Abstract
Poly-(ADP-ribose) polymerase-2 (PARP-2) belongs to a large family of enzymes that synthesize and transfer ADP-ribose polymers to acceptor proteins, modifying their functional properties. PARP-2-deficient (Parp-2−/−) cells, similar to Parp-1−/− cells, are sensitive to both ionizing radiation and alkylating agents. Here we show that inactivation of mouse Parp-2, but not Parp-1, produced a two-fold reduction in CD4+CD8+ double-positive (DP) thymocytes associated with decreased DP cell survival. Microarray analyses revealed increased expression of the proapoptotic Bcl-2 family member Noxa in Parp-2−/− DP thymocytes compared to littermate controls. In addition, DP thymocytes from Parp-2−/− have a reduced expression of T-cell receptor (TCR)α and a skewed repertoire of TCRα toward the 5′ Jα segments. Our results show that in the absence of PARP-2, the survival of DP thymocytes undergoing TCRα recombination is compromised despite normal amounts of Bcl-xL. These data suggest a novel role for PARP-2 as an important mediator of T-cell survival during thymopoiesis by preventing the activation of DNA damage-dependent apoptotic response during the multiple rounds of TCRα rearrangements preceding a positively selected TCR.
Keywords: apoptosis, BH3-only proteins, Noxa, PARP-2, TCRα rearrangement
Introduction
Poly(ADP-ribose) polymerases (PARP) constitute a family of enzymes that, using NAD+ as a substrate, synthesize and transfer ADP-ribose polymers onto glutamic acid residues of acceptor proteins (Schreiber et al, 2006). PARP-1, the founding member of the family, and PARP-2 are the only known members whose activity is stimulated by DNA strand interruptions. Their targets are mainly involved in chromatin structure and DNA metabolism such as histones, DNA repair proteins and transcription factors as well as PARP-1 and PARP-2 themselves (Schreiber et al, 2006). In cellular extracts, poly-ADP-ribosylation fully stimulated by DNA strand breaks is mediated mainly by PARP-1, while PARP-2 is responsible for 10–15% of the total activity (Schreiber et al, 2002). As described for Parp-1-deficient (Parp-1−/−) mice, Parp-2−/− mice are sensitive to both ionizing radiation and alkylating agents, thus supporting a role of both proteins in the cellular response to DNA damage (Menissier de Murcia et al, 2003). Likewise, both proteins can heterodimerize and heteromodify each other and can accumulate at similar nuclear sites such as nucleoli (Meder et al, 2005). Moreover, double mutant mice are not viable (Menissier de Murcia et al, 2003), confirming that both proteins possess overlapping functions, although they could also have non-redundant functions that have not been established yet.
We report here that thymocyte numbers in Parp-2−/− mice were diminished to half of those found in age-matched wild-type (WT) or Parp-1−/− mice. Intrathymic T-cell development and homeostasis of thymocytes is ordered by sequential steps involving cell proliferation and apoptosis, which require changes in the gene expression patterns. After arriving as precursors from the bone marrow (BM), highly immature CD4−CD8− double-negative (DN) thymocytes undergo extensive T-cell receptor β (TCRβ) gene rearrangement. A successfully rearranged and expressed TCRβ associates with the pre-TCR and CD3 chains to form the pre-TCR complex (von Boehmer and Fehling, 1997). Consequences of pre-TCR signalling include proliferation and phenotypic alteration to yield the CD4+CD8+ double-positive (DP) thymocytes, the most abundant population in thymus (von Boehmer et al, 1999). DP thymocytes undergo multiple rounds of TCRα gene rearrangements that maximize the chances of forming a functional αβ TCR complex (Petrie et al, 1993; Hawwari et al, 2005). It has been suggested that most DP thymocytes die by neglect because they fail to form a TCR that interacts productively with self-MHC/self-peptide complexes (Werlen et al, 2003). DP thymocytes expressing TCRs that bind self-MHC/self-peptide molecules with low affinity are rescued through positive selection, while negative selection eliminates those with high-affinity TCRs for self-MHC/self-peptide ligands. DP thymocytes rescued through positive selection differentiate into either CD4+ or CD8+ single-positive (SP) thymocytes, which then will seed the peripheral lymphoid tissues (Werlen et al, 2003).
The mechanisms that mediate and regulate thymocyte apoptosis are not fully understood, but it is thought that thymocyte death is controlled by a genetic program that is induced within the dying cell (Ellis et al, 1991; DeRyckere et al, 2003; Schmitz et al, 2003; Zucchelli et al, 2005). This transcriptional alteration could influence the balance in the Bcl-2 family of proteins, which have been involved in apoptosis regulation (Adams and Cory, 2001). Members of the Bcl-2 family fall into three different classes of proteins based on conservation of Bcl-2 homology (BH1–4) domains: multidomain death antagonist proteins (Bcl-2, Bcl-xL, Mcl1, A1, Bcl-w), multidomain death agonist proteins (Bax, Bak, Bok) and BH3-only death agonist proteins (Bad, Bim, Bik, Hrk, Bmf, Noxa, Puma, Bid) (Strasser, 2005). The proapoptotic BH3-only proteins are the most apical regulators of the Bcl-2-mediated death signalling cascade, and are activated by multiple stimuli from inside or outside the cell to initiate the apoptotic response. Some BH3-only proteins such as Puma and Noxa are regulated transcriptionally, whereas others are regulated by post-translational modifications (Puthalakath and Strasser, 2002).
Our results show that Parp-2−/− mice have a reduced number of thymocytes associated with decreased DP cell survival rather than a problem with DN maturation or cell proliferation. Indeed, Parp-2−/− thymocytes were strikingly susceptible to apoptosis induced by in vivo CD3 stimulation. Microarray data revealed an increased expression of the proapoptotic BH3-only protein Noxa in Parp-2−/− DP thymocytes compared to WT. Likewise, we have found a reduced TCRα expression in Parp-2−/− DP thymocytes compared to WT, which is associated with an inefficient Vα to Jα rearrangement. The data suggest a model whereby in the absence of PARP-2, the physiological DNA breaks associated with Vα-to-Jα recombination activate DNA damage-dependent cellular pathways in DP thymocytes leading to transcriptional upregulation of the proapoptotic BH3-only protein Noxa and also, to a lesser extent, the BH3-only protein Puma, which are critical initiators of cell death. Our results provide a novel role for PARP-2 as an important mediator of T-cell survival during thymopoiesis probably participating in the repair of strand breaks made during TCRα rearrangement.
Results
PARP-1 and PARP-2 are expressed in the thymus
The expression and distribution of PARP-1 and PARP-2 in the thymus were evaluated using in situ hybridization experiments with antisense specific probes. Expression of both Parp-1 and Parp-2 genes was particularly high in the cortex and subcapsular region. Expression was also found in the medulla, although it seems to decrease as thymocytes mature. Distribution of the recombinase activating gene-1 (Rag-1) transcript was used for comparison (Figure 1A). Western blot on cellular extracts from DP and SP thymocytes confirms the expression of both proteins in WT mice, with a decreased expression in SP thymocytes compared to DP thymocytes (Figure 1B). As expected, mutant thymocytes were completely devoid of their corresponding proteins (Figure 1B). To evaluate the contribution of PARP-1 and PARP-2 enzymes to the PARP activity in thymocytes, we compared the incorporation of 3H-NAD+ into proteins using permeabilized thymocytes from Parp-1−/−, Parp-2−/− and WT mice. While in vitro oligonucleotide-stimulated PARP activity was not affected in Parp-2−/− thymocytes compared to WT cells, we observed a marked decrease of 3H-NAD+ incorporation into proteins in Parp-1−/− thymocytes (Figure 1C), in agreement with Schreiber et al (2002), who showed similar findings in mouse embryonic fibroblasts lacking either Parp-1 or Parp-2.
Figure 1.
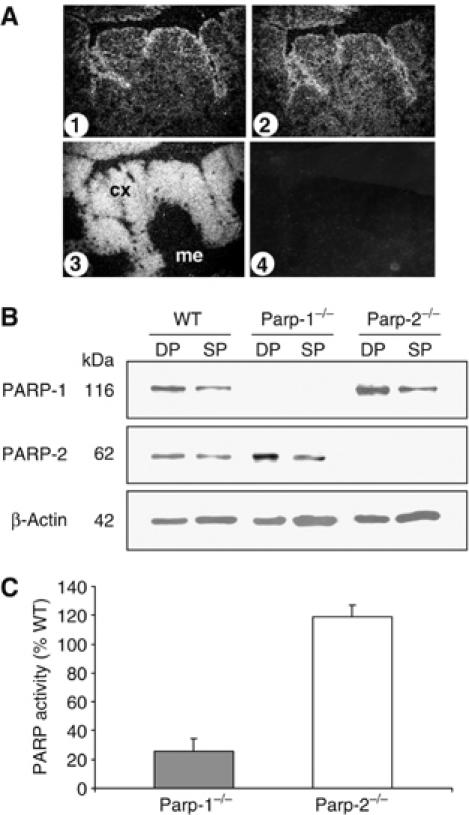
PARP expression and activity in thymocytes. (A) In situ analysis of the distribution of Parp-1 (1) and Parp-2 (2) transcripts in thymus and comparison with the distribution of Rag-1 (3) transcript. (4) Negative staining control. Cx, cortex; me, medulla. (B) Western blot of PARP-1 and PARP-2 protein levels in DP and SP thymocytes from control (WT), Parp-1−/− and Parp-2−/− mice. (C) Relative PARP activity in Parp-1−/− and Parp-2−/− thymocytes. Permeabilized thymocytes were incubated with 3H-NAD+ and a palindromic oligonucleotide for 10 min. Activity is expressed as the percentage of the radioactivity of acid-insoluble material produced by thymocytes from Parp-1−/− or Parp-2−/− compared to WT thymocytes.
Parp-2-deficient mice display reduced thymic cellularity
Based on macroscopic inspection, thymi from Parp-2−/− mice are smaller than those from Parp-1−/− or WT mice (Figure 2A). Total thymocytes from Parp-1−/−, Parp-2−/− and WT mice were counted by hemocytometer, and thymocyte subsets were analyzed by flow cytometry using antibodies specific for CD4 and CD8 T-cell surface antigens. We observed a reproducible and significant two-fold reduction in thymus cellularity in Parp-2−/− mice compared to age-matched Parp-1−/− or WT mice (Figure 2B and C). Figure 2B shows the distribution of thymocytes among DN, DP, CD4SP and CD8SP compartments. The percentage of cells in DP and SP compartments was mostly similar in all mice. However, the percentage of DN thymocytes was slightly increased in Parp-2−/− mice compared to Parp-1−/− or WT mice (Figure 2B). Analysis of absolute number of thymocytes in each population revealed a decrease in the total number of DP, and a decrease in CD4SP and CD8SP cells in Parp-2−/− mice compared with the two other genotypes (Figure 2C). Further examination of CD4SP and CD8SP compartments by analysis of TCRβ expression revealed no differences between WT and Parp-2−/− thymocytes (Figure 2D). Likewise, we found similar expression levels of CD3, CD24, CD5 and MHC class I in the CD8SP compartment of WT and Parp-2−/− thymocytes, indicating normal thymocyte development in Parp-2−/− mice at the immature single-positive (ISP) stage (CD8+CD4−CD3−CD24hiMHC class IloCD5lo) (Figure 2E). In contrast to the reduction of DP and SP thymocyte numbers in Parp-2−/− mice, the absolute number of DN cells was similar to those found in WT and Parp-1−/− mice (Figure 2C). In fact, a closer look at the DN compartment showed a similar distribution at DN1 (CD25−CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44−) and DN4 (CD25−CD44−) stages in both Parp-2−/− and WT mice (Figure 2F). Interestingly, Parp-2−/− BM cells displayed a competitive disadvantage in thymocyte reconstitution compared to Parp-2+/+ counterparts in a Parp-2+/+ thymic environment (Supplementary data and Supplementary Table S-I).
Figure 2.
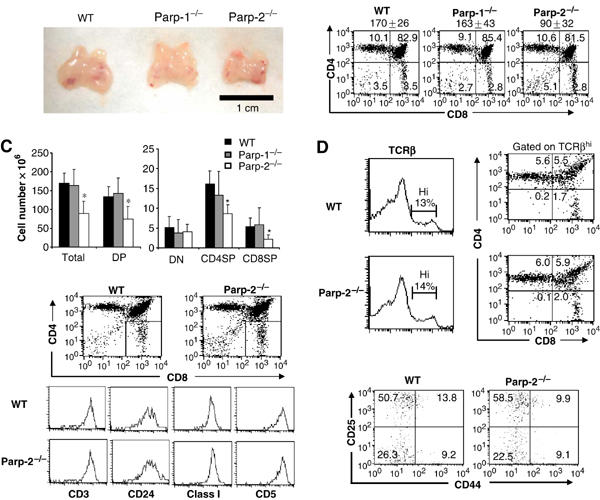
T-cell development. (A) Macroscopic comparison of control (WT), Parp-1−/− and Parp-2−/− thymuses. (B) Single cell suspensions of thymocytes from 6-week-old WT, Parp-1−/− and Parp-2−/− mice were counted, stained with PE-anti-CD4 and FITC-anti-CD8 mAbs and analyzed by flow cytometry. The mean±s.d. of total number of thymocytes in WT (n=45), Parp-1−/− (n=9) and Parp-2−/− (n=45) mice is indicated in the top of each graph. Representative dot plots are shown. Percentage of cells in the individual subpopulations is indicated in each quadrant. (C) Absolute number of thymocyte populations. The number of thymocytes in each population was calculated by multiplying the percentage of each population by the total number of thymocytes. Values represent the mean±s.d. (D) Representative analysis of thymic TCRβ expression in age-matched Parp-2−/− and WT littermates. Thymocytes with high expression of TCRβ were gated and further analyzed for the expression of CD4 and CD8. (E) ISP cells analysis in Parp-2−/− and WT mice. CD8SP thymocytes were gated and analyzed for the expression of CD3, CD24, CD5 and MHC class I. (F) DN thymocytes analysis in Parp-2−/− and WT mice. Thymocytes were stained with mAbs against CD4, CD8, CD25, CD44 and lineage markers (CD3, Gr-1, B220 and CD11b). Thymocytes negative for CD4, CD8 and lineage markers were gated and analyzed for CD25 and CD44 expression. Representative dot plots are shown. The percentage of cells in each quadrant represents the mean from at least six mice in each group. *Statistically significant difference (P<0.05).
We analyzed whether the reduced thymic cellularity in Parp-2−/− mice influences peripheral T-cell population. Parp-2−/− mice had a slightly higher number of total splenocytes than WT mice. Splenic CD4SP and CD8SP T-cell numbers in Parp-2−/− mice were comparable to those in littermate controls (Supplementary Figure S1).
Reduced thymus cellularity in Parp-2−/− mice is not caused by a lower cell proliferation rate
To study whether the deficit in thymocytes was secondary to proliferation defects in cells deficient in Parp-2, we measured in vivo 5-bromo-2′-deoxyuridine (BrdU) incorporation by thymocytes in Parp-2−/− and WT mice. Thymocytes were isolated 5 h after BrdU intraperitoneal (i.p.) injection, stained for surface expression of CD4 and CD8 and the percentage of cells in each subset that had incorporated BrdU was determined by using an antibody against BrdU. As indicated in Figure 3, similar percentages of dividing thymocytes in DN, CD4SP and CD8SP populations are seen in both Parp-2−/− and WT thymocytes. Furthermore, Parp-2−/− mice show a statistically significant increase in both total and DP BrdU-positive thymocytes when compared to WT mice (Figure 3).
Figure 3.
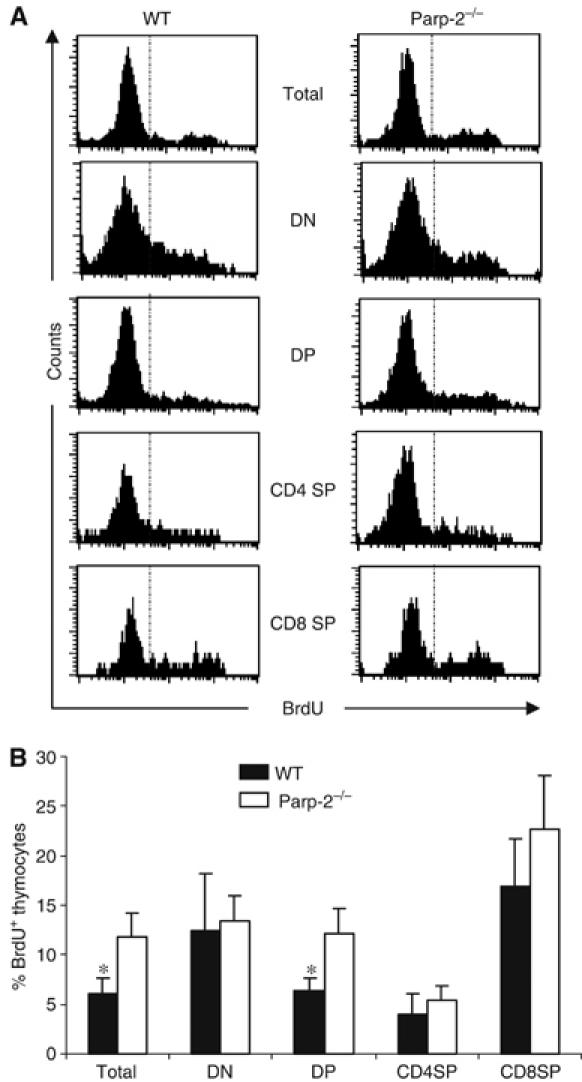
Thymocyte proliferation in control (WT) and Parp-2−/− mice. Six-week-old mice were i.p. injected with BrdU (1.5 mg/mouse) and thymocytes were analyzed by flow cytometry, 5 h after injection, for CD4 and CD8 expression and for the incorporation of BrdU. (A) Representative histograms are shown. (B) Percentage of BrdU+ cells in total, DN, DP, CD4SP and CD8SP thymocytes. Results shown represent the mean±s.d. values obtained from two independent experiments including WT (n=5 per experiment) and Parp-2−/− (n=6 per experiment) mice. *Statistically significant difference (P<0.05).
Parp-2-deficient thymocytes are highly sensitive to apoptosis
An alternative explanation for the decrease in the number of DP thymocytes in Parp-2−/− mice is that DP cells in these mice might be more sensitive to apoptosis, resulting in increased susceptibility to death in response to physiological stimuli. We attempted to establish the intrinsic sensitivity of thymocytes from Parp-2−/− and WT mice to physiological induction of apoptosis by in situ TUNEL assay. As shown in Figure 4, Parp-2−/− thymuses exhibit a significantly higher number of apoptotic cells than WT thymuses. However, the apoptosis detected in vivo or on freshly isolated thymocytes is usually very low because of continuous clearance of apoptotic cells by macrophages (Surh and Sprent, 1994; Savill et al, 2002).
Figure 4.
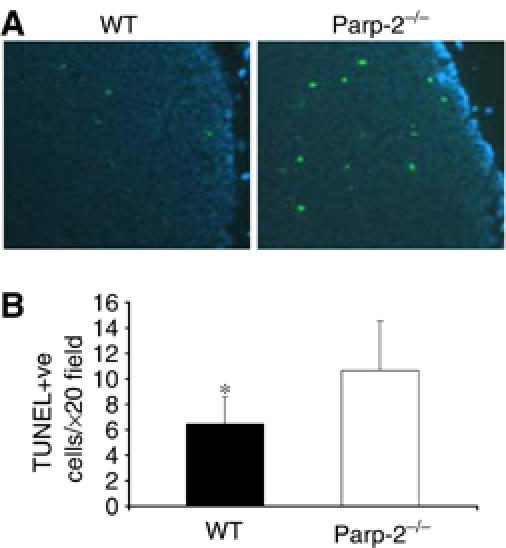
Apoptosis in thymus glands. (A) In situ apoptosis detection in Parp-2−/− and WT mice by the TUNEL assay. Apoptotic cells are labelled in green and cell nuclei are stained with DAPI (blue). (B) Numerical representation of TUNEL-positive (TUNEL +ve) cells. Values represent mean±s.d. for 10 randomly selected cortical fields (n=6 mice per experimental group) (magnification, × 20). *Statistically significant difference (P<0.05).
Therefore, we analyzed the susceptibility of thymocytes from Parp-2−/− and WT mice to cell death by in vivo i.p. administration of anti-CD3 antibody, which has been described to induce rapid depletion of DP thymocytes by different mechanisms such as overstimulation of the TCR and cytokine production (Shi et al, 1991; Page et al, 1998). Thymocyte subsets were assessed by flow cytometry at different times after anti-CD3 injection. As shown in Figure 5A, the percentages of DP thymocytes in Parp-2−/− mice 24 and 48 h after anti-CD3 stimulation were drastically reduced compared to those in WT mice. Analysis of the absolute number of thymocytes in each population revealed a greatly decreased total number of DP cells in Parp-2−/− compared to control mice, while the absolute number of DN thymocytes was increased. CD4SP and CD8SP cells were similar in both mice after stimulation with anti-CD3 (Figure 5A).
Figure 5.
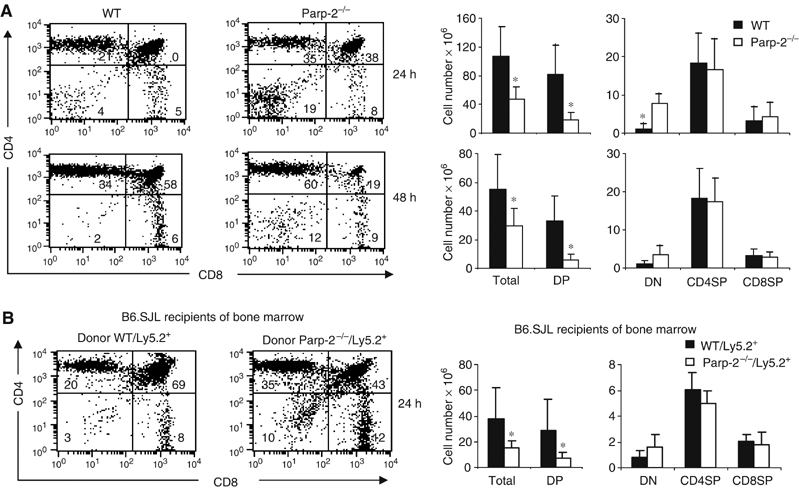
Increased anti-CD3-mediated deletion of Parp-2−/− DP thymocytes. WT and Parp-2−/− mice (A) or irradiated B6.SJL (Ly5.1+) recipient mice of WT or Parp-2−/− BM cells (Ly5.2+), 30 days after transplantation (B) were i.p. injected with anti-CD3 mAb (0.25 μg/g). Freshly isolated thymocytes were analyzed by flow cytometry for CD4 and CD8 expression at different times after anti-CD3 administration. In the case of B6.SJL recipient mice, all cells were additionally stained and gated for the marker of donor cells (Ly5.2). Representative dot plots are shown. The percentage of cells in the individual subpopulations is indicated in each quadrant. Absolute numbers of thymocytes were calculated by multiplying the percentage of each population by the total number of thymocytes. Results shown represent the mean±s.d. values obtained from at least six mice in each group. *Statistically significant difference (P<0.05).
To determine whether the survival defect of Parp-2−/− thymocytes is present within a Parp-2+/+ thymic structure, we also analyzed the susceptibility of thymocytes to anti-CD3-mediated depletion in lethally irradiated Ly5.1+ mice recipients of WT (Ly5.2+) or Parp-2−/− (Ly5.2+) BM cells (Figure 5B). At 30 days after transplantation, the vast majority of thymocytes are derived from the donor animals in both groups (92±7% Ly5.2+ thymocytes among recipients of Parp-2−/− BM cells and 97±2% Ly5.2+ thymocytes among recipients of WT BM cells). As happens with Parp-2−/− mice, recipients of Parp-2−/− BM cells had a two-fold reduction in thymic cellularity compared with the recipients of WT BM cells (data not shown). Twenty-four hours after anti-CD3 injection, the percentage of Ly5.2+Parp-2−/− DP thymocytes was reduced compared to Ly5.2+WT DP thymocytes. Analysis of the absolute number of thymocytes in each population revealed a decrease in the total number of DP cells in the recipients of Parp-2−/− BM cells compared to the recipients of WT BM cells. The absolute number of DN thymocytes was slightly increased whereas CD4SP and CD8SP cells were similar in both recipient mice after stimulation with anti-CD3 (Figure 5B).
Gene expression microarray analysis in DP thymocytes from WT and Parp-2−/− mice
In order to determine whether dis-regulation of a genetic pathway could explain the increased apoptosis of Parp-2−/− DP thymocytes, the gene expression profiles in DP thymocytes derived from WT and Parp-2−/− mice were analyzed using Affymetrix oligonucleotide chips (mouse genome 430 2.0). Only those changes in RNA abundance that were reproducible in three independent experiments using different batches of DP thymocytes and exhibited a >2-fold change between WT and Parp-2−/− were considered. Of over 39 000 genes or expressed sequence tags (ESTs) represented on the array, only 13 transcripts are expressed differentially in the two genotypes (Table I). These included seven known full-length genes and six differentially expressed ESTs. Identification of full cDNAs encoding for these unknown genes, as well as their biological relevance, will be addressed in additional studies. Parp-2 was found to be underexpressed by only 2.5 times in Parp-2−/− DP thymocytes. Reverse transcription (RT)–PCR analysis showed that Parp-2−/− DP thymocytes produced a truncated Parp-2 transcript including at least exons 13–16 (recognized by the Affymetrix probe), which was less abundant than the transcript found in WT DP thymocytes (data not shown). However, Parp-2−/− mice were completely devoid of PARP-2 protein, as judged by Western blot analysis of thymocytes, using a mouse monoclonal antibody against mouse PARP-2 raised in Parp-2−/− mice (Monreal et al, 2006) (Figure 1B) or polyclonal antibodies raised against the catalytic domain or the N-terminal domain of mouse PARP-2 (data not shown). Of the other six known genes differentially expressed between WT and Parp-2−/− DP thymocytes, three genes (Tcra, Gmfb and Rpgrip1) were downregulated in the absence of PARP-2 while the other three genes (Noxa, Supt16 h and Phlda3) were overexpressed in Parp-2−/− compared to WT.
Table 1.
Differential gene expression profile in Parp-2−/− versus WT DP thymocytesa
| Gene name | Description/function | Accession no. | Fold changeb |
|---|---|---|---|
| Noxa | Apoptosis | NM_021451.1 | 3.3 |
| Supt16h | Transcription | BC012433.1 | 2.9 |
| Phlda3 | Death receptor pathway | NM_013750.1 | 2.4 |
| 9030617O03Rik | Unknown | BC021385.1 | 2.5 |
| EST | Unknown | BB184171 | 3.6 |
| EST | Unknown | BC005512 | 2.6 |
| EST | Unknown | BB748708 | 4.3 |
| EST | Unknown | BG066504 | 3.8 |
| Gmfb | Growth factor | BG228815 | −2.2 |
| Parp-2 | Poly-ADP-ribose polymerase | AF072521.1 | −2.5 |
| Rpgrip1 | Protein binding | NM_023879.1 | −18.2 |
| Tcra | Antigen receptor | AK077765 | −10.9 |
| EST | Unknown | AI452290 | −4.7 |
| aGene expression profiles were generated using Affymetrix mouse genome 430 2.0 microarrays. Genes whose expression was increased or decreased (negative value) >2-fold in Parp-2−/− relative to WT DP thymocytes are indicated. | |||
| bResults represent the mean value of three independent microarray experiments using different batches of DP thymocytes. |
The BH3-only protein Noxa is overexpressed in Parp-2−/− DP thymocytes
Our microarray data revealed that Parp-2−/− DP thymocytes have increased transcription of Noxa (3.3-fold) (Table I), a member of the BH3-only Bcl-2 protein family essential as an initiator of apoptosis (Oda et al, 2000). However, the microarray analysis could not detect an altered transcription of other Bcl-2 family members between the genotypes, although there was a mild induction of Puma (1.7-fold increase) in Parp-2−/− compared to WT. Phlda3 mRNA, also known as T-cell death-associated gene 51 (TDAG51), upregulated in Parp-2−/− compared to WT DP thymocytes, has been previously implicated in the regulation of activation-induced cell death in a murine T-cell hybridoma (Oberg et al, 2004). However, the expression of other genes involved in death receptor-mediated apoptosis, including Fas/CD95/APO-1, Fas ligand, TRAIL and TRAIL-receptor DR3, was not altered in our array data.
To validate our array results, we evaluated the expression of Noxa and several other Bcl-2 family members by quantitative real-time PCR in freshly isolated DP thymocytes. We detected increased expression levels (2.6-fold) of Noxa and a small induction of Puma (1.7-fold) in Parp-2−/− versus WT DP thymocytes without changes in other Bcl-2 members analyzed. Thus, our quantitative PCR data correlate well with the differential gene expression data produced using Affymetrix GeneChips (Figure 6A). The increase in the mRNA levels of Noxa in Parp-2−/− DP thymocytes also correlated with a parallel increase in the amount of Noxa protein detected by Western blot (Figure 6B). In contrast, spleen T cells from Parp-2−/− and WT mice had undetectable mRNA expression levels of Noxa (data not shown).
Figure 6.
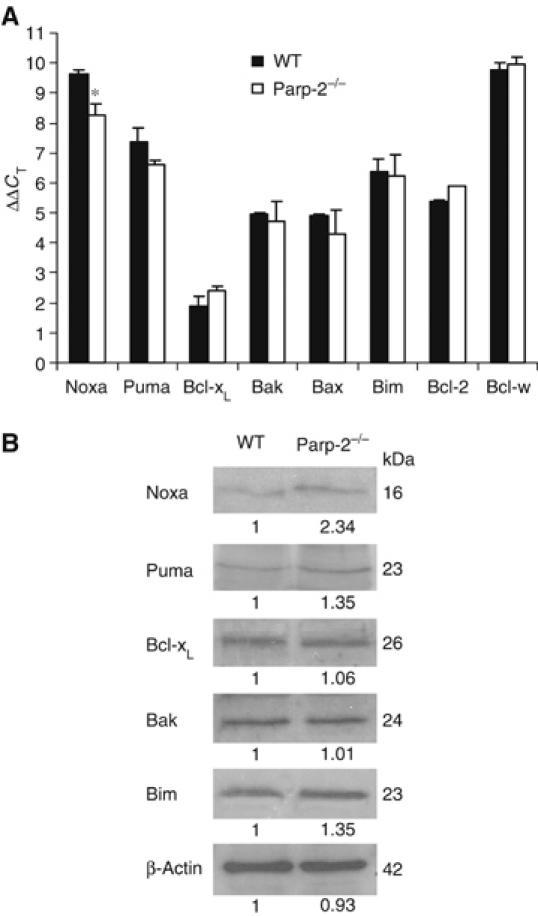
Increased Noxa expression in Parp-2−/− DP thymocytes. (A) Expression of Noxa as well as other Bcl-2 family members in freshly isolated DP thymocytes from WT and Parp-2−/− mice by real-time PCR analysis. Data shown are the average ΔΔCT of three independent experiments carried out in triplicate (n=6 mice per experimental group). * represents a >2-fold difference in expression between WT and Parp-2−/−. (B) Western blot expression analysis of Bcl-2 family member proteins in DP thymocytes from WT and Parp-2−/− mice. The numbers below each band indicate densitometry analysis of the relative amount of each protein taking into account the abundance of β-actin in both samples. The data are representative of two independent experiments.
TCRα expression is impaired and the Jα usage is skewed in Parp-2−/− DP thymocytes
The microarray analyses also revealed a profound reduction in the levels of TCRα mRNA (10.9-fold decrease) in Parp-2−/− DP thymocytes without any change in TCRβ expression levels (Table I). Quantitative real-time PCR from purified DP thymocytes confirmed the array data showing a 4.9-fold decrease of TCRα transcripts in Parp-2−/− DP thymocytes compared to WT, again with no effect on the expression levels of TCRβ (Figure 7A).
Figure 7.
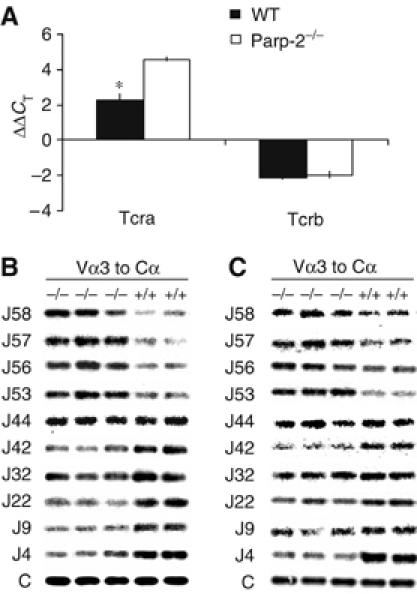
Impaired TCRα expression and skewed Jα usage in Parp-2−/− DP thymocytes. (A) Real-time PCR expression analysis of Tcra and Tcrb in freshly isolated DP thymocytes from WT and Parp-2−/− mice. Data shown are the average ΔΔCT of three independent experiments carried out in triplicate (n=6 mice per experimental group). * represents a >2-fold difference in expression between WT and Parp-2−/−. (B) Profiles of Jα usage in DP thymocytes from two WT and three Parp-2−/− mice. RT–PCR was performed with primers specific for Vα3 and Cα using RNA from purified DP thymocytes. PCR products were probed sequentially with specific Jα oligonucleotide probes as indicated. Probing with an internal Cα probe was used to normalize the input RNA. (C) Profiles of Jα usage in splenic T cells from WT and Parp-2−/− mice.
Assembly of the gene encoding TCRα is characterized by an orderly progression of primary Vα to Jα rearrangements, followed by several rounds of secondary recombination events. Jα rearrangements proceed 5′ to 3′, whereas Vα rearrangements proceed 3′ to 5′ (Thompson et al, 1990; Guo et al, 2002). Rearrangements of the closest segments are attempted first (the most 3′ Vαs rearranging to the most 5′ Jαs), progressing to more and more distant Vα to Jα recombination (Huang and Kanagawa, 2001; Pasqual et al, 2002; Hawwari et al, 2005). To define the effect of Parp-2 deletion on Vα–Jα rearrangement, we compared the pattern of Jα usage in transcripts from Parp-2−/− and WT DP thymocytes. TCRα-specific transcripts were reverse transcribed and PCR amplified using primers specific for the Vα3 family in conjunction with a Cα. Southern blots of the PCR products obtained from two WT and three Parp-2−/− mice were hybridized with a series of oligonucleotide probes, each representing a specific Jα segment ranging from the most 5′ to the most 3′ Jα. An internal Cα probe was used to assess the quantity of total Vα–Cα PCR products (Figure 7B). Comparison of Jα usage between Parp-2−/− and WT DP thymocytes revealed that the Jα usage in Parp-2−/− mice is skewed toward the 5′ Jα segments. Thus, the most 5′ Jα segments, 58 through 53, were over-represented in Parp-2−/− mice compared to controls. In contrast, the Jα segment 44 usage was comparable in both Parp-2−/− and WT mice, whereas usage of Jα segments 42 to 4 was under-represented in Parp-2−/− mice (Figure 7B). Altogether, these data suggest that Parp-2 deletion results in reduced expression of TCRα and a skewed repertoire of TCRα toward the 5′ Jα segments. Analysis of Jα usage in Parp-2−/− splenic T cells gave similar results to those found in DP thymocytes (Figure 7C).
Parp-2−/− DP thymocytes are highly susceptible to apoptosis induced by p53-dependent but not by p53-independent stimuli
The Vα to Jα recombination process is mediated by the RAG-1 and RAG-2 recombinases and involves the generation of DNA double-strand breaks (DSBs) between TCR encoding gene segments and flanking recombination signal sequences (Bassing et al, 2002). DSBs trigger a p53-mediated signalling pathway that halts the cell cycle and thus allows DNA repair. In thymocytes, p53 is mandatory for the induction of an early apoptotic response to DNA damage, as indicated by the resistance of thymocytes derived from p53-null mice to irradiation-induced apoptosis (Clarke et al, 1993; Lowe et al, 1993). However, apoptosis is observed in p53−/− thymocytes responding to phorbol ester (phorbol 12-myristate 13-acetate) or calcium ionophore (Clarke et al, 1993; Lowe et al, 1993), indicating that at least two independent apoptosis-inducing pathways are present in developing thymocytes. We analyzed the sensitivity of Parp-2−/− and WT DP thymocytes to apoptosis induced in vitro by both p53-dependent (X-ray irradiation and etoposide) and p53-independent (ionomycin, staurosporine) stimuli. Parp-2−/− DP thymocytes were markedly more susceptible to apoptosis induced by irradiation and, to a lesser extent, to etoposide, than control thymocytes. No differences were observed in the apoptosis susceptibility to p53-independent stimuli (Figure 8).
Figure 8.
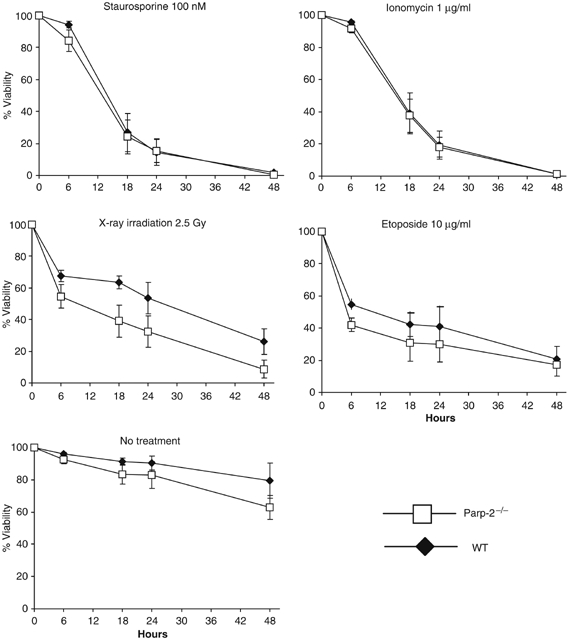
p53-dependent and p53-independent cell death response in DP thymocytes. Isolated DP thymocytes from 6-week-old WT and Parp-2−/− mice were subjected to cell death stimuli and cultured for the indicated times. Apoptosis was detected by FITC-Annexin-V and propidium iodide staining and flow cytometric analysis. Each point represents the mean±s.d. values obtained from three independent experiments including WT (n=4 in each experiment) and Parp-2−/− (n=4 in each experiment) mice.
Discussion
Here, we have found that genetic inactivation of Parp-2, but not of Parp-1, affects thymic development, providing the first evidence for an important and not redundant role of the PARP-2 protein during thymocyte maturation. A two-fold reduction in DP thymocyte numbers without any change in DN numbers, normal distribution of DN subtypes and normal ISP population suggest that disruption of PARP-2 function does not affect either DN population or the DN to DP transition. Interestingly, Parp-2−/− thymocytes show increased BrdU incorporation, suggesting that the reduction of thymocyte numbers in Parp-2−/− thymi would be associated with a decreased DP cell survival instead of a proliferation defect. This observation supports previous data where enhanced thymocyte apoptosis was accompanied by an increase in the number of cycling thymocytes (Brady et al, 1996; Luke et al, 2003). In the attempt to increase the number of thymocytes, the increased proliferation could contribute to the increased proportion of both DP cycling Parp-2−/− thymocytes in homeostatic conditions and DN Parp-2−/− population in anti-CD3-treated mice.
Our results indicate increased in vivo apoptosis in thymocytes from Parp-2−/− compared to WT mice. In addition, using a BM transplantation approach, we observed that the defect in survival is present in the Parp-2−/− thymocytes within a Parp-2+/+ thymic structure. These data point to an abnormal regulation of programmed cell death of thymocytes in the absence of PARP-2. Here, we have used a large-scale gene expression analysis to identify differentially expressed genes between DP thymocytes from WT and Parp-2−/− mice that might function as components of the death signalling pathway. Our microarray analyses revealed differences in 13 transcripts between the two genotypes from which seven corresponded to known genes. Interestingly, the microarray analyses revealed a decreased expression of TCRα in Parp-2−/− DP thymocytes. Defects in TCRα expression have been ascribed to DP thymocyte death soon after the initiation of TCRα rearrangement (Guo et al, 2002; Xi and Kersh, 2004), hinting at the possibility that inefficient TCRα rearrangement could result from the impaired survival of thymocytes at the DP stage in the absence of PARP-2.
DP cells have an intrinsic lifespan of 3–4 days during which they undergo multiple rounds of TCRα rearrangements to maximize the chance of forming a functional TCR that allows positive selection (Petrie et al, 1993; Wang et al, 1998). The skewed repertoire of TCRα toward the 5′ Jα proximal segments in Parp-2−/− DP thymocytes suggested that these cells have an inefficient Vα to Jα rearrangement and therefore a shorter lifespan. This results in fewer chances to produce a functional TCRα chain that pairs with the already rearranged TCRβ chain. Although a skewed repertoire of TCRα toward the 5′ Jα proximal segments and a reduced lifespan of DP thymocytes has also been described in other mouse models such as RORγ-null mice (Guo et al, 2002) or transgenic mice that constitutively express Egr3 (Xi and Kersh, 2004), a distinct mechanism seem to operate in Parp-2−/− mice that is not mediated by differences in the expression of Bcl-xL. Accordingly, the number of ISP thymocytes was unaffected by the absence of PARP-2.
Interestingly, one of the genes overexpressed in Parp-2−/− DP thymocytes is the proapoptotic BH3-only protein Noxa. Noxa is a member of the Bcl-2 family important for the initiation of apoptotic cell death in response to DNA damage and its expression is regulated at transcriptional level by both p53-dependent (Oda et al, 2000; Shibue et al, 2003; Villunger et al, 2003) and p53-independent (Sun and Leaman, 2005) pathways. Therefore, we have studied the susceptibility of Parp-2−/− DP thymocytes to several p53-dependent or p53-independent proapoptotic stimuli. Our results suggest that the absence of PARP-2 made DP thymocytes much more susceptible to in vitro-induced apoptosis by p53-dependent pathways. Noxa binds to and inhibits prosurvival Bcl-2-family members such as A1 and Mcl1. Engagement of Mcl-1 by Noxa then displaces Bak from Mcl-1, which activates the subsequent cascade of apoptotic events. While Noxa is not essential for efficient apoptosis in response to p53-dependent signals since Noxa−/− thymocytes undergo apoptosis as efficiently as WT thymocytes in response to several proapoptotic p53-dependent signals (Shibue et al, 2003; Villunger et al, 2003), our data suggest a role for Noxa in mediating the PARP-2-dependent apoptotic pathway. In contrast, loss of Puma, which binds at least five different antiapoptotic relatives (Gélinas and White, 2005; Willis and Adams, 2005), renders thymocytes resistant to γ-radiation and etoposide (Villunger et al, 2003). Interestingly, Puma mRNA was upregulated 1.7-fold in Parp-2−/− DP thymocytes as compared with littermates. Thymocyte survival seems to be regulated by the balance between BH3-only proteins and their prosurvival relatives. Therefore, the increased expression of Noxa and, to a lesser extent, Puma in Parp-2−/− DP thymocytes might tilt the balance toward apoptosis and reduced lifespan.
The increased expression of Noxa and Puma is associated with a clear impairment of the TCRα rearrangements, perhaps as a consequence of defects in the repair of a particularly heavy burden of DSBs generated in the DP thymocyte population by the physiological Vα to Jα sequential rearrangement events. DNA rearrangement is a very dangerous process, and it must be carefully controlled to prevent the generation of random breaks in the DNA. Regulation takes place at different levels: expression of the RAG1/2 recombinase, intrinsic biochemical properties of the cleavage reaction, the post-cleavage DNA repair stage of the process and accessibility of the substrate to the recombinase (Oettinger, 2004). In fact, it has been reported that DP thymocytes are more resistant than mature T cells to DNA damage intercalating agents, suggesting that this could reflect their ‘tolerance' to DSBs that occur normally during antigen receptor gene rearrangement (Bhandoola et al, 2000).
We propose a model whereby PARP-2 might participate as a component of molecular complexes with other molecules involved in the tolerance to the constant threat for induced DNA breaks in DP thymocytes, by mechanisms that could involve chromatin accessibility and/or DNA repair pathways. In the absence of PARP-2, the ‘tolerance' to DSBs induced by Vα to Jα rearrangements is altered and the physiological DNA damage-induced cellular pathway leads to transcriptional upregulation of the proapoptotic protein Noxa and to a lesser extent Puma, critical initiators of cell death. The absence of secondary rearrangements on the TCRβ gene might explain why DN to DP differentiation was not impaired or DN half-life was not reduced in Parp-2−/− mice. Therefore, increased apoptosis would preferentially affect the lifespan of DP thymocytes and as a consequence would impair secondary Vα to Jα rearrangements (Figure 9). Thus, Parp-2−/− mice may prove useful in future studies that aim to dissect the molecular events that regulate the mechanisms responsible for the progression of primary to secondary Vα to Jα recombination events and their link with the control of cell programmed death in DP thymocytes.
Figure 9.

Model for increased apoptosis of DP thymocytes in Parp-2−/− mice. In the absence of PARP-2, the repair of DSBs generated during Vα to Jα rearrangements could be affected. This could activate a physiological DNA damage-induced cellular apoptotic pathway leading to transcriptional upregulation of the proapoptotic BH3-only proteins Noxa and, to a lesser extent, Puma, which are critical initiators of cell death. Increased apoptosis would affect the lifespan of DP thymocytes and as a consequence impair secondary Vα to Jα rearrangements.
In summary, we describe here the effect of PARP-2 deficiency in thymic development, which results in reduced thymic cellularity as a result of decreased DP cell survival. The developmental defect in DP thymocytes is mediated by increased expression of the proapoptotic protein Noxa and to a lesser extent Puma. Our data provide the first link between PARP-2, Vα to Jα rearrangement and apoptosis mediated by the BH3-only protein Noxa, and open new avenues for exploration into how this PARP family member interacts to control apoptosis associated to V(D)J recombination and the role of Noxa in the control of lymphocyte development and homeostasis.
Materials and methods
Mice
The generation of Parp-1−/− and Parp-2−/− mouse strains (129/Sv × C57BL/6) has been described previously (Menissier de Murcia et al, 1997, 2003). Genotyping was performed by PCR analysis using tail DNA, as described previously (Menissier de Murcia et al, 2003; Corral et al, 2005). B6.SJL mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All animal studies were carried out in accordance with the European Union regulations for animal experimentation.
Western blot
Western blot analysis was performed as described previously (Menissier de Murcia et al, 1997).
In situ hybridization
Detection of Parp-1, Parp-2 and Rag-1 transcripts by in situ hybridization was performed on serial sections (10 μm thick) of frozen organs dissected from 16-week-old CD1 mice, as described previously (Schreiber et al, 2002). The Rag-1 probe was kindly provided by Dr N Labreque (IGBMC, Illkirch, France). Exposure was for 1 week for Rag-1 probe and 4 weeks for Parp-1 and Parp-2 probes.
PARP assay
PARP activity in freshly isolated thymocytes was determined as described previously (Muiras et al, 1998).
TUNEL
Thymuses from 6-week-old mice were fixed in 4% paraformaldehyde, and 4-μm-thick paraffin-embedded sections were mounted on glass slides, deparaffinized and dehydrated. The TUNEL assay was performed using a Dead-End TUNEL kit (Promega, Madison, WI) according to the manufacturer's protocol.
BM transplantation
BM cells (5 × 106 cells/recipient) from WT or Parp-2−/− mice (Ly5.2+) were infused into B6.SJL mice (Ly5.1+) that received two doses of 4.25 Gy, spaced 24 h, using an MG325 X-ray equipment (300 kV, 12.8 mA; Philips, Hamburg, Germany).
Flow cytometry
Cell suspensions were washed in PBS, resuspended in PBS containing 0.5% BSA and incubated with antibodies on ice for 30 min. Incubation with biotin-labelled antibodies was followed by incubation with streptavidin-Tri-color conjugate. All antibodies used are given in Supplementary data. For in vivo BrdU labelling experiments, mice were i.p. injected with BrdU (Pharmigen, San Diego, CA) (1.5 mg/mouse). Five hours after injection, thymocytes were isolated and surface stained with anti-CD8-biotin, washed in PBS and incubated with anti-CD4-PE and streptavidin-Tri-color. Cells were then fixed, permeabilized and intracellularly stained with anti-BrdU-FITC using a BrdU flow kit (Pharmigen). In experiments where apoptosis was triggered in vivo, mice were challenged with a single i.p. injection of anti-CD3 mAb (0.25 μg/g). All cell analysis was performed with a FACSort flow cytometer and CellQuest software (BD).
Cell sorting
Thymocytes were isolated and surface stained with anti-CD8 and anti-CD4 as indicated above. CD4+/CD8+ DP cells or CD4SP plus CD8SP cells were collected by fluorescence-activated cell sorting using a MoFlo® cell sorter (Cytomation Inc., Fort Collins, CO). All reagents used during the isolation procedure were at 4°C and cells were sorted into a receptacle on ice. In all cases, purity of collected cells was more than 99%.
Mature T cells were isolated from the spleen of Parp-2−/− and WT mice by magnetic depletion of non-T cells using a MACS Pan-T Cell isolation kit, according to the manufacturer's instructions (Mintenyi Biotec, Bergisch Gladbach, Germany).
Apoptosis detection
DP thymocytes were resuspended in DMEM medium (Biowhittaker, Verviers, Belgium) supplemented with 10% fetal bovine serum (Sigma, St Louis, MO), treated or not with X-ray irradiation (2.5 Gy) and plated in a 24-well tissue culture plate. No irradiated cells were treated with staurosporine (100 nM), ionomycin (1 μg/ml) or etoposide (10 μg/ml) (all from Sigma). At the indicated time points, cells were washed with PBS and the presence of apoptotic cells was analyzed by flow cytometry using the annexin V-FITC apoptosis detection kit (Pharmigen) according to the manufacturer's instructions.
Microarrays
Total RNA was isolated from WT and Parp-2−/− DP thymocytes by using the Rneasy Total RNA Isolation kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Total RNA was subjected to reverse transcription using the One Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA), containing a T77-(dT)24 primer and a T7 RNA polymerase promoter site. Biotinylated cRNA was obtained, purified and fragmented as per the instructions of Affymetrix. Fifteen micrograms of the in vitro transcripts with appropriate controls and spikes was hybridized at 45°C for 16 h with rotation in the Affymetrix GeneChip Hyb Oven 640 to an Affymetrix mouse genome 430 2.0 microarray, which contains probes for over 39 000 known genes and ESTs. Chips were washed and stained with streptavidin-phycoerythrin in the Fluidics Station 450 (Affymetrix) using the protocol EukGE-WS2-v5 provided by Affymetrix. Chips were scanned in an Agilent G3000 GeneArray Scanner. CEL files were imported into the ArrayAsist package (Stratagene) and pre-processed using the robust multiarray analysis algorithm with the default parameters (Irizarry et al, 2003). Only those differences in RNA abundance that were reproducible in three independent experiments and represented a change of two-fold or greater were considered.
Quantitative RT–PCR
RNA isolation, cDNA synthesis and quantitative real-time PCR were performed as described previously (Carrillo et al, 2004). Specific primers to the different genes are described in Supplementary Table S-II. Each gene was normalized to the housekeeping Gapdh gene. The relative changes in gene expression were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Briefly, cycle threshold (CT) value means the number of PCR cycles required for the detection of fluorescence signal to exceed a fixed threshold. The data were analyzed using ΔΔCT=CT target–CT gapdh. With this calculation, ΔΔCT is equal to log2 ratio; therefore, value 1 of ΔΔCT corresponds to a two-fold change. Only genes that showed over a two-fold change were considered significant.
Southern blot analysis
Amplifications of TCRα transcripts were performed from WT and Parp-2−/− DP thymocytes and splenic T cells cDNA using the MVAF3 (sense Vα3) and MCA1 (antisense Cα) primers as previously indicated (Villey et al, 1996). PCR products were electrophoresed on 1.5% agarose gel and transferred to a Hybond N+ nylon membrane (Amersham Biosciences, Buckinghamshire, UK). Hybridizations were carried out with γ32P-end-labelled Jα-specific oligonucleotide probes previously described (Villey et al, 1996; Riegert and Gilfillan, 1999). Analysis was performed on a PhosphorImager using ImageQuant 5.2 software (Amersham Biosciences). For sequential hybridization, blots were stripped by boiling the membranes three times for 5 min. Each was confirmed by exposing the membranes to the PhosphorImager.
Statistical analysis
Data are expressed as mean±s.d. Statistical differences in the results for the different parameters between groups were evaluated by the two-tailed Student's t-test. A probability (P) value of <0.05 was considered significant.
Supplementary Material
Supplementary data
Acknowledgments
We thank Dr C Rada for critical reading of the manuscript, A Huber for assistance with mice, Juan A Bueren for useful comments and F Núñez for assistance with the microarray analysis. This work was supported by Fundación Séneca (grants PB01FS02, 00603/PI/04), Instituto de Salud Carlos III (grants C03/02, PI051013), Spanish Ministerio de Educación y Ciencia (grants HF2004-0032, SAF2003-310, BIO-2005-01393) and funds from Centre National de la Recherche Scientifique, Association pour la Recherche contre le Cancer, Electricité de France, Comité du Haut-Rhin de la Ligue Nationale Contre le Cancer and Commissariat à l'Energie Atomique. JY and EA are Investigators from the Ramón y Cajal Program.
Conflict of interest The authors have no conflict of interest relative to the data reported in this manuscript.
References
- Adams JM, Cory S (2001) Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci 26: 61–66 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109: S45–S55 [DOI] [PubMed] [Google Scholar]
- Bhandoola A, Dolnick B, Fayad N, Nussenzweig A, Singer A (2000) Immature thymocytes undergoing receptor rearrangements are resistant to an Atm-dependent death pathway activated in mature T cells by double-stranded DNA breaks. J Exp Med 192: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady HJM, Gil-Gomez G, Kirberg J, Berns AJ (1996) Bax alpha perturbs T cell development and affects cell cycle entry of T cells. EMBO J 15: 6991–7001 [PMC free article] [PubMed] [Google Scholar]
- Carrillo A, Monreal Y, Ramírez P, Marín L, Parrilla P, Oliver F J, Yélamos J (2004) Transcription regulation of TNF-α-early response genes by Poly(ADP-ribose) polymerase-1 in murine heart endothelial cells. Nucleic Acids Res 32: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH (1993) Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362: 849–852 [DOI] [PubMed] [Google Scholar]
- Corral J, Yélamos J, Hernández-Espinosa D, Monreal Y, Mota R, Arcas I, Miñano A, Parrilla P, Vicente V (2005) Role of lipopolysaccharide and cecal ligation and puncture on blood coagulation and inflammation in sensitive and resistant mice models. Am J Pathol 166: 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRyckere D, Mann DL, DeGregori J (2003) Characterization of transcriptional regulation during negative selection in vivo. J Immunol 171: 802–811 [DOI] [PubMed] [Google Scholar]
- Ellis RE, Yuan JY, Horvitz HR (1991) Mechanisms and functions of cell death. Annu Rev Cell Biol 7: 663–698 [DOI] [PubMed] [Google Scholar]
- Gélinas C, White E (2005) BH3-only proteins in control: specificity regulates MCL-1 and BAK-mediated apoptosis. Genes Dev 19: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW (2002) Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat Immunol 3: 469–476 [DOI] [PubMed] [Google Scholar]
- Hawwari A, Bock C, Krangel MS (2005) Regulation of T cell receptor alpha gene assembly by a complex hierarchy of germline Jalpha promoters. Nat Immunol 6: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Kanagawa O (2001) Ordered and coordinated rearrangement of the TCR α locus: role of secondary rearrangement in thymic selection. J Immunol 166: 2597–2601 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362: 847–849 [DOI] [PubMed] [Google Scholar]
- Luke JJ, van de Wetering C I, Knudson CM (2003) Lymphoma development in Bax transgenic mice is inhibited by Bcl-2 and associated with chromosomal instability. Cell Death Differ 10: 740–748 [DOI] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, Schreiber V (2005) PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci 118: 211–222 [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G (1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA 94: 7303–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22: 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monreal Y, Menissier de Murcia J, Yélamos J (2006) Anti-Poly-ADP-ribose polymerase-2 (PARP-2) mouse mAb 4G8. Hybridoma 25: 102 [Google Scholar]
- Muiras ML, Muller M, Schachter F, Bürkle A (1998) Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J Mol Med 76: 346–354 [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Oberg HH, Sipos B, Kalthoff H, Janssen O, Kabelitz D (2004) Regulation of T-cell death-associated gene 51 (TDAG51) expression in human T-cells. Cell Death Differ 11: 674–684 [DOI] [PubMed] [Google Scholar]
- Oettinger MA (2004) How to keep V(D)J recombination under control. Immunol Rev 200: 165–181 [DOI] [PubMed] [Google Scholar]
- Page DM, Roberts EM, Peschon JJ, Hedrick SM (1998) TNF receptor-deficient mice reveal striking differences between several models of thymocyte negative selection. J Immunol 160: 120–133 [PubMed] [Google Scholar]
- Pasqual N, Gallagher C, Aude-Garcia C, Loiodice M, Thuderoz F, Demengeot J, Ceredig R, Marche PN, Jouvin-Marche E (2002) Quantitative and qualitative changes in V–J α rearrangements during murine thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J Exp Med 196: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN, Shortman K (1993) Multiple rearrangements in T cell receptor α chain genes maximize the production of useful thymocytes. J Exp Med 178: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A (2002) Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 9: 505–512 [DOI] [PubMed] [Google Scholar]
- Riegert P, Gilfillan S (1999) A conserved sequence block in the murine and human TCR J alpha region: assessment of regulatory function in vivo. J Immunol 162: 3471–3480 [PubMed] [Google Scholar]
- Savill J, Dransfield I, Gregory C, Haslett C (2002) A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2: 965–975 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier de Murcia J, de Murcia G (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277: 23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Amé JC, de Murcia G (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Shi Y, Bissonnette RP, Parfrey N, Szalay M, Kubo RT, Green DR (1991) In vivo administration of monoclonal antibodies to the CD3T cell receptor complex induces cell death (apoptosis) in immature thymocytes. J Immunol 146: 3340–3346 [PubMed] [Google Scholar]
- Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, Morishita Y, Akira S, Taniguchi T, Tanaka N (2003) Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 17: 2233–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz I, Clayton LK, Reinherz EL (2003) Gene expression analysis of thymocyte selection in vivo. Int Immunol 15: 1237–1248 [DOI] [PubMed] [Google Scholar]
- Strasser A (2005) The role of BH3-only proteins in the immune system. Nat Rev Immunol 5: 189–200 [DOI] [PubMed] [Google Scholar]
- Sun Y, Leaman DW (2005) Involvement of noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J Biol Chem 280: 15561–15568 [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J (1994) T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 372: 100–103 [DOI] [PubMed] [Google Scholar]
- Thompson SD, Pelkonen J, Hurwitz JL (1990) First T cell receptor alpha gene rearrangements during T cell ontogeny skew to the 5′ region of the J alpha locus. J Immunol 145: 2347–2352 [PubMed] [Google Scholar]
- Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP (1996) Defect in rearrangement of the most 5′TCR-Jα following targeted deletion of T early α (TEA): implications for TCRα locus accessibility. Immunity 5: 331–342 [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Müllauer F, Böck G, Ausserlechner MJ, Adams JM, Strasser A (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Aifantis HI, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, Azogui O (1999) Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol 11: 135–142 [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Fehling HJ (1997) Structure and function of the pre-T cell receptor. Annu Rev Immunol 15: 433–452 [DOI] [PubMed] [Google Scholar]
- Wang F, Huang CY, Kanagawa O (1998) Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha–Jalpha segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA 95: 11834–11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werlen G, Hausmann B, Naeher D, Palmer E (2003) Signaling life and death in the thymus: timing is everything. Science 299: 1859–1863 [DOI] [PubMed] [Google Scholar]
- Willis SN, Adams JM (2005) Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol 17: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Kersh GJ (2004) Sustained early growth response gene 3 expression inhibits the survival of CD4/CD8 double-positive thymocytes. J Immunol 173: 340–348 [DOI] [PubMed] [Google Scholar]
- Zucchelli S, Holler P, Yamagata T, Roy M, Benoist C, Mathis D (2005) Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 22: 385–396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


