of full-length AtBRCA1 consisting of 4485 bp and 15 exons. The first exon postulated by Lafarge and Montane (2003) is part of another gene rather than the ORF of AtBRCA1.
In plants it was demonstrated before that some genes coding for proteins involved in nucleotide metabolism and DNA repair can be induced by DNA damage, among them AtBRCA1 (e.g. Chen et al, 2003; Lafarge and Montane, 2003). To characterise a possible correlation between the expression of AtBRCA1 and AtBARD1 2-week-old seedlings were irradiated by γ-ray (100 Gy) and the transcript amount of both genes was measured after 1 h by quantitative real-time PCR (Figure 2A). As reported previously, a strong induction of the AtBRCA1 transcript could be detected. In contrast, no significant change of the mRNA level of AtBARD1 was found. Additionally, the expression of both genes in different tissues of 6–8-week-old Arabidopsis plants was analysed. RNA from roots, rosette leaves, inflorescence, young cauline leaves, flowers and siliques was isolated and the transcript amount of both genes was measured via real-time PCR. Higher amounts of mRNA of both AtBRCA1 and AtBARD1 could be detected in flowers and siliques. The expression in roots, rosette leaves, inflorescence and young cauline leaves was low (Figure 2B). Thus, in contrast to the application of genotoxic stress, the expression pattern of both genes in different organs correlated well, hinting to a functional interaction.
Figure 2.
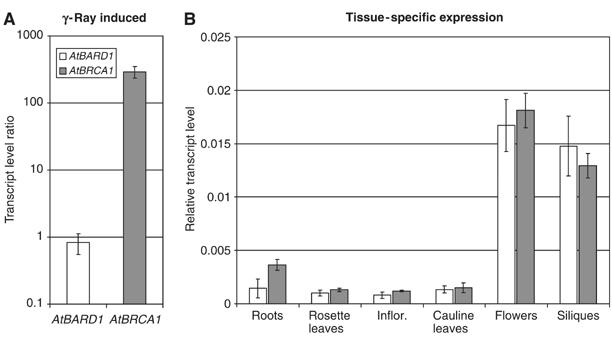
Expression analysis of AtBARD1 and AtBRCA1 in Arabidopsis. (A) The expression of AtBARD1 and AtBRCA1 was analysed by relative quantification using real-time PCR 1 h after irradiation by γ-ray. Transcription level ratio of AtBARD1 and AtBRCA1 is given in relation to actin mRNA and the mRNA of the respective untreated seedlings, and is the mean of six different reactions ±s.d. White bars, AtBARD1; grey bars, AtBRCA1. (B) The expression pattern of AtBARD1 and AtBRCA1 in different plant tissues was analysed by relative quantification using real-time PCR. RNA from roots, rosette leaves, inflorescence, cauline leaves, flowers and siliques of soil-grown plants was analysed. Expression of AtBARD1/AtBRCA1 is given relative to actin mRNA levels and is the mean of six different reactions±s.d. Similar results were obtained in independent experiments.
Protein–protein interaction between AtBARD1 and AtBRCA1
To test whether AtBRCA1 and the putative AtBARD1 protein are also able to interact directly, a two-hybrid analysis was performed.
First, it was tested whether AtBRCA1 and AtBARD1 contained an activation domain. It was previously demonstrated that this is the case for the hBRCA1 protein, whereas so far this has not been reported for the hBARD1 protein (Welcsh et al 2002). With the help of the LexA-based yeast two-hybrid system, we could clearly demonstrate that the full-length AtBARD1 protein contained an autoactivation domain (Figure 3A). Unfortunately, no consistent results were obtained using the full-length AtBRCA1 protein. This might reflect the presence of a weak transcriptional activation domain. Thus, in this assay it was only possible to use truncated versions of AtBRCA1 or AtBARD1 as bait. An N-terminal fragment of AtBRCA1 coding for the first 59 aa and containing the RING domain did not display any autoactivation and was used as bait. As prey the full-length AtBARD1 protein and a C-terminal AtBARD1 fragment containing the BRCT repeats but missing the RING domain were constructed. Indeed, an interaction of the RING domain of AtBRCA1 with the full-length AtBARD1 protein could be demonstrated (Figure 3A), whereas no interaction of the RING domain of AtBRCA1 with the C-terminal part of the AtBARD1 protein was found. Unfortunately, we failed to detect an interaction using the RING domain of AtBARD1 as bait and the complete AtBRCA1 protein (result not shown). However, it is not uncommon in two-hybrid analysis that only certain bait and prey combinations result in detectable interactions (Uetz et al, 2000).
Figure 3.
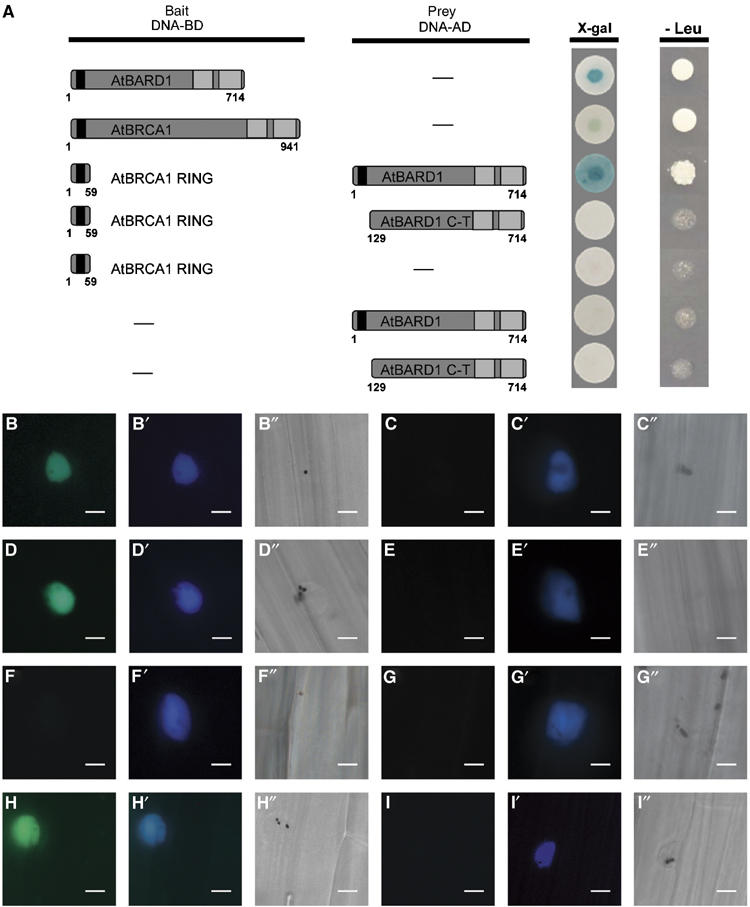
Characterisation of the AtBARD1 and AtBRCA1 interaction by yeast two-hybrid assay and by BiFC in transiently transfected mustard seedlings. (A) Yeast two-hybrid assay. Different constructs of AtBRCA1 or AtBARD1 were used either as bait (DNA-BD) or prey (DNA-AD) and tested for their ability to activate the lacZ reporter gene (X-gal) and the nutritional marker gene leucine (Leu). The full-length protein AtBARD1 was, when fused to a DNA-binding domain (DNA-BD), able to activate the lacZ reporter gene as well as the leucine reporter gene. The use of AtBRCA1 as bait led to inconsistent results. A truncated version of AtBRCA1, containing the first 59 N-terminal aa (representing the RING domain) interacted with the complete AtBARD1 protein fused to the activation domain and resulted in the activation of the lacZ reporter gene and the leucine reporter gene. No interaction could be demonstrated between the AtBRCA1 RING domain and a C-terminal part of AtBARD1 (AtBARD1 C-T). Furthermore, none of the single used constructs was able to activate transcription. Blue staining of the yeast colonies appeared within 30 min for the AtBARD1 protein, to up to 2 h for the AtBRCA1 AtBARD1 interaction, whereas the growth of yeast colonies on LEU lacking medium was determined after 2 days. (B–I) (B′–I′) and (B″–I″) BiFC analysis in transiently transfected mustard seedlings. The pictures B–I show an YFP signal in case of a protein interaction, in the nucleus of a representative cell, owing to the restoration of the YFP complex. The pictures B′–I′ show a CFP signal from a cotransfected nuclear marker. The pictures B″–I″ show the same cells as in (B–I) and (B′–I′), respectively, by bright field microscopy. Bars=20 μm. B, B′ and B″ AtBRCA1 (YC) and AtBARD1 (YN); C, C′ and C″ AtBRCA1 (YC) and empty vector pMAV-GW-YN; D, D′ and D″ AtBRCA1 RING (YC) and AtBARD1 (YN); E, E′ and E″ AtBRCA1 RING (YC) and empty vector pMAV-GW-YN; F, F′ and F″ AtBRCA1 C-terminus (YC) and AtBARD1 (YN); G, G′ and G″ AtBARD1 (YN) and empty vector pMAV-GW-YC; H, H′ and H″ ASK1 (YN) and EID1 (YC), positive control; I, I′ and I″ ASK1 (YN) and EID1ΔF (YC), negative control.
Interaction of AtBARD1 and AtBRCA1 in planta
To further sustain our observation that AtBRCA1 and AtBARD1 interact, in vivo studies were carried out. We used a well-established method of bimolecular fluorescence complementation (BiFC; Hu et al, 2002) for the in vivo detection of protein–protein interactions, namely the split YFP system (Stolpe et al, 2005). Briefly, the assay is based on the observation that a N- (YN) and a C-terminal (YC) fragment of the yellow fluorescent protein (YFP) can only reconstitute a functional fluorophore when they are brought into tight contact. Two ORFs, driven by a double 35S promoter, are fused on separate plasmids to the respective YFP fragments; next, both constructs are brought into a plant cell for expression and the interaction of the fusion proteins can be monitored via epifluorescence microscopy.
To confirm the possible interaction between AtBRCA1 and AtBARD1, the full-length AtBRCA1 ORF, the first N terminal 88 aa coding for the AtBRCA1 RING domain and the last 797 C-terminal aa of the ORF of AtBRCA1 were fused to the C-terminal part of the YFP protein. The full-length AtBARD1 ORF was fused to the N-terminal part of the YFP protein. Next, the different constructs, together with a plasmid containing the CPRF2 protein (common plant regulatory factor 2) fused to CFP as nuclear marker (e.g. Figure 3B′ and C′), were transiently expressed after particle bombardment in etiolated mustard seedlings. As positive control, the ASK1 protein fused to the N-terminus of YFP, and an EID1-YFP-C-terminal fusion was used (Figure 3H). EID1 and ASK1 are interacting proteins of the Skp1-Cullin-F-box-protein ubiquitin ligase that targets proteins for degradation and functions as a negative regulator in phytochrome A-specific light signalling. The negative control was a deleted version of the EID1 (EID1ΔF) protein not able to interact with ASK1 (Figure 3I; Stolpe et al, 2005). As further controls AtBRCA1 or AtBARD1 constructs fused with the N- or C-terminal part of the YFP protein, respectively, were used together with the respective pMAV-GW-YN and pMAV-GW-YC empty vectors. After an overnight incubation period, the seedlings were screened for the presence of an YFP signal. Routinely, 1–5 transfected cells per seedling were obtained. The results are based on at least two independent experiments using four mustard seedlings for each transfection. For each single combination, the results were uniform, that is, besides the CFP signal, either in all or in none of the transfected cells an YFP signal could be detected.
Not only in case of the full-length ORF of AtBRCA1 combined with the complete AtBARD1 protein an YFP signal could be detected (Figure 3B), but also in the AtBRCA1 RING domain and the AtBARD1 protein (Figure 3D). No YFP signal was observed when the C-terminus of AtBRCA1 was coexpressed with the AtBARD1 protein (Figure 3F). No YFP signal could be obtained when combinations of the single constructs of AtBRCA1 and AtBARD1 with the pMAV-GW-YN and pMAV-GW-YC empty vectors were used (Figure 3C, E and G). Taken together, our experiments clearly demonstrate that AtBARD1 and AtBRCA1 are able to interact, and that this interaction is mediated by the AtBRCA1 RING domain. This is in line with our experiments from the two-hybrid system.
Mutant atbard1 plants are phenotypically normal but sensitive to mitomycin C
Functional studies were necessary in order to elucidate the biological role of the BARD1 homologue in plants. The putative AtBARD1 gene sequence was used to screen the sequence database of T-DNA insertion mutants on the SIGnAL webpage (Salk Institute Genomic Analysis Laboratory; Alonso et al, 2003). Two atbard1 T-DNA mutant lines were identified. The respective plants were obtained, propagated, and homozygous individuals of the respective insertions could be identified. The insertion sites were determined in detail by PCR. Figure 4 provides a detailed characterisation of the T-DNA insertions of AtBARD1.
Figure 4.
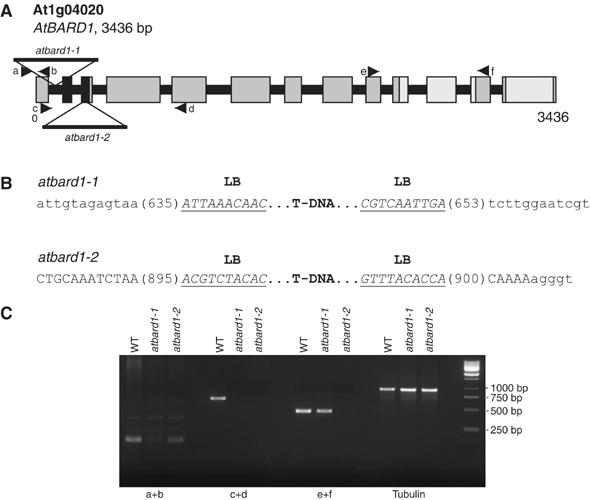
Schematic structure of the AtBARD1 gene and its T-DNA insertions. (A) The AtBARD1 gene consists of 13 exons. Regions coding for the RING and BRCT domain are indicated in black and light grey, respectively. Two T-DNA insertions were identified. One insertion is located in the first intron, and denominated atbard1-1 whereas the second insertion is located in the third exon, and denominated atbard1-2. (B) An overview of the precise locations of the T-DNA inserts in the AtBARD1 gene. Intron sequences are displayed as lower-case letters, exon sequences as capital letters, and T-DNA border sequences are underlined (LB: left border). (C) Semiquantitative RT–PCR on different regions of the AtBARD1 gene. Primer pairs were used that bind in front of (a+b), across (c+d) and after (e+f) the T-DNA insertions. The β-tubulin gene was taken as control. WT: wild type.
The two atbard1 T-DNA insertions are located at the beginning of the gene. Both insertions carry left T-DNA borders at their ends, indicating the integration of a double T-DNA insert in inverted orientation. The first insertion, SALK_097601, atbard1-1, is present in the first intron and results in a deletion of 18 nucleotides. The second insertion, SALK_031862, atbard1-2, is located in the third exon, which codes for the N-terminal RING domain and leads to a deletion of 5 nucleotides within the coding sequence (Figure 4B).
In order to assess the expression level of AtBARD1 in the homozygous T-DNA lines, reverse transcription–polymerase chain reaction (RT–PCR) experiments were performed with homozygous mutants using primer pairs binding in front, across and after the insertions (Figure 4A and C). In case of both lines, expression of an mRNA before the insertion could be demonstrated (Figure 4C). With primers spanning across the insertions, we were not able to amplify any product for both alleles. An expression after the insertion was detected for atbard1-1. In contrast, no expression could be found for atbard1-2, indicating that this allele most probably represents a ‘true' null atbard1 mutation.
In comparison to wild-type plants, all plant lines homozygous for the respective insertions did not differ in their phenotypes when grown under standard conditions. However, when challenged with the DNA crosslinking agent mitomycin C (MMC), the T-DNA insertion mutants showed a more sensitive phenotype as compared to wild-type seedlings (Figure 5). The mutant seedlings were smaller and less viable. Interestingly, the line atbard1-2 showed a slightly stronger phenotype after treatment with MMC than atbard1-1. Other mutagenic treatments with bleomycin or UV radiation did not display an increased sensitivity in the mutant background (results not shown).
Figure 5.
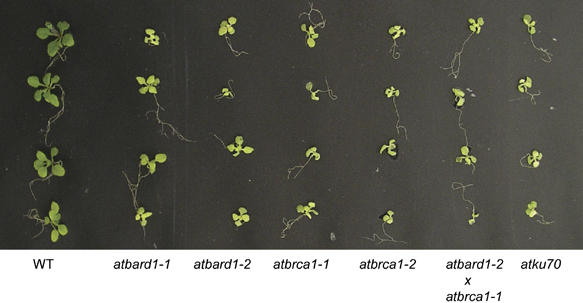
Hypersensitivity of different Arabidopsis mutants to the DNA-damaging agent MMC. Arabidopsis seeds from the mutant lines atbard1-1, atbard1-2, atbrca1-1, atbrca1-2 as well as the double-mutant atbard1-2/atbrca1-1 were tested for their sensitivity to MMC. Wild-type seeds (Columbia) and atku70 (a sensitive control line; Bundock et al, 2002) were used as controls. Seeds were plated on GM medium containing 30 μg MMC/ml, 17 days later seedlings were analysed for their sensitivity. WT: wild type.
AtBARD1 is dispensable for meiosis
We checked whether the selfed progeny of the T-DNA mutant atbard1 plants was fertile in order to test whether the AtBARD1 protein plays a role during meiosis. Both atbard1 T-DNA insertion mutants produced viable seeds at similar numbers as the wild-type plants. As minor meiotic defects are often correlated with reduced viability of male gametes, pollen of both mutants were analysed with Alexander (1969) staining. However, a similar number of viable pollen could be detected in wild-type and mutant anthers, indicating that AtBARD1 is not necessarily required for the progression of meiosis in plants (data not shown).
Intrachromosomal HR is reduced in atbard1 mutant plants and less inducible by genotoxic stress
To test the frequency of somatic HR in planta, a well-established recombination assay using the transgenic line 651 was performed (Swoboda et al, 1994). The recombination substrate within the transgene consists of two overlapping fragments of the β-glucuronidase gene (GUS; uidA) interrupted by a hygromycin selectable marker gene. The separated uidA sequences share a common overlap of 566 bp in inverted orientation. HR between the two overlapping DNA sequences produces a functional uidA gene. Cell clusters expressing β-glucuronidase activity can be detected as blue sectors after histochemical staining, and it was shown before that these sectors indeed arise from recombination events (Swoboda et al, 1994). The homozygous atbard1-1 and 1-2 mutants were crossed with a transgenic line carrying the 651 transgene and selfed again to obtain plants homozygous with respect to the atbard1 insertion as well as the 651 transgene. Seedlings were incubated in liquid germination medium (GM) with and without bleomycin (10 μg/ml). Bleomycin is a radiomimeticum causing single-stranded breaks (SSB) and DSB (Harsch et al, 2000). Next, recombination events were counted in 12-day-old seedlings. For both mutant atbard1 lines, the distribution and frequency of recombination events were determined. Figure 6 shows a representative individual experiment for each mutant line. The significance of the differences of the HR events between mutants and segregated control plants was confirmed by the pair-wise nonparametric Mann–Whitney U-test. The experiment was repeated three times for each line (Table I). In all three independent experiments, a significant reduction of HR was found in the mutant backgrounds, either with or without genotoxic stress. A comparison between the untreated segregated control plants and the untreated atbard1-1 and atbard1-2 homozygous plants showed that the frequency of recombination events in the mutant plants was 2–3 times and about 10 times lower, respectively, as compared to the control line (Figure 6A and B; Table I).
Figure 6.
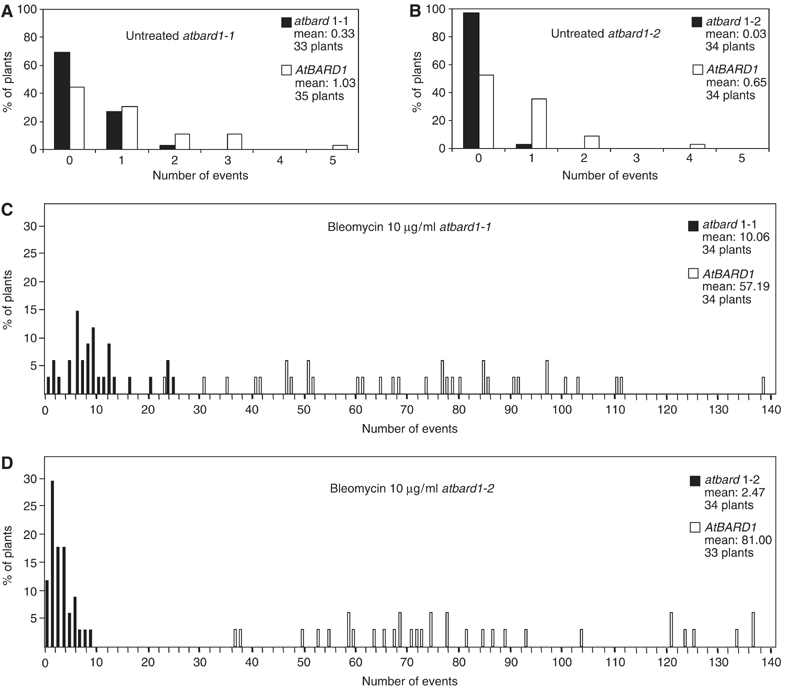
HR events in 651/atbard1 seedlings. The diagrams show the percentage of seedlings with a given number of blue spots. (A) Untreated atbard1-1, (B) untreated atbard1-2, (C) bleomycin-treated atbard1-1 and (D) bleomycin-treated atbard1-2. atbard1 seedlings are displayed as black bars, segregated control plants homozygous for AtBARD1 are shown as white bars.
Table 1.
Somatic HR in atbard1-1 and segregated control plants (A), and in atbard1-2 and segregated control plants (B)
| Control | atbard1 | Relation | ||||
|---|---|---|---|---|---|---|
| n | N | m1 | n | N | m2 | m2/m1 |
| (A) No genotoxic stress | ||||||
| 35 | 16 | 0.46 | 34 | 7 | 0.21 | 0.46 |
| 36 | 22 | 0.61 | 33 | 6 | 0.18 | 0.30 |
| 35 | 36 | 1.03 | 33 | 11 | 0.33 | 0.32 |
| Mean | 0.70±0.20 | 0.24±0.08 | 0.36* | |||
| Bleomycin induction (10 μg/ml) | ||||||
| 33 | 2733 | 77.97 | 34 | 803 | 23.61 | 0.30 |
| 33 | 1947 | 61.97 | 34 | 393 | 11.56 | 0.19 |
| 34 | 2431 | 57.19 | 34 | 342 | 10.06 | 0.18 |
| Mean | 65.71±10.88 | 15.08±7.43 | 0.22* | |||
| (B) No genotoxic stress | ||||||
| 33 | 16 | 0.48 | 34 | 2 | 0.06 | 0.13 |
| 31 | 20 | 0.65 | 32 | 3 | 0.09 | 0.14 |
| 34 | 22 | 0.65 | 34 | 1 | 0.03 | 0.04 |
| Mean | 0.59±0.10 | 0.06±0.03 | 0.10* | |||
| Bleomycin induction (10 μg/ml) | ||||||
| 34 | 1422 | 41.82 | 34 | 28 | 0.82 | 0.02 |
| 34 | 3063 | 90.09 | 34 | 65 | 1.91 | 0.02 |
| 33 | 2673 | 81.00 | 34 | 84 | 2.47 | 0.03 |
| Mean | 70.97±25.65 | 1.73±0.84 | 0.22* | |||
| Data are numbers of plants tested (n), total blue stained recombination spots (N), and the mean number of spots per plant per chromosomal recombination assay (m1: control; m2: atbard1) in three different experiments (*calculated from the means of the three experiments). | ||||||
When both mutant atbard1 lines were challenged with bleomycin (10 μg/ml), the frequency of recombination events increased by about two orders of magnitude in the control lines, whereas the induction was significantly lower in both atbard1-1 and atbard1-2 lines (Figure 6C and D; Table I). This is also demonstrated by the fact that in all cases the relation between the mean recombination frequencies of mutant (m2) and segregated control plants (m1) was lower with than without application of genotoxic stress (see Table I last column m2/m1). Taking into account the enhanced sensitivity to MMC of atbard1-2 in comparison to atbard1-1, the differences between the two mutants in HR can be taken as a hint that only in case of atbard1-2 the insertion of the T-DNA into the gene resulted in a ‘true' null mutation.
Independent of the different degrees of deficiency found in the two mutant lines, our results clearly demonstrate that AtBARD1 is not only required for the repair of DSBs by HR under standard growth conditions, but also for the regulation of HR induction after application of genotoxic stress.
AtBRCA1 and AtBARD1 are epistatic for cross-link repair
Our two-hybrid data as well as in planta experiments indicated that AtBARD1 and AtBRCA1 physically interact. To demonstrate a genetic interaction we screened the sequence databases of T-DNA insertion mutants on the SIGnAL (Salk Institute Genomic Analysis Laboratory; Alonso et al, 2003) and Garlic (Syngenta) webpages (Sessions et al, 2002). Two atbrca1 T-DNA mutant lines were identified. The insertion sites were determined in detail by PCR. Figure 7 provides a precise characterisation of the T-DNA insertions in the AtBRCA1 gene. The first insertion in the AtBRCA1 gene, SALK 014731, atbrca1-1, is located in the fourth exon. Left borders of T-DNA were found at both ends of the insert, indicating the integration of T-DNAs in tandem inverted orientation. The integration led to the deletion of 18 nucleotides of the fourth exon (Figure 7A and B). The second insertion, GARLIC 916_C09, atbrca1-2, was located in the fifth intron more to the middle of the gene, and the insert is flanked by a right and a left T-DNA border. This insertion led to the deletion of eight nucleotides of the fifth intron (Figure 7A and B).
Figure 7.

Schematic structure of the AtBRCA1 gene and its T-DNA insertions. (A) The AtBRCA1 gene consists of 14 exons. The RING and BRCT domains are indicated as black and light grey regions, respectively. Two T-DNA insertions were identified. One insertion is located in the fourth exon and denominated atbrca1-1, whereas the second insertion is located in the fifth intron and denominated atbrca1-2. (B) An overview of the precise locations of the T-DNA inserts in the AtBRCA1 gene. Intron sequences are displayed as lower-case letters, exon sequences as capital letters, and T-DNA border sequences are underlined. (LB: left border; RB: right border). (C) Semi-quantitative RT–PCR on different regions of the AtBRCA1 gene. Primer pairs were used in front of (a+b), across (c+d) and after (e+f) the T-DNA insertion. The β-tubulin gene was taken as control. WT: wild type.
A precise analysis of the AtBRCA1 expression level in the atbrca1 mutant lines demonstrated that the expression level of the mRNA in front of the T-DNA inserts was unchanged (Figure 7C). With primers spanning the insertion sites, we were not able to amplify any product, neither from the atbrca1-1 nor from the atbrca1-2 allele. Primers downstream of the insertion demonstrated that the expression in the atbrca1-1 line is drastically reduced as compared to the wild-type AtBRCA1 expression, whereas the expression in the atbrca1-2 line is moderately increased.
Both insertion lines were viable and fully fertile. Moreover, both lines were sensitive to MMC (Figure 5). The atbrca1-1 mutant line was crossed with the mutant atbard1-2 line to create an atbard1-2/atbrca1-1 double mutant. To elucidate whether both proteins act in the same DNA repair pathway, we quantified in repeated experiments the degree of MMC sensitivity by fresh weight determination of the double mutant in comparison with both single mutants (Figure 8). Indeed, the double mutant was not more sensitive than each of the single mutants, indicating that AtBRCA1 and AtBARD1 are epistatic for DNA cross-link repair.
Figure 8.
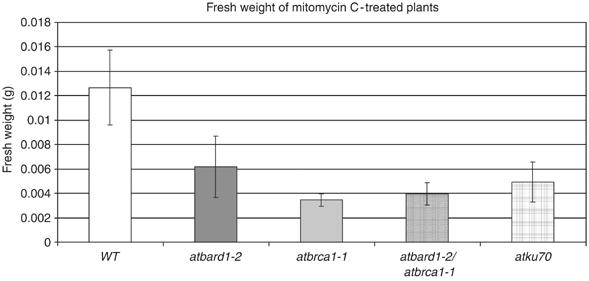
Fresh weight determination of different atbard1 and atbrca1 mutants treated with the DNA-damaging agent MMC. Fresh weight of atbard1-2, atbrca1-1, as well as the double-mutant line atbard1-2/atbrca1-1 and atku70 (a sensitive control line) mutant seedlings grown on GM medium containing 30 μg MMC/ml for 17 days. Values shown are means from 10 seedlings (±s.d.). WT: wild type.
Discussion
Sequence analysis has revealed that Arabidopsis contains orthologues of genes involved in human genetic diseases as well as in cancer (the Arabidopsis genome initiative). Interestingly, two genes, the mutations of which were found to be frequently associated with breast cancer, BRCA1 and BRCA2, are present in plant genomes, too. BRCA2 is involved in HR and seems to nucleate RAD51 filament formation at dsDNA–ssDNA junctions (e.g. Yang et al, 2005). In Arabidopsis, two recently duplicated BRCA2 homologues are present. An RNAi approach demonstrated that knocking down the expression of the genes strongly impairs meiosis (Siaud et al, 2004). It has been reported before that Arabidopsis contains a BRCA1 homologue, the expression of which is induced after DNA damage (Lafarge and Montane, 2003). We have now also been able to detect an ORF for a BARD1 homologue in the Arabidopsis genome and to characterise its function and the relation to AtBRCA1 in plants.
AtBARD1 is not induced by genotoxic stress
The expression levels of AtBARD1 and AtBRCA1 were similar in most investigated plant tissues (Figure 2), consistent with the hypothesis of their functional interaction. However, under genotoxic stress, the AtBARD1 transcript level remained constant, whereas AtBRCA1 was strongly induced. Interestingly, in mice the BARD1 expression also correlated with the BRCA1 expression. However, during the ovulatory cycle, BRCA1 and BARD1 are modulated differently in the uterus (Irminger-Finger et al, 1998). Furthermore, upon genotoxic stress both BRCA1 and BARD1 mRNAs are induced (Aunoble et al, 2001; Irminger-Finger et al, 2001). Therefore, it seems that the regulatory mechanism of AtBARD1 and AtBRCA1 in plants differs from humans.
AtBARD1 and AtBRCA1 interact and are involved in cross-link repair
hBARD1 has been reported to form a dimer with hBRCA1 (Wu et al, 1996). If the two ORFs in plants had similar functions as in mammals, we would expect the respective proteins to interact. Indeed, we were able to demonstrate a specific interaction by the use of the split YFP system in planta and the yeast two-hybrid assay (Figure 3). This result can be taken as a strong hint that both proteins function in a common complex in a similar way as the E3 ubiquitin ligase in humans. Further support for a genetic association of the two proteins could be obtained by mutant analysis. Various studies in mammals have shown that mutations in genes important for the DNA repair lead to chromosomal instabilities and increased sensitivity to DNA-damaging agents, such as radiation, or cross-linking agents, like MMC (e.g. Cui et al, 1999). Similar effects were also reported for brca1 and bard1 mutant cells (Mamon et al, 2003; Westermark et al, 2003). To decipher whether the putative AtBARD1 and AtBRCA1 homologues were involved in DNA repair, we investigated the sensitivity of the insertion mutants to DNA damage-inducing chemicals. When challenged with the cross-linking agent MMC, we could observe an increased sensitivity for atbard1 and atbrca1 mutants. The fact that the atbard1-2/atbrca1-1 double mutant was not more sensitive than the single mutants indicates that both factors are involved in the same pathway of DNA cross-link repair in plants (Figure 8). Interestingly, the fact that the mutants are not sensitive to bleomycin is reminiscent to the behaviour of certain plant mutants involved in HR. It has been reported that the RAD51 paralogues AtRAD51B, AtRAD51C and AtXRCC3 are sensitive to MMC but not to γ-rays (Bleuyard and White, 2004; Abe et al, 2005; Osakabe et al, 2005).
BARD1 might be involved in transcriptional regulation
Recent studies showed that hBRCA1 associates with the upstream stimulatory factor 2 and is a component of a DNA-binding complex (Cable et al, 2003). These findings suggest a role for the endogenous BRCA1 protein complex in transcription through a defined DNA-binding sequence and indicate that one function of BRCA1 is to coregulate the expression of genes involved in various cellular processes. It is noteworthy to state that in our two-hybrid experiments, it was demonstrated that the complete AtBARD1 protein contains an activation domain (Figure 3A). In contrast to BRCA1 (Monteiro et al, 1996), a similar finding has not been reported before for other eukaryotic BARD1 ORFs. The activation domain of AtBARD1 could be taken as a hint that similar to hBRCA1 BARD1 might also be involved in transcriptional regulation in eukaryotes in general. It will be interesting to determine to what extent changes in the transcriptome of Arabidopsis occur in the atbard1 mutants.
The role of AtBARD1 and AtBRCA1 in meiosis
Although key factors in the mechanisms and in the regulation of HR are conserved between different eukaryotes, very important differences can be detected between yeast, plants and animals (Hartung and Puchta, 2004). Saccharomyces cerevisiae does not contain a number of factors involved in the regulation of HR, such as BRCA1, BARD1, BRCA2 and p53 that are present in animals. Interestingly, plants do contain both BRCA homologues and BARD1, although no indication for a structural or functional p53 homologue exists. Nevertheless, indications for an UV-induced apoptosis pathway present in Arabidopsis were recently reported (Danon et al, 2004).
A row of viable mutant defects in certain steps of HR could be isolated in plants in contrast to other higher eukaryotes owing to embryo lethality. Among these genes are AtRAD51, AtMRE11, AtRAD50 and the RAD51 paralogues. Owing to the fact that plant mutants are viable, Arabidopsis is an ideal object to analyse the role of these factors in meiosis. Indeed, mutations of AtRAD51, AtMRE11, AtRAD50, AtXRCC3 and AtRAD51C resulted in sterility owing to aberrant meiosis (Bleuyard and White, 2004; Bleuyard et al, 2004, 2005; Li et al, 2004, 2005; Puizina et al, 2004; Abe et al, 2005). A sterile phenotype was also reported for a mouse BRCA1 mutant with a deletion of intron 11 that was obtained in a p53+/− background (Xu et al, 2003). To our knowledge, no studies have been published on the role of BARD1 in meiosis of mammals. In C. elegans depletion of BRCA1 or BARD1 by a RNAi approach, which does not lead to a complete depletion of the protein, resulted in a reduction of germ cell viability of about 20% (Boulton et al, 2004). Thus, the fact that atbrca1 and atbard1 plants seem to be as fertile as wild types was surprising to us. Alexander staining of the pollen from both mutants showed no differences to wild-type pollen, demonstrating that both proteins do not have an indispensable role in meiosis. However, we cannot, of course, exclude minor effects on pollen viability in the percent range. In principle, AtBRCA1 and AtBARD1 could have an influence on meiotic recombination. However, as no functional p53 homologue seems to be present in plants, it might well be that the respective meiocytes survive, and completion of meiotic recombination is achieved by other factors. It will be interesting to test with a recently developed assay system if crossover rates are changed in the mutants (Melamed-Bessudo et al, 2005).
The role of AtBARD1 in homologous DNA recombination in somatic cells
The data presented in this study are, to our knowledge, the first direct proof using bard1−/− mutants that BARD1 is involved in homologous DSB repair in somatic eukaryotic cells. bard1-null mice generated by targeted mutagenesis display a phenotype of early embryonic lethality (McCarthy et al, 2003), eliminating the possibility of a detailed study to the effect on HR. Experiments performed with mouse cells expressing truncated mouse or human BARD1 peptides, capable of interacting with BRCA1, indeed resulted in a deficiency of homologous DNA repair. Repair was mildly reduced in BRCA1 wild-type cells and severely reduced in cells that harbour a BRCA1 splice product deleted for exon 11 (Westermark et al, 2003; Stark et al, 2004). However, strictly speaking this approach of negative complementation disturbed the function of the BRCA1/BARD1 heterodimer and not by directly blocking the function of the BARD1 protein.
Interestingly, both atbard1 mutants show a defect in HR with and without induction of genotoxic stress. This is reminiscent to studies on AtERCC1 (Dubest et al, 2004) and AtRAD51C (Abe et al, 2005). AtERCC1 is part of an exonuclease that is involved in removing nonhomologous ends from DSBs during HR in somatic cells (Dubest et al, 2004). AtRAD51C is most probably involved in the resolution of intermediates arising during DSB repair by HR. Whereas these proteins are directly involved in the recombination process by their enzymatic activities, BARD1 as part of an ubitiquin ligase might be more indirectly involved in the process by regulating the activity of respective factor(s) involved in the mechanisms of the reaction. Recently, a defect in the induction of recombination after Flagelin and H2O2 activity was correlated with a mutation in the AtSNM1 gene (Molinier et al, 2004). However, no defect of induction could be observed with bleomycin, so that there must be at least two different regulation cascades involved in the induction of HR after stress, one induced by a pathogen attack, and the other by DNA damage. It will be a challenge of future experiments to define the cascades in detail.
Materials and methods
Characterisation of the A. thaliana insertion mutants
The atbard1 T-DNA insertion lines (SALK_097601 and SALK_031862) as well as the atbrca1 insertion line SALK_014731 were obtained from the Arabidopsis T-DNA collection in Nottingham. The atbrca1 insertion line, Garlic line 916_C09, was obtained from the T-DNA collection of Syngenta Biotechnology Inc. (SBI).
To obtain 651/atbard1 plants, homozygous lines for atbard1 were crossed with plants homozygous for the transgene 651, carrying a scorable recombination substrate (Puchta et al (1995) in C24 background). Siblings homozygous for 651 transgene and homozygous for the AtBARD1 wild-type alleles were used as control lines.
Growth conditions and mutagen test
Seeds of A. thaliana were surface sterilised in 6% sodium hypochlorite for 8 min and rinsed several times with sterile water. Plants were grown in growth chambers at 23°C under white light (16 h light/8 h dark). Sterilised seeds were spread on GM agar containing different concentrations of MMC. Two weeks later, plants were screened for their sensitivity.
Detection of recombination events and calculation of recombination frequency in wild-type and mutant Arabidopsis seedlings
One-week-old seedlings were transferred to Petri dishes containing liquid GM medium. The next day bleomycin was added (Duchefa) to a concentration of 10 μg/ml, 5 days later the seedlings were used for GUS staining. Histochemical staining was performed as described by Swoboda et al (1994). Plants were destained in 70% ethanol. Blue spots were counted under a binocular.
Two-hybrid analysis
Yeast two-hybrid experiments were performed with the yeast strain EGY48 (MATα his3, trp1, ura3 LexAop*6-LEU2), which had been transformed by integration of the linearised reporter plasmid p8op-lacZ into the genome (carrying a lacZ reporter gene under the control of eight LexA operators) (Estojak et al, 1995).
Arabidopsis AtBRCA1 and AtBARD1 sequences, either full-length or truncated, were amplified by PCR as EcoR1 and Xho1/Sal1 fragments and cloned into the EcoR1–Xho1 sites of the bait and prey vectors pGildaBD and pB42AD in order to create an in-frame fusion protein. Primer pairs are listed in the Supplementary data.
Two-hybrid assays were performed as described by Estojak et al (1995) and according to the manufacturer's instructions (Clontech).
BiFC analysis
AtBRCA1 or fragments of AtBRCA1 were fused with the C-terminal part of the YFP protein whereas AtBARD1 was fused to the N-terminal part of YFP. AtBRCA1 full-length, RING and C-terminal fragments as well as the AtBARD1 full-length fragment were cloned into the pMAV-GW-YN and pMAV-GW-YC vectors, respectively, via two-step Gateway cloning (Invitrogen). Primer pairs are listed in the Supplementary data. Constructs were introduced by biolistic transformation into mustard seedlings as described by Holweg et al (2004). Vector combinations (samples and controls) were used (1–2 μg of each plasmid) as described by Stolpe et al (2005). Images were captured on a Zeiss Axioscope 2 microscope with a Zeiss Axiocam video camera and enhanced using Adobe Photoshop 6.0 Software.
RNA isolation and real-time PCR analysis
Total RNA was isolated with the help of the Qiagen Total RNA isolation kit. Next, RNA was treated with RNAse free DNAse, followed by a reverse transcription with MuMLV reverse transcriptase and polydT as first-strand synthesis primer. Real-time PCR analysis was performed as described (Chen et al, 2003). Primers for quantitative RT–PCR are listed in the Supplementary data.
Database screening
Sequence searches were performed using TAIR BLASTP 2.0. Protein sequences were aligned by pileup. Sequence files were exported to ESPript 2.0 at http://prodes.toulouse.inra.fr/ESPript/cgi-bin/nph-ESPript_exe.cgi for box-shading analysis.
Supplementary Material
Supplementary section
Acknowledgments
We thank Sabine Buss for excellent technical assistance, Manfred Focke and Daniela Kobbe for fruitful discussions. Frank Hartung and I-Peng Chen for thorough reading of the manuscript. We also want to thank Rebecca Müller and Thomas Kretsch (both Universität Freiburg) as well as Carola Holweg and Peter Nick (both Universität Karlsruhe) for their assistance in setting up the split YFP assay. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the European Arabidopsis Stock Centre (NASC) as well as Syngenta for providing seeds.
References
- Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, Ichikawa H, Toki S (2005) Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aunoble B, Bernard-Gallon D, Bignon YJ (2001) Regulation of BRCA1 and BRCA2 transcript in response to cisplatin, adriamycin, taxol and ionising radiation is correlated to p53 functional status in ovarian cancer cell lines. Oncol Rep 8: 663–668 [PubMed] [Google Scholar]
- Baer R, Ludwig T (2002) The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev 12: 86–91 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogues in meiosis and DNA repair. Plant J 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2004) Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113: 197–203 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23: 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M (2004) BRCA1/BARD1 orthologues required for DNA repair in Caenorhabditis elegans. Curr Biol 14: 33–39 [DOI] [PubMed] [Google Scholar]
- Bundock P, van Attikum H, Hooykaas P (2002) Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res 30: 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable PL, Wilson CA, Calzone FJ, Rausher FJ III, Scully R, Livingston DM, Li L, Blackwell CB, Futreal PA, Afshari CA (2003) Novel consensus DNA-binding sequence for BRCA1 protein complexes. Mol Carcinog 38: 85–96 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP (1997) From BRCA1 to RAP1:a widespread BRCT module closely associated with DNA repair. FEBS Lett 400: 25–30 [DOI] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H (2003) The transcriptional response of Arabidopsis to genotoxic stress—a high-density colony array study (HDCA). Plant J 35: 771–786 [DOI] [PubMed] [Google Scholar]
- Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin EH, Chen DJ (1999) The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res 434: 75–88 [DOI] [PubMed] [Google Scholar]
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004) Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against apoptotic death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Dubest S, Gallego ME, White CI (2004) Roles of the AtErcc1 protein in recombination. Plant J 39: 334–342 [DOI] [PubMed] [Google Scholar]
- Estojak J, Brent R, Golemis EA (1995) Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol 15: 5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH (2000) Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet 12: 191–194 [DOI] [PubMed] [Google Scholar]
- Harsch A, Marzilli LA, Bunt RC, Stubbe J, Vouros P (2000) Accurate and rapid modeling of iron-bleomycin-induced DNA damage using tethered duplex oligonucleotides and electrospray ionization ion trap mass spectrometric analysis. Nucleic Acids Res 28: 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2004) What comparative genomics tells us about the evolution of the eukaryotic recombination machinery. Curr Genomics 5: 109–121 [Google Scholar]
- Holweg C, Susslin C, Nick P (2004) Capturing in vivo dynamics of the actin cytoskeleton stimulated by auxin or light. Plant Cell Physiol 45: 855–863 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Leung WC, Li J, Dubois-Dauphin M, Harb J, Feki A, Jefford CE, Soriano JV, Jaconi M, Montesano R (2001) Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol Cell 8: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Soriano JV, Vaudan G, Montesano R, Sappino AP (1998) In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J Cell Biol 143: 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ (1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16: 1097–1112 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Joukov V, Chen J, Fox EA, Green JB, Livingston DM (2001) Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc Natl Acad Sci USA 98: 12078–12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Altschul SF, Bork P (1996) BRCA1 protein products… Functional motifs…. Nat Genet 13: 266–268 [DOI] [PubMed] [Google Scholar]
- Lafarge S, Montane MH (2003) Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1:AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res 31: 1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol 138: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamon HJ, Dahlberg W, Azzam EI, Nagasawa H, Muto MG, Little JB (2003) Differing effects of breast cancer 1, early onset (BRCA1) and ataxia-telangiectasia mutated (ATM) mutations on cellular responses to ionizing radiation. Int J Radiat Biol 79: 817–829 [DOI] [PubMed] [Google Scholar]
- McCarthy EE, Celebi JT, Baer R, Ludwig T (2003) Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol 23: 5056–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Yehuda E, Stuitje AR, Levy AA (2005) A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J 43: 458–466 [DOI] [PubMed] [Google Scholar]
- Meza JE, Brzovic PS, King M-C, Klevit RE (1999) Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem 274: 5659–5665 [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- Molinier J, Stamm ME, Hohn B (2004) SNM-dependent recombinational repair of oxidatively induced DNA damage in Arabidopsis thaliana. EMBO Rep 5: 994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H (1996) Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci USA 93: 13595–13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, Kato T, Tabata S, Kurei S, Yoshioka Y, Machida Y, Seki M, Kobayashi M, Shinozaki K, Ichikawa H, Toki S (2005) Arabidopsis Rad51B is important for double-stranded DNA breaks repair in somatic cells. Plant Mol Biol 57: 819–833 [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Puchta H (2005) The repair of double-strand breaks in plants:mechanisms and consequences for genome evolution. J Exp Bot 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Puchta H, Swoboda P, Gal S, Blot M, Hohn B (1995) Somatic intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol Biol 28: 281–292 [DOI] [PubMed] [Google Scholar]
- Puizina J, Sirokyc J, Mokrosc P, Schweizera D, Rihaa K (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Au WW, Henderson BR (2004) Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res 293: 14–21 [DOI] [PubMed] [Google Scholar]
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM (2001) Cancer-predisposing mutations within the RING domain of BRCA1:loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA 98: 5134–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM (1997b) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM (1997a) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy E, Gerard E, Takvorian N, Doutriaux MP (2004) Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Pastink A, Jasin M (2004) Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe T, Susslin C, Marrocco K, Nick P, Kretsch T, Kircher S (2005) In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146 [DOI] [PubMed] [Google Scholar]
- Swoboda P, Gal S, Hohn B, Puchta H (1994) Intrachromosomal homologous recombination in whole plants. EMBO J 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo CI, Argonza R, King M-C (1996) Human, canine and murine BRCA1 genes:sequence comparison among species. Hum Mol Genet 5: 1289–1298 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King M-C (2002) BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA 99: 7560–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME (2003) BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol Cell Biol 23: 7926–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MCW, Hwang LY, Bowcock AM, Baer R (1996) Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 14: 430–440 [DOI] [PubMed] [Google Scholar]
- Xu X, Aprelikova O, Moens P, Deng CX, Furth PA (2003) Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130: 2001–2012 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Hooloman WK, Pavletich NP (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433: 653–657 [DOI] [PubMed] [Google Scholar]
- Yarden RI, Brody LC (2001) Identification of proteins that interact with BRCA1 by Far-Western library screening. J Cell Biochem 83: 521–531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary section


