Abstract
Global cellular responses induced by epidermal growth factor (EGF) receptor (EGFR) occur immediately with a less than 1% occupancy among tens of thousands of EGFR molecules on single cell surface. Activation of EGFR requires the formation of a signaling dimer of EGFR bound with a single ligand to each molecule. How sufficient numbers of signaling dimers are formed at such low occupancy rate is still not known. Here, we have analyzed the kinetics of EGF binding and the formation of the signaling dimer using single-molecule imaging and mathematical modeling. A small number of EGFR on the cell surface formed dimeric binding sites, which bound EGF two orders of magnitude faster than the monomeric binding sites. There was a positive cooperative binding of EGF to the dimeric binding sites through a newly discovered kinetic intermediate. These two mechanisms facilitate the formation of signaling dimers of EGF/EGFR complexes.
Keywords: dimerization, EGF, mathematical modeling, signal transduction
Introduction
Epidermal growth factor (EGF) receptor (EGFR) belongs to the ErbB family of receptor tyrosine kinases responsible for cell proliferation, differentiation and migration in embryogenesis and carcinogenesis (Carpenter, 1987; Schlessinger, 2000). Binding of the ligand and formation of the signaling dimer are the initial and critical events in the EGF signaling pathway. In the signaling dimer of EGFR composed of two ligand and two receptor molecules, the intrinsic tyrosine kinase activity of EGFR is enhanced and induces the phosphorylation of tyrosine residues in the carboxy-terminal domain of EGFR, providing scaffolds for various cytoplasmic signaling proteins (Schlessinger, 2002).
The process of formation of EGF/EGFR complexes is not fully understood. Cells sensitive to EGF typically express several tens of thousands of EGFRs on the plasma membrane. EGF-induced global responses in cells, like the calcium response or loss of affinity to the ligand, were observed within 1 min after application of EGF at less than 1% occupancy of EGFRs (Wiley et al, 1989; Uyemura et al, 2005). Recently, it was demonstrated that the EGFR has the ability to form dimers independently of ligand binding (predimer) on the cell surface (Moriki et al, 2001; Yu et al, 2002). Predimers appear to be responsible for the effective formation of signaling dimers. However, assuming simple association reactions, the probability of signaling dimer formation with an occupancy rate of less than 1% would be too low to induce global cellular responses. Under simple association, even if all EGFR molecules present as predimers, only a few signaling dimers will be formed on cells expressing 50 000 EGFR molecules with a 1% occupancy rate.
In fact, the association between EGF and EGFR is not simple. Many studies have demonstrated that EGFRs on the cell surface exhibit two apparently different affinities to EGF in the Scatchard plot analysis; the majority have low affinity, whereas only a small number exhibit high affinity (Shoyab et al, 1979; King and Cuatrecasas, 1982; Krupp et al, 1982; Boonstra et al, 1985). Recent crystallographic studies on the extracellular domain of EGFR have shown the presence of two configurations (Cho and Leahy, 2002; Garrett et al, 2002; Ogiso et al, 2002; Ferguson et al, 2003): the tethered conformation is thought to be a low-affinity binding state to EGF and cannot form dimers, whereas the extended conformation is thought to form the signaling dimers. However, there appears to be no direct or static correlation between the affinity difference in the Scatchard plot analysis and the conformational differences in the crystallographic studies. Some mathematical models for the dimerization and activation of the EGFRs, where the different affinity sites represent receptors in different conformations, could not explain the experimental results obtained (Wofsy et al, 1992; Chung et al, 1997; Klein et al, 2004). Therefore, to understand the mechanism of EGF signaling, kinetic analysis of the association between EGF and EGFR is important, in which the dynamic changes in the affinity between EGF and EGFR should be correlated with the formation of preformed and signaling dimers.
In this study, the formation of EGF/EGFR complex and its dimerization, especially, in the early stages of EGF signaling on the living cell surface, was quantitatively analyzed using single-molecule imaging. Single-molecule analyses in living cells have been effectively used to analyze intracellular reaction kinetics and protein dynamics (Sako and Yanagida, 2004). Examples include kinetic analysis of receptor–ligand interaction (Ueda et al, 2001; Uyemura et al, 2005), measurement of protein dynamics and reactions in the nucleus (Yang et al, 2004) or plasma membrane (Sako et al, 2000; Schütz et al, 2000; Iino et al, 2001; Hibino et al, 2003; Ichinose et al, 2004; Douglass and Vale, 2005). Here, analysis of single-molecule process of EGF binding to the cell surface revealed two association rate constants, the presence of a binding intermediate not previously identified and the rate constant for the formation of this intermediate. Global fitting of the binding curves to a simple model containing the kinetic intermediate with experimentally obtained rate constants provides new insight into the formation of signaling dimers of EGF/EGFR complexes.
Results and discussion
Visualization of single molecules of EGF bound to the surface of living cells
Rh-EGF was applied to the medium of HeLa cells at a final concentration of 0.1–0.5 nM under an oblique illumination microscope (Figure 1A and B). After the application of Rh-EGF, fluorescence spots appeared on the cell surface. Individual spots emitted almost constant fluorescence, then suddenly disappeared (Figure 1C, red). The disappearance was most likely caused by the photobleaching of Rh and not by the dissociation of EGF molecules, as dissociation of EGF from EGFR is known to be slow. Single-step photobleaching indicated that a single molecule of Rh-EGF had bound to the EGFR (Sako et al, 2000). Spots that disappeared in two steps were also observed (Figure 1C, black), indicating that these spots contained two molecules of Rh-EGF.
Figure 1.
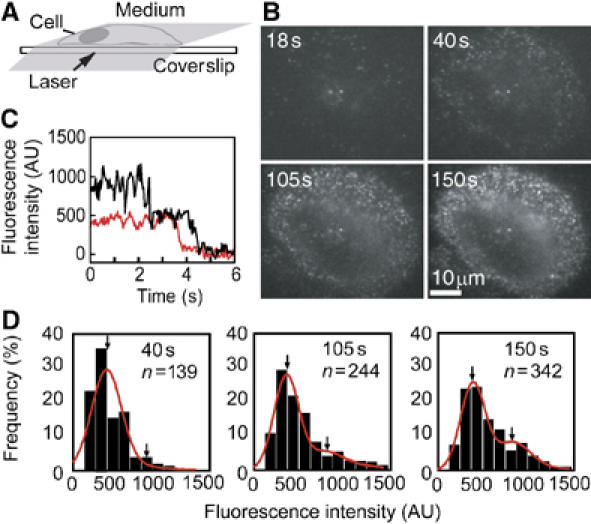
Single-molecule visualization of Rh-EGF bound on the living cell surface. (A) Oblique illumination with a thin laser beam through the objective lens of a microscope allows single-molecule visualization on the apical cell surface. (B) Once the apical surface near the edge of a cell was brought into focus, Rh-EGF was added to the extracellular medium. Pictures taken 18, 40, 105 and 150 s after the addition of Rh-EGF (0.5 nM final concentration) are shown. (C) A typical fluorescent spot on the cell was photobleached in a single step (red) or double steps (black), indicating that the spot contained a single molecule or two molecules of Rh-EGF, respectively. (D) Distribution of the fluorescence intensity of Rh-EGF spots bound on the cell shown in (B) analyzed 40, 105 and 150 s after the addition of Rh-EGF. The distributions were fitted to a sum of two Gaussian functions (red line). Arrows indicate the mean of the fractions containing one and two Rh-EGF molecules. n: total number of spots; AU: arbitrary unit.
The distribution of the fluorescence intensity of Rh-EGF spots bound to the cell surface was obtained at different time points after the addition of Rh-EGF (Figure 1D). The distributions were fitted to the sum of two Gaussian functions, which allowed the ratio of the Rh-EGF monomer and the dimer to be calculated. As the mean of the first component was the same as that for the photobleaching step, it is most likely that this component contained a single Rh-EGF molecule. The second component, which emitted a signal twice as intense as the first component, should represent spots containing two Rh-EGF molecules. This component possibly represents two Rh-EGF molecules bound to a single EGFR dimer or to two EGFR monomers positioned closer than the spatial resolution of the microscope. The latter case is less likely to occur because of the low density of fluorescent spots and the synchronized lateral diffusion movements of two Rh-EGF molecules forming a single spot (Supplementary data). Fractions that contained more than three Rh-EGF molecules were small.
The total number of bound molecules of Rh-EGF per cell was subdivided into monomers and dimers according to the distribution of fluorescence intensity at each time point (Figure 1D), and plotted as a function of time (Figure 2). As shown here, using single-molecule visualization, it is possible to separately analyze the formation of monomeric and dimeric binding sites of EGF. Significant internalization of Rh-EGF did not take place until 180 s after the application of Rh-EGF to the medium (Supplementary data). Data during this period were used in the mathematical modeling of EGFR dimerization in the later sections.
Figure 2.
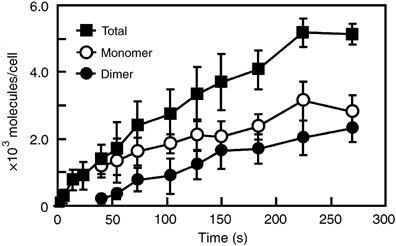
Time course of Rh-EGF binding on the cell surface. The total number (closed square) of Rh-EGF molecules bound on individual cells was calculated at the indicated times. The numbers were then subdivided into number of monomers (open circle) and dimers (closed circle) of Rh-EGF according to fluorescence intensity distributions (Figure 1D). The average and standard deviation of 10 cells are shown. In the initial stages of EGF binding (∼20 s), the total numbers of EGF molecules bound to the cell surface were quite small (<70), so they were not subdivided into groups. These results were obtained in the presence of 0.5 nM Rh-EGF. The average of the cell surface areas was 4500 μm2.
Single-molecule analysis of the association of the first EGF molecule
The fluorescent spots that appeared on the cell surface after the addition of Rh-EGF to the cell culture medium (Figure 3A) were due to the binding of single Rh-EGF molecules to EGFRs in the plasma membrane. Molecules in solution cannot be observed as spots owing to the rapid Brownian movement in solution. The time taken for the appearance of individual spots was measured immediately after the addition of Rh-EGF to the medium (Figure 3B).
Figure 3.
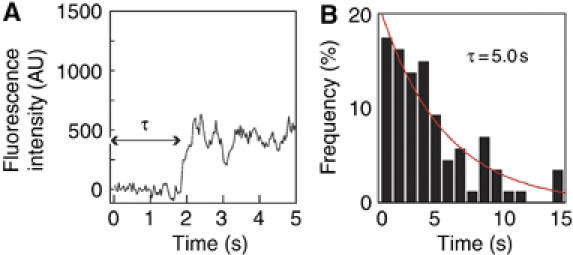
Kinetic analysis of the first molecule of Rh-EGF binding. (A) A typical fluorescence intensity change in the binding site of a single Rh-EGF molecule. Sudden step-like increases in fluorescence intensity indicate the binding of single Rh-EGF molecules. (B) Histogram of the duration (τ) of each single-molecule binding event after the addition of 0.5 nM Rh-EGF to the medium bathing the cells. The total number of events was 102. The histogram was fitted to the single exponential function (red lines) as described in equation (1), where kon=4.0 × 108 M−1 s−1. AU: arbitrary unit.
The association reaction between the ligand (L) and the receptor oligomer (Rx) forming a complex (LRx) can be written as  where kon is the second-order association rate constant and x is the number of receptor molecules contained in a single binding site. In our experiments, the number of EGFR molecules in each individual binding site should be one (monomer) or two (predimer) in most cases as a functional binding site (Figure 2). Scheme 1 represents the association of the first Rh-EGF molecule in the formation of a signaling dimer. The histogram of the duration until the first binding event could be fitted to the following function (Supplementary Scheme 1):
where kon is the second-order association rate constant and x is the number of receptor molecules contained in a single binding site. In our experiments, the number of EGFR molecules in each individual binding site should be one (monomer) or two (predimer) in most cases as a functional binding site (Figure 2). Scheme 1 represents the association of the first Rh-EGF molecule in the formation of a signaling dimer. The histogram of the duration until the first binding event could be fitted to the following function (Supplementary Scheme 1): 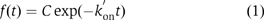 Here, k′on is the first-order association rate constant (k′on=kon[L]; [L] is the concentration of L). The histogram obtained (Figure 3B) was fitted with this function, where kon=4.0 × 108 M−1 s−1.
Here, k′on is the first-order association rate constant (k′on=kon[L]; [L] is the concentration of L). The histogram obtained (Figure 3B) was fitted with this function, where kon=4.0 × 108 M−1 s−1.
Single-molecule analysis of the association of the second EGF molecule
Occasionally, the fluorescence intensity of single spots increased suddenly by a factor of approximately two (Figure 4A). After the increase of fluorescence intensity, these spots moved as a unit along the cell membrane. These events are most likely the result of the association of a second Rh-EGF molecule to the complexes, which already contained single Rh-EGF and more than one vacant EGFR molecule. Figure 4B shows the histograms of the durations between the first and second EGF binding to the same site. The histograms show a peak at around 1 s (Figure 4B). The presence of the peak suggests that the process did not arise from a simple ligand–receptor association reaction but had two sequential transitions. We propose that a kinetic intermediate exists during EGFR dimerization, which has not been identified in previous studies.
Figure 4.
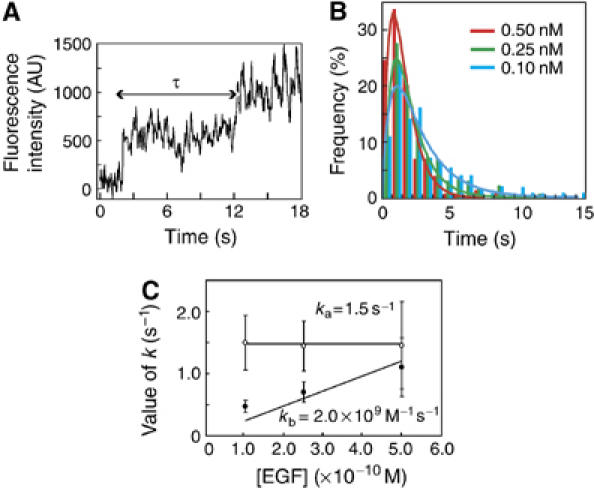
Kinetic analysis of the second Rh-EGF binding event. (A) A typical change in the fluorescence intensity at the site of the formation of an EGFR dimer. Each step represents the first and the second binding events of Rh-EGF to an EGFR dimer. (B) Histograms of the duration (τ) between the binding of the first and second Rh-EGF molecules to the same binding sites in the presence of 0.10 nM (blue bars), 0.25 nM (green bars) and 0.50 nM (red bars) of Rh-EGF (total number of events was 163, 147 and 157, respectively). These histograms were fitted to equation (2) representing a tandem reaction. The results of the fitting are shown as lines. (C) Kinetic parameters ka (open circles) and kb′ (=[Rh-EGF]kb; filled circles) in equation (2) obtained by the fitting are plotted as a function of the concentration of Rh-EGF.
The reaction containing a kinetic intermediate can be generally written as  Here, LRx (x=1 or 2) can contain either one (LR) or two (LR2) receptor molecule(s). LR2* was the kinetic intermediate. The reaction in Scheme 2 could be expressed as (Supplementary Scheme 2)
Here, LRx (x=1 or 2) can contain either one (LR) or two (LR2) receptor molecule(s). LR2* was the kinetic intermediate. The reaction in Scheme 2 could be expressed as (Supplementary Scheme 2) 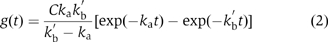 where k′b=kb[L] considering the EGF binding as a pseudo-first-order reaction. The dissociation reaction in the second step is not the subject of this analysis, as only binding events have been observed. The reverse reaction of the first step in Scheme 2 has been ignored (Supplementary data).
where k′b=kb[L] considering the EGF binding as a pseudo-first-order reaction. The dissociation reaction in the second step is not the subject of this analysis, as only binding events have been observed. The reverse reaction of the first step in Scheme 2 has been ignored (Supplementary data).
The histograms shown in Figure 4B were fitted with equation (2). The reaction was analyzed under three different concentrations of Rh-EGF (0.10, 0.25 and 0.50 nM; Figure 4C). One parameter ka was independent of the concentration of Rh-EGF, indicating that it is for the first and EGF-independent step. In contrast, parameter kb′ increased in proportion to the concentration of Rh-EGF, indicating that it was for the second and EGF-dependent process, that is, the association of the second EGF molecule. The values for ka and kb were 1.5 s−1 and 2.0 × 109 M−1 s−1, respectively. Thus, single-molecule analysis revealed the rate constants of the association between EGF and EGFR and the existence of a kinetic intermediate, which could not be identified using conventional methods.
Models for the formation of signaling dimers of EGF/EGFR complexes
A simple model has been considered to explain the formation of the signaling dimer of EGF/EGFR complexes including a kinetic intermediate (Model A in Figure 5A). This model was based on the reports that EGFR molecules present on the cell surface either as monomeric or dimeric binding sites before application of EGF to cells (Moriki et al, 2001; Yu et al, 2002). In this model, there are three possible routes for the formation of the signaling dimer. First, two EGF molecules bind sequentially to a preformed dimeric binding site. Second, an EGF binds to a monomeric binding site and couples with a vacant binding site by lateral diffusion and collision, then the second EGF binds to the vacant binding site. Third, two EGF/EGFR complexes laterally diffuse and collide to form a signaling dimer. The intermediate in Figure 4 was found between successive bindings of the first and the second ligands from the solution. In Model A, the intermediate is LR2.
Figure 5.

Fitting of the time course of Rh-EGF binding to the models for the formation of signaling dimers. (A) Two simple schemes for the formation of a signaling dimer (EGF/EGFR–EGF/EGFR). Model A includes the intermediate EGF/EGFR–EGFR formed by the association of EGF/EGFR and EGFR. In Model B, the intermediate is EGF/EGFR–EGFR*. (B) Changes in the number of Rh-EGF monomers (open circles) and dimers (filled circles) bound to the cell surface (the same plots shown in Figure 2) have been fitted using functions obtained form Models A (red lines) and B (solid black lines). In fitting, three of the rate constants were fixed to the experimentally obtained values. Model A could not explain the experiments. Results of fitting using Model B floating every parameter are also shown (black broken lines). The total number of bound Rh-EGF molecules before the 40 s time period is shown as closed squares for reference.
The changes in the number of monomeric and dimeric EGF molecules bound to the cell surface by 180 s after the addition of Rh-EGF (Figure 2) were fitted with the functions obtained by solving the coupled differential equations according to Model A (Supplementary data) using nonlinear least-squares method (Figure 5B). In the fitting, the rate constants obtained from the single-molecule experiments (Figures 3 and 4) were fixed as k1=kon (4.0 × 108 M−1 s−1), k4=ka (1.5 s−1) and k5=kb (2.0 × 109 M−1 s−1). Other parameters (k3, k6, R(0) and R2(0)) were floated. R(0) and R2(0) are the numbers of monomers and dimers of EGFR per cell before the addition of EGF. The best-fit results according to Model A could not explain the experiments (Figure 5B; Supplementary data). There is a possibility that kon represents k3 (and not k1), that is the association rate constant between EGF and monomeric EGFR. However, substitution of 4.0 × 108 (M−1 s−1) for k3 instead of k1 also did not agree with the experimental data (data not shown). Thus, Model A cannot explain EGF binding on the cell surface.
Model A was extended to contain a separated intermediate state (LR2*) between LR2 and L2R2 (Model B in Figure 5A). Model B assumes a conformational change of the dimeric binding sites bound with a single EGF molecule. The experimental results shown in Figure 2 were fitted with the functions obtained from Model B (Supplementary data). In the fitting, the rate constants obtained from the single-molecule experiments (Figures 3 and 4) were fixed as k1=kon (4.0 × 108 M−1 s−1), k2=ka (1.5 s−1) and k5=kb (2.0 × 109 M−1 s−1). Other parameters (k3, k4, k6, R(0) and R2(0)) were floated. The result of fitting agreed with the experiment within the range of experimental errors (Figure 5B). The best-fit parameters are shown in Table I. Parameters predicted from the fitting but not determined directly in this study (k3, the association rate constant of EGFR monomer; R(0)+2R2(0), the total number of receptor molecules) were consistent with the values reported in previous studies. Substitution of 4.0 × 108 (M−1 s−1) for k3 instead of k1 resulted in physically meaningless fitting (a negative value for k4). If k4 was constrained to be positive, the best-fit results did not agree with the experiments at all (data not shown).
Table 1.
Kinetic parameters obtained from Model B and experiments
| Parameter | Reaction | Fitting with Model B | Experiments | |
|---|---|---|---|---|
| Fixeda | Floatedb | |||
| k1 (M−1 s−1) | L+R2 → LR2 | 4.0 × 108c | 2.0 × 108 | 4.0 × 108c |
| k2 (s−1) | LR2 → LR2* | 1.5c | 1.0 | 1.5c |
| k3 (M−1 s−1) | L+R → LR | 4.0 × 106 | 2.3 × 106 | 3.3–9.8 × 106d |
| k4 (s−1)e | LR+R → LR2 | 3.0 × 10−3 | 3.0 × 10−3 | ND |
| k5 (M−1 s−1) | LR2*+L → L2R2 | 2.0 × 109c | 2.0 × 109 | 2.0 × 109 c |
| k6 (s−1)e | LR+LR → L2R2 | 3.3 × 10−3 | 3.3 × 10−3 | ND |
| R(0) (/cell) | 25 000 | 27 000 | 47 000f | |
| R2(0) (/cell) | 100 | 200 | ||
| Kinetic parameters obtained from fitting the time course of Rh-EGF-binding (Figure 2) to Model B in Figure 5 are listed. L and R represent EGF and EGFR, respectively. R(0) and R2(0) are the number of monomeric and preformed dimeric biding sites of EGFR per cell in the absence of EGF. Averages of the best-fit values of parameters from two sets of experimental data at different concentrations of [Rh-EGF] (0.1 and 0.5 nM) are shown. ND: not determined. | ||||
| ak1, k2 and k5 were fixed to the experimental values determined in this study. | ||||
| bAll parameters were floated. | ||||
| cExperimental values determined in this study. | ||||
| dBellot et al (1990), Berkers et al (1991), Felder et al (1992), Chung et al (1997) and Wilkinson et al (2001). | ||||
| eThese are first-order rate constants specific at the receptor density in HeLa cells. | ||||
| fTotal number of EGFR molecules (Berkers et al, 1991). | ||||
Fittings resulted in fundamentally the same conclusion when all the parameters were floated (Figure 5B and Table I). The parameters k1=2.0 × 108 M−1 s−1, k2=1.0 s−1 and k5=2.0 × 109 M−1 s−1 obtained from this fitting were consistent with experimentally obtained values. Probably, some of the dimeric spots of Rh-EGF were pseudo-dimers constructed by two monomers accidentally presented nearby. However, the analysis of movements of dimeric spots suggests that almost all (98%) of the molecules in dimeric spots were moving synchronously (Supplementary data) and the presence of pseudo-dimers up to 50% of the observed dimeric fraction does not essentially change the results of fitting (Supplementary Table II). Fitting to Model A could not explain the experiments even when all of the parameters were floated (Supplementary data). Models containing the reverse reactions are too complex to be solved analytically. However, numerical analysis (Sasagawa et al, 2005) of the models confirmed that all reverse reactions can be ignored (Supplementary data).
Kinetics of the formation of signaling dimer
The formation of signaling dimers of EGF/EGFR complexes could be well explained using Model B. In this model, a conformational change in EGFR dimer bound with a single EGF molecule (EGF/EGFR–EGFR*) was taken as the kinetic intermediate. The result provides some insight into the formation of signaling dimer of EGF/EGFR complexes:
(1) About 1–2% of EGFR molecules (2R2(0)/{R(0)+2R2(0)}; Table I) on the cell surface formed dimeric binding sites independent of EGF binding (preformed dimeric binding sites). If monomeric and preformed dimeric binding sites on the cell surface were in a state of equilibrium,  the dissociation constant would be quite large (KD=R2/R2∼4 × 106 sites/cell). In cells expressing high amounts of EGFR, such as A431 (3 × 106 EGFR/cell; Defize et al, 1988), the amount of preformed dimeric binding site should be 7 × 105 sites/cell. This number is similar to that suggested previously (Gadella and Jovin, 1995; Martin-Fernandez et al, 2002).
the dissociation constant would be quite large (KD=R2/R2∼4 × 106 sites/cell). In cells expressing high amounts of EGFR, such as A431 (3 × 106 EGFR/cell; Defize et al, 1988), the amount of preformed dimeric binding site should be 7 × 105 sites/cell. This number is similar to that suggested previously (Gadella and Jovin, 1995; Martin-Fernandez et al, 2002).
It is expected that the number of EGFR predimer decreases proportionally to the square of the receptor density. As a result, in cells expressing smaller amounts of EGFR, the formation of signaling dimers will be slow and diffusion and collision between EGF/EGFR monomers will become more important. In such cells, the number of EGF molecules required to induce cellular responses will be increased.
(2) EGF bound to the predimeric binding sites ( ; k1=2.0–4.0 × 108 M−1 s−1) two orders of magnitude faster than to the monomeric binding sites (
; k1=2.0–4.0 × 108 M−1 s−1) two orders of magnitude faster than to the monomeric binding sites ( ; k3=2.3−4.0 × 106 M−1 s−1). This is most likely important in facilitating the formation of signaling dimers of EGF/EGFR complexes at low concentrations of EGF. The formation of signaling dimers from predimeric sites would be more efficient than association of two EGF/EGFR complexes, which is limited by diffusion along the plasma membrane, as predicted by Mayawala et al (2005) based on our previous results (Sako et al, 2000). The association rate constant to monomeric sites (k3) is consistent with the association rate constants between EGF and EGFR, as reported previously (Bellot et al, 1990; Berkers et al, 1991; Felder et al, 1992; Chung et al, 1997; Wilkinson et al, 2001). Our calculation suggests that approximately 98–99% of EGFRs exist as monomeric sites on the cell surface. The association rate constant obtained using conventional biochemical techniques should be close to that for monomeric sites.
; k3=2.3−4.0 × 106 M−1 s−1). This is most likely important in facilitating the formation of signaling dimers of EGF/EGFR complexes at low concentrations of EGF. The formation of signaling dimers from predimeric sites would be more efficient than association of two EGF/EGFR complexes, which is limited by diffusion along the plasma membrane, as predicted by Mayawala et al (2005) based on our previous results (Sako et al, 2000). The association rate constant to monomeric sites (k3) is consistent with the association rate constants between EGF and EGFR, as reported previously (Bellot et al, 1990; Berkers et al, 1991; Felder et al, 1992; Chung et al, 1997; Wilkinson et al, 2001). Our calculation suggests that approximately 98–99% of EGFRs exist as monomeric sites on the cell surface. The association rate constant obtained using conventional biochemical techniques should be close to that for monomeric sites.
(3) The association rate constant for the second molecules of EGF to the dimeric binding sites (k5=2.0 × 109 M−1 s−1) was one order of magnitude higher than that for the first EGF molecule to the preformed dimeric binding sites (k1=2.0−4.0 × 108 M−1 s−1). This indicates positive cooperativity of EGF binding to the dimeric binding sites, as predicted in our previous study (Uyemura et al, 2005). The first molecules of bound EGF induce a conformational change in the dimeric sites, which increases the association rate for the second EGF. The positive cooperative binding of EGF guarantees the selective binding of the second ligand to the first binding sites, which further accelerates the formation of signaling dimers at low concentrations of EGF.
(4) The formation of signaling dimer through association of two EGF/EGFR complexes was very slow in the presence of low concentrations of the ligand. EGF has been thought to promote dimerization between EGFRs according to the scheme 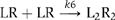 where two EGF/EGFR complexes coupled to each other by lateral diffusion and collision on the cell membrane. According to our model, this process is much slower than the formation of signaling dimer from the predimeric binding sites (k2 was 1.0–1.5 s−1 whereas k6 was 3.3 × 10−3 s−1) at least under our experimental conditions ([Rh-EGF]=0.1–0.5 nM). In the initial stages of EGF signaling and/or in the presence of low concentrations of EGF, the association following lateral diffusion may not be important.
where two EGF/EGFR complexes coupled to each other by lateral diffusion and collision on the cell membrane. According to our model, this process is much slower than the formation of signaling dimer from the predimeric binding sites (k2 was 1.0–1.5 s−1 whereas k6 was 3.3 × 10−3 s−1) at least under our experimental conditions ([Rh-EGF]=0.1–0.5 nM). In the initial stages of EGF signaling and/or in the presence of low concentrations of EGF, the association following lateral diffusion may not be important.
Thus, sensitivity of cells to EGF was improved in the initial stages of signaling pathway. Signal amplification has been known in the later stages, for example, secondary phosphorylation of EGFR (Reynolds et al, 2003; Ichinose et al, 2004) and calcium-induced calcium release. However, simple high-gain systems amplify noise at the same time as signal amplification (Shibata and Fujimoto, 2005). Improvement of signals in the earlier stages is desirable to avoid excess signal amplification in the later stages that generates extra noise.
Relation between the kinetics and the structural information
The extracellular EGF-binding domain of EGFR contains four subdomains (subdomains I–IV). Subdomains I and III participate in EGF binding (Lax et al, 1989; Garrett et al, 2002; Ogiso et al, 2002). The crystals of the extracellular region of EGFR prepared in low-affinity conditions have revealed a monomeric form of the EGF/EGFR complex that exists in an autoinhibited configuration. In this configuration, specific intramolecular interactions between subdomains II and IV tether the structure and EGF interacts with only subdomain I (Cho and Leahy, 2002; Ferguson et al, 2003). However, in the dimeric form of EGF/EGFR complex, EGF binds simultaneously to subdomains I and III to induce an extended conformation of EGFR. In this configuration, the dimerization arm in subdomain II is available for mediating interactions with a neighboring extended EGFR molecule (Garrett et al, 2002; Ogiso et al, 2002). This extended conformation is thought to be a high-affinity conformation. Interactions between two EGF molecules or crosslinking of two receptor molecules by EGF has not been found in the crystals (Garrett et al, 2002; Ogiso et al, 2002).
Although the conformation of EGFRs in the predimers is not yet known, large increases in the association rate of EGF to the predimeric binding sites suggest that the conformation resembles the extended form. In addition, the positive cooperativity suggests that the association of EGF with one of the EGFR molecules in the dimeric sites induced an allosteric conformational change in the EGF-binding site in the other EGFR molecule. Fluctuations of the vacant receptor molecule may be suppressed and almost fixed to the high-affinity conformation state by binding of EGF to the neighboring molecules in the dimers (Figure 6).
Figure 6.
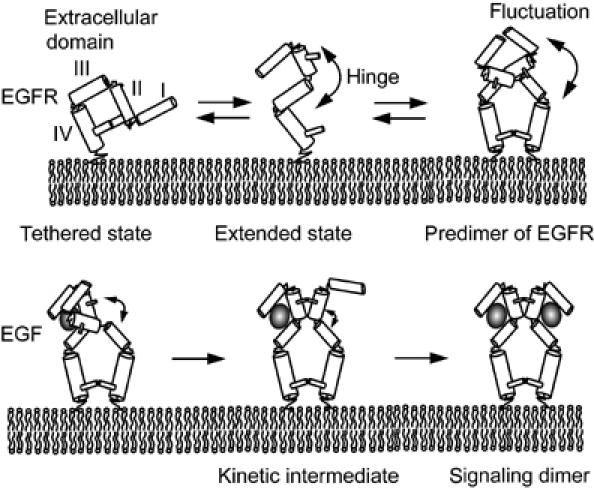
A model for the formation of signaling dimer of EGF/EGFR complexes. The extracellular EGF-binding domain of EGFR contains subdomains I–IV. Subdomains I and III participate in EGF binding (Lax et al, 1989). An EGFR molecule exists in the extended low-affinity (low-association rate) state or in the extended high-affinity (high-association rate) state and can switch between the states by dynamic fluctuations mainly at the hinge between subdomains II and III (Cho and Leahy, 2002; Garrett et al, 2002; Ogiso et al, 2002; Ferguson et al, 2003). Monomers of EGFR exist primarily in the tethered state, and the formation of predimers biases the structure of EGFR toward extended state-like conformations with high association rates to EGF. EGF bound to one of the EGFR molecules in a predimer induces an allosteric conformational change in another EGFR molecule to a kinetic intermediate structure with a considerably high association rate to EGF. Thus, a dynamic conformational change in the predimer facilitates the formation of signaling dimer of EGF/EGFR complexes. Only the portion of EGFR outside the cell surface is shown.
Single-molecule kinetics has revealed that two functionally different populations of binding sites exist on the cell surface to which EGF is associated as a monomer or a dimer. However, the dimerization state of EGFR was not observed directly; thus, it is possible that each binding site is involved in a larger cluster rather than a single site. In addition, there may be EGFR dimers behaving as two individual monomeric binding sites. We cannot exclude the possibility that the dimeric binding sites of Rh-EGF were EGFR molecules confined in small membrane domains and do not interact with each other directly. However, in that case, there must be a mechanism to increase the association rate of EGF to the confined receptors. Such a mechanism has not been proposed as far as we know. The assumption of direct interaction (predimerization of EGFR) is simpler and seems to be preferable at the present time.
Conclusion
The mechanisms that guarantee the rapid and sensitive signal transduction of EGF remain to be determined. The model for the formation of the signaling dimer proposed in the present study suggests that a small fraction of EGFR formed predimeric binding sites for EGF, and that the high association rate for the first binding event and the positive cooperativity of the binding of two EGF to the predimeric sites facilitated the formation of signaling dimers. The present model is in good agreement with previous kinetic (Bellot et al, 1990; Berkers et al, 1991; Felder et al, 1992; Chung et al, 1997; Wilkinson et al, 2001; Uyemura et al, 2005) and structural (Cho and Leahy, 2002; Garrett et al, 2002; Ogiso et al, 2002; Ferguson et al, 2003) studies. Formation of the predimeric binding sites of EGFR makes the EGF signaling rapid and very sensitive even when the concentration of EGF is low, and the expression of excess amounts of EGFR on the cell surface is required for the formation of predimeric binding sites of EGFR.
Materials and methods
Cell culture
HeLa cells were cultured on glass coverslips in Dulbecco's modified Eagle's medium (Nissui, Tokyo, Japan) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Before the experiment, the culture medium was changed to that without serum and phenol red overnight.
Optical settings
An objective type total internal reflection fluorescence microscope system (Tokunaga et al, 1997; Sako et al, 2000) was modified for oblique illumination. The microscope was based on an inverted microscope (IX-70, Olympus, Tokyo, Japan) equipped with an oil-immersion objective (PlanApo × 100, NA 1.45, Olympus) and an excitation laser (532 nm; Compass 315M, Coherent, Santa Clara, CA). Fluorescence images of the specimen were intensified with an image intensifier (C8600-03; Hamamatsu photonics, Hamamatsu, Japan) and acquired with an electron bombardment CCD camera (C7190; Hamamatsu photonics) at video rate.
Measurement of bound EGF molecules on the cell surface
Tetramethylrhodamine-conjugated mouse EGF (Rh-EGF) was purchased from Molecular Probes (Eugen, OR). In this probe, EGF was conjugated with a single Rh molecule at the N terminus. The ratio of dye to protein of Rh-EGF was 0.98 (Uyemura et al, 2005). Rh-EGF was applied to the culture medium of HeLa cells at a final concentration of 0.1–0.5 nM while cells were being observed under the microscope. To study the binding time courses of Rh-EGF onto the cells, images of cells were recorded for a few seconds at each indicated time point after the application of Rh-EGF. Exposure time to acquire images was kept to a minimum to avoid photobleaching. Any slight effect of photobleaching was corrected (Hibino et al, 2003). In the single-molecule tracking experiments of EGF binding, images of cells were recorded continuously. All experiments were performed at 25°C. The apical surface of cells was observed to avoid rate determination of the binding kinetics by the diffusion of Rh-EGF between the basal cell surface and the coverslip. The HeLa cells on the glass coverslip were convex in appearance; thus, only fluorescent spots bound on the apical surface near the edge of cells were observed. The density of bound Rh-EGF molecules in the focus plane was calculated from the area of the plane, the sum of the intensity of every fluorescent spot in the plane and the intensity of single molecules, and then, scaled to the total number of bound molecules per cell with respect to the averaged surface area of the cell (4500 μm2/cell).
Data analysis
Parameter estimation was carried out using a nonlinear least-squares fitting routine program originally developed by our group (Uyemura et al, 2005). This routine program is based on a finite difference, Levenberg–Marquart algorithm. Numerical calculations of coupled differentiation equations were carried out using GENESIS simulator (version 2.2) with a Kinetikit interface (Sasagawa et al, 2005).
Supplementary Material
Supplementary Infromation
Acknowledgments
We thank Shinya Kuroda and Yu-ichi Ozaki at the University of Tokyo for numerical analysis of the models, and Takeshi Uyemura and other members of our laboratory for useful discussion. YT was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. HT is supported by Leading Project (Bio Nano Process) MEXT, Japan.
References
- Bellot F, Moolenaar W, Kris R, Mirakhur B, Verlaan I, Ullrich A, Schlessinger J, Felder S (1990) High-affinity epidermal growth factor binding is specifically reduced by a monoclonal antibody, and appears necessary for early responses. J Cell Biol 110: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers JAM, Van Berger en Henegouwen PMP, Boonstra J (1991) Three classes of epidermal growth factor receptors on HeLa cells. J Biol Chem 266: 922–927 [PubMed] [Google Scholar]
- Boonstra J, Mummery CL, van der Saag PT, de Laat SW (1985) Two receptor classes for epidermal growth factor on pheochromocytoma cells, distinguishable by temperature, lectins, and tumor promoters. J Cell Physiol 123: 347–352 [DOI] [PubMed] [Google Scholar]
- Carpenter G (1987) Receptors for epidermal growth factor and other polypeptide mitogens. Annu Rev Biochem 56: 881–914 [DOI] [PubMed] [Google Scholar]
- Cho HS, Leahy DJ (2002) Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297: 1330–1333 [DOI] [PubMed] [Google Scholar]
- Chung JC, Sciaky N, Gross DJ (1997) Heterogeneity of epidermal growth factor binding kinetics on individual cells. Biophys J 73: 1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defize LHK, Arndt-Jovin DJ, Jovin TM, Boonstra J, Meisenhelder J, Hunter T, de Hey HT, de Laart SW (1988) A431 cell variants lacking the blood group A antigen display increased high affinity epidermal growth factor-receptor number, protein-tyrosine kinase activity, and receptor turnover. J Cell Biol 107: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Vale RD (2005) Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell 121: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S, LaVin J, Ullrich A, Schlessinger J (1992) Kinetics of binding endocytosis, and recycling of EGF receptor mutants. J Cell Biol 117: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho H-S, Leahy DJ, Lemmon MA (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell 11: 507–517 [DOI] [PubMed] [Google Scholar]
- Gadella TWJ Jr, Jovin TM (1995) Oligomerization of epidermal growth factor receptors on A431 cells studies by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol 129: 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TPJ, McKern NM, Low M, Ellman TC, Adams TE, Lovrecz GO, Zhu H-J, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell 110: 763–773 [DOI] [PubMed] [Google Scholar]
- Hibino K, Watanabe T, Kozuka J, Iwane AH, Okada T, Kataoka T, Yanagida T, Sako Y (2003) Single- and multiple-molecule dynamics of the signaling from H-Ras to c-Raf1 visualized on the plasma membrane of living cells. Chem Phys Chem 4: 748–753 [DOI] [PubMed] [Google Scholar]
- Ichinose J, Murata M, Yanagida T, Sako Y (2004) EGF signaling amplification induced by dynamic clustering of EGFR. Biochem Biophys Res Commun 324: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Iino R, Koyama I, Kusumi A (2001) Single molecule imaging of green fluorescent proteins in living cells: E-cadhern forms oligomers on the free cell surface. Biophys J 80: 2667–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, Cuatrecasas P (1982) Resolution of high and low affinity epidermal growth factor receptors. Inhibition of high affinity component by low temperature, cycloheximide, and phorbol esters. J Biol Chem 257: 3053–3060 [PubMed] [Google Scholar]
- Klein P, Mattoon D, Lemmon MA, Schlessinger J (2004) A structure-based model for ligand binding and dimerization of EGF receptors. Proc Natl Acad Sci USA 101: 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp MN, Connolly DT, Lane MD (1982) Synthesis, turnover, and down-regulation of epidermal growth factor receptors in human A431 epidermoid carcinoma cells and skin fibroblasts. J Biol Chem 257: 11489–11496 [PubMed] [Google Scholar]
- Lax I, Bellot F, Howk R, Ullrich A, Givol D, Schlessinger J (1989) Functional analysis of the ligand binding site of EGF-receptor utilizing chimeric chicken/human receptor molecules. EMBO J 8: 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fernandez M, Clarke DT, Tobin MJ, Jones SV, Jones GR (2002) Preformed oliogomeirc epidermal growth factor receptors undergo an ectodomain structure change during signaling. Biophys J 82: 2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayawala K, Vlachos DG, Edwards JS (2005) Computational modeling reveals molecular details of epidermal growth factor binding. BioMed Central 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriki T, Maruyama H, Maruyama IN (2001) Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol 311: 1011–1026 [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fuakai S, Yamanaka M, Kim J-H, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110: 775–787 [DOI] [PubMed] [Google Scholar]
- Reynolds AR, Tischer C, Verveer PJ, Rocks O, Bastiaens PIH (2003) EGFR activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat Cell Biol 5: 447–453 [DOI] [PubMed] [Google Scholar]
- Sako Y, Minoguchi S, Yanagida T (2000) Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol 2: 168–172 [DOI] [PubMed] [Google Scholar]
- Sako Y, Yanagida T (2004) Single-molecule visualization in cell biology. Nat Rev Mol Cell Biol 4 (Suppl): SS1–SS5 [PubMed] [Google Scholar]
- Sasagawa S, Ozaki Y, Fujita K, Kuroda S (2005) Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol 7: 365–373 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103: 211–225 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110: 669–672 [DOI] [PubMed] [Google Scholar]
- Schütz GJ, Kada G, Pastushenko VP, Schindler H (2000) Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J 19: 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Fujimoto K (2005) Noisy signal amplification in ultrasensitive signal transduction. Proc Natl Acad Sci USA 102: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoyab M, De Larco JE, Todaro GJ (1979) Biologically active phorbol esters specifically alter affinity of epidermal growth factor receptors. Nature 279: 387–391 [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T (1997) Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem Biophys Res Commun 235: 47–53 [DOI] [PubMed] [Google Scholar]
- Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T (2001) Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science 294: 864–867 [DOI] [PubMed] [Google Scholar]
- Uyemura T, Takagi H, Yanagida T, Sako Y (2005) Single-molecule analysis of epidermal growth factor signaling that leads to ultrasensitive calcium response. Biophys J 88: 3720–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS, Walsh BJ, Lund KA (1989) Global modulation of the epidermal growth factor receptor is triggered by occupancy of only a few receptors: evidence for a binary regulatory system in normal human fibroblasts. J Biol Chem 264: 18912–18920 [PubMed] [Google Scholar]
- Wilkinson JC, Stein RA, Guyer CA, Beechem JM, Staros JV (2001) Real-time kinetics of ligand/cell surface receptor interactions in living cells: binding of epidermal growth factor to the epidermal growth factor receptor. Biochemistry 40: 10230–10242 [DOI] [PubMed] [Google Scholar]
- Wofsy C, Goldstein B, Lund K, Wiley HS (1992) Implications of epidermal growth factor (EGF) induced egf receptor aggregation. Biophys J 63: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Gelles J, Musser SM (2004) Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA 101: 12887–12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell 13: 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Infromation


