Abstract
Background
Mitochondrial (mt) gene arrangement is highly variable among molluscs and especially among bivalves. Of the 30 complete molluscan mt-genomes published to date, only one is of a heterodont bivalve, although this is the most diverse taxon in terms of species numbers. We determined the complete sequence of the mitochondrial genomes of Acanthocardia tuberculata and Hiatella arctica, (Mollusca, Bivalvia, Heterodonta) and describe their gene contents and genome organisations to assess the variability of these features among the Bivalvia and their value for phylogenetic inference.
Results
The size of the mt-genome in Acanthocardia tuberculata is 16.104 basepairs (bp), and in Hiatella arctica 18.244 bp. The Acanthocardia mt-genome contains 12 of the typical protein coding genes, lacking the Atpase subunit 8 (atp8) gene, as all published marine bivalves. In contrast, a complete atp8 gene is present in Hiatella arctica. In addition, we found a putative truncated atp8 gene when re-annotating the mt-genome of Venerupis philippinarum. Both mt-genomes reported here encode all genes on the same strand and have an additional trnM. In Acanthocardia several large non-coding regions are present. One of these contains 3.5 nearly identical copies of a 167 bp motive. In Hiatella, the 3' end of the NADH dehydrogenase subunit (nad)6 gene is duplicated together with the adjacent non-coding region. The gene arrangement of Hiatella is markedly different from all other known molluscan mt-genomes, that of Acanthocardia shows few identities with the Venerupis philippinarum. Phylogenetic analyses on amino acid and nucleotide levels robustly support the Heterodonta and the sister group relationship of Acanthocardia and Venerupis. Monophyletic Bivalvia are resolved only by a Bayesian inference of the nucleotide data set. In all other analyses the two unionid species, being to only ones with genes located on both strands, do not group with the remaining bivalves.
Conclusion
The two mt-genomes reported here add to and underline the high variability of gene order and presence of duplications in bivalve and molluscan taxa. Some genomic traits like the loss of the atp8 gene or the encoding of all genes on the same strand are homoplastic among the Bivalvia. These characters, gene order, and the nucleotide sequence data show considerable potential of resolving phylogenetic patterns at lower taxonomic levels.
Background
Metazoan mitochondrial genomes are typically conserved in gene content and length. They are usually circular, 14 to 20 kb long, and encode for 13 proteins of the respiratory chain [cytochrome c oxidase subunits I-III (cox I – cox III), apocytochrome b (cytb), atpase subunits 6 and 8 (atp6, atp8), and NADH dehydrogenase subunits 1–6 and 4L (nad 1–6, nad 4L)] and 24 RNA genes of the translation system [small (S) and large (L) subunit ribosomal RNA (rrn) and 22 transfer RNAs] [1]. The high number of possible arrangements makes it very unlikely that identical gene orders arise by chance [2]. Such a complex character combined with a low frequency of gene rearrangements is highly valuable for reconstructing palaeozoic or even pre-Cambrian phylogenetic events. Examples for this situation are Vertebrata (over 540 species sequenced) and Arthropoda (over 100 species sequenced): both show few rearrangements within the phylum [3].
In contrast, only 30 complete mitochondrial genomes of Mollusca are published: ten Gastropoda, nine Bivalvia, one Polyplacophora, two Scaphopoda and eight Cephalopoda. However, even this small taxonomic sample reveals much greater variability of gene arrangements compared to vertebrates and arthropods and notable differences in rearrangement frequencies between phyla and also within the Mollusca [3]. Whereas the order of the protein coding and the rRNA (rrn) genes in the mt genomes of the polyplacophoran Katharina tunicata, the vetigastropod Haliotis rubra and the cephalopods Octopus vulgaris and Octopus ocellatus are identical and the apogastropod Ilyanassa obsoleta and the other cephalopods can be related to them, the euthyneuran gastropods, the scaphopods and the bivalves are highly rearranged.
An additional complication in the Bivalvia, termed doubly uniparental inheritance (DUI), is the existence of distinct male and female mitochondrial lineages [4-10]. It is not clear whether this mode of inheritance is characteristic for all bivalves, or if it contributes to the accelerated rearrangement rate in this group. There are yet more special features of molluscan mt genes. Hoffmann et al. [11] described an additional trn-Met in Mytilus edulis; Katharina tunicata has two additional tRNAs [12]. Some pulmonate gastropods have unusual tRNA s lacking the T-stem or the D-stem, similar to nematode mt tRNAs [13]. The atp6 and atp8 genes are separated in scaphopods [14,15] and most gastropods (only the prosobranch Littorina saxatilis, the vetigastropd Haliotis rubra and the apogastropod Ilyanassa obsoleta have adjacent atp6 and atp8). The published heterodont and pteriomorph bivalve sequences lack the atp8 gene altogether. This is unusual because the atp6 – atp8 cluster is common to most animal mitochondrial genomes, often with overlapping reading frames [3]. It is, thus, not clear for which molluscan taxa and on which systematic levels mitochondrial gene order data and genomic characters like those mentioned above are phylogenetically informative.
The phylogenetic relationships of the major taxa of the heterodont bivalves are only partly resolved. Molecular phylogenetic analyses [16,17] agreed on the exclusion of the Hiatellidae from the Myoida placing this taxon close to the base of the higher Heterodonta ("unnamed clade I" in [16] fig. 3.6). The latter clade also contains the Cardiidae and Veneridae. With the complete mitochondrial sequences of one species of each Hiatellidae, Carditidae and Veneridae available for the present study we are able to test the monophyly and sistergroup relationships of the higher heterodonts, Acanthocardia and Venerupis.
Results
Genome size, genes, base composition and codon usage
The size of the complete mt-genome of Acanthocardia tuberculata is 16.104 basepairs (bp) and has an overall A+T content of 59.6 %. All genes are on one strand (Fig. 1). The Acanthocardia genome features 1.751 non-coding bp. The largest non-coding region (Table 1), of 1.103 bp is located between trn-Met and trn-His. It contains a 599 bp fragment composed of 3.5 nearly identical copies of a 167 bp motive (Fig. 2). This repeat has an A+T content of 60 %. The other 23 non-coding regions range between 1 and 128 bp.
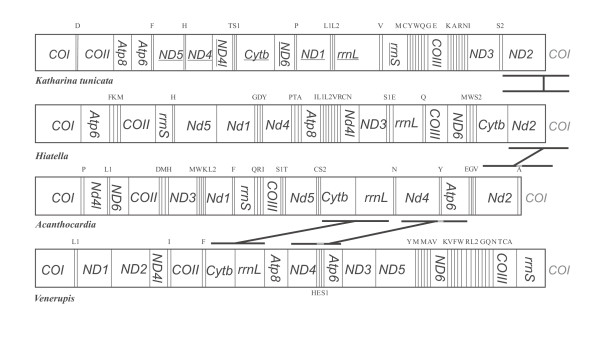
Figure 1.
Gene order of Katharina, Acanthocardia, Hiatella and Venerupis. Linearized representation of the mitochondrial gene arrangement in Acanthocardia tuberculata and Hiatella arctica, in comparison with the near-plesiomorphic condition in the polyplacophoran Katharina tunicata and the third heterodont bivalve,Venerupis philipinarium. Genes encoded on the opposite strand are underlined. The bars indicate identical gene junctions.
Table 1.
Non-coding regions and overlaps in the mitochondrial genome of Acanthocardia tuberculata
| Position | between | between |
| Non-coding | ||
| 1594–1598 | COI/tRNA-Pro | 5 |
| 1663–1699 | tRNA-Pro/ND4l | 37 |
| 1982–2000 | ND4l/tRNA-Leu | 19 |
| 2528–2531 | ND6/COII | 4 |
| 3318–3326 | tRNA-Asp/tRNA-MetI | 9 |
| 3395–4497 | tRNA-MetI/tRNA-His | 1103 |
| 4559–4571 | tRNA-HisI/ND3 | 13 |
| 4920–4927 | ND3/tRNA-MetII | 8 |
| 4992–4999 | tRNA-MetII/tRNA-Trp | 8 |
| 5064 | tRNA-Trp/tRNA-Lys | 1 |
| 5197–5226 | tRNA-Leu/ND1 | 30 |
| 6139–6148 | ND1/tRNA-Phe | 10 |
| 7041–7070 | 12S rRNA/tRNA-Gln | 30 |
| 8099–8104 | COIII/tRNA-SerI | 6 |
| 8174 | tRNA-SerI/tRNA-Thr | 1 |
| 8238–8293 | tRNA-Thr/ND5 | 56 |
| 10005–10006 | tRNA-Cys/tRNA-SerII | 2 |
| 12521–12648 | tRNA-Asn/ND4 | 128 |
| 13990–13994 | ND4/tRNA-Tyr | 5 |
| 14060–14064 | tRNA-Tyr/Atp6 | 5 |
| 14777–14884 | Atp6/tRNA-Glu | 108 |
| 14944–14995 | tRNA-Glu/tRNA-Gly | 52 |
| 15067–15099 | tRNA-Val/ND2 | 33 |
| 16027–16104 | ND2/COI | 78 |
| Overlapping | ||
| 7132–7133 | tRNA-Gln/tRNA-Arg | 2 |
Figure 2.
Alignment of the large duplicated regions in Acanthocardia tuberculata and Hiatella arctica.
All but one (atp8) of the 37 typical mitochondrial genes are present, with an additional copy of the trn-Met (Fig. 3). The Acanthocardia mt-genome encodes for a total of 3.647 amino acids. The most frequent codon is TTT (Phe; n = 264), followed by TTA (Leu; n = 172). An A or T nucleotide is present at the third position in 2.269 codons (61.13%). Five of the 12 protein coding genes start with ATG or ATA (Table 2), six starts with the alternative start codon ATT (Isoleucine). The atp6 gene starts with a GTG codon. Eight genes are terminated by TAA and four by TAG. An incomplete stop codon is inferred from the alignment of the atp6 gene. The genes for trn-Gln and trn-Arg overlap by two 2 bp.
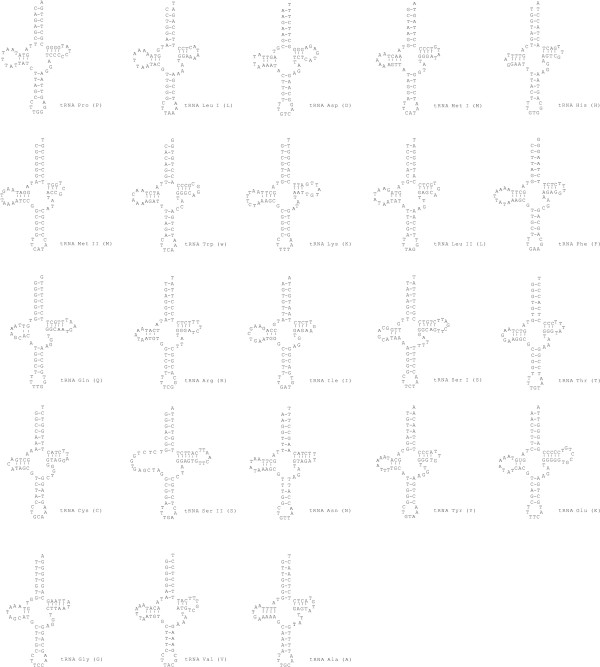
Figure 3.
Cloverleaf structures of the 23 tRNA genes in the mitochondrial genome of Acanthocardia tuberculata.
Table 2.
Organisation of the mitochondrial genome of Acanthocardia tuberculata
| Gene | Position | Strand | Start | Stop |
| COI | 1–1593 | + | ATA | TAG |
| tRNA-Pro | 1599–1662 | + | ||
| ND4l | 1700–1981 | + | ATT | TAA |
| tRNA-LeuI | 2001–2065 | + | ||
| ND6 | 2066–2527 | + | ATT | TAA |
| COII | 2532–3254 | + | ATT | TAA |
| tRNA-Asp | 3255–3317 | + | ||
| tRNA-MetI | 3327–3394 | + | ||
| tRNA-His | 4498–4558 | + | ||
| ND3 | 4572–4919 | + | ATG | TAA |
| tRNA-MetII | 4928–4991 | + | ||
| tRNA-Trp | 5000–5063 | + | ||
| tRNA-Lys | 5065–5132 | + | ||
| tRNA-LeuII | 5133–5196 | + | ||
| ND1 | 5227–6138 | + | ATG | TAG |
| tRNA-Phe | 6149–6216 | + | ||
| 12S rRNA | 6217–7040 | + | ||
| tRNA-Gln | 7071–7133 | + | ||
| tRNA-Arg | 7132–7196 | + | ||
| tRNA-Ile | 7197–7261 | + | ||
| COIII | 7262–8098 | + | ATG | TAA |
| tRNA-SerI | 8105–8173 | + | ||
| tRNA-Thr | 8175–8237 | + | ||
| ND5 | 8294–9940 | + | ATT | TAA |
| tRNA-Cys | 9941–10004 | + | ||
| tRNA-SerII | 10007–10074 | + | ||
| Cytb | 10075–11232 | + | ATT | TAG |
| 16S rRNA | 11233–12455 | + | ||
| tRNA-Asn | 12456–12520 | + | ||
| ND4 | 12649–13989 | + | ATA | TAA |
| tRNA-Tyr | 13995–14059 | + | ||
| Atp6 | 14065–14776 | + | GTG | T incomplete |
| tRNA-Glu | 14885–14943 | + | ||
| tRNA-Gly | 14996–15007 | + | ||
| tRNA-Val | 15008–15066 | + | ||
| ND2 | 15100–16026 | + | ATT | TAA |
| tRNA-Ala | 16029–16092 | + |
The mt-genome of Hiatella arctica is 18.204 bp in length and has an A+T content of 66.35 %. As in Acanthocardia, all genes are on the same strand (Fig. 1). The longest of the 30 non-coding regions (Tab 3) has 614 bp and is located between the genes for trn-Ala and atp8. The others range between 1 and 376 bp in length.Hiatella has two copies of a 121 bp motive (Fig. 4) starting in the 3' end of the nad6 gene and extending into the non-coding region before the tRNA-Trp gene. The genes for trn-Leu I and trn-LeuII overlap by one nucleotide.
Table 3.
Non-coding regions and overlaps in the mitochondrial genome of Hiatella arctica
| Position | between | Length |
| Non-coding | ||
| 1654–1819 | COI/Atp6 | 166 |
| 2603–2609 | Atp6/tRNA-Phe | 7 |
| 2674–2678 | tRNA-Phe/tRNA-Lys | 5 |
| 2743 | tRNA-Lys/tRNA-Met | 1 |
| 2809 | tRNA-Met/COII | 1 |
| 3894–4270 | COII/12S rRNA | 376 |
| 5235–5294 | tRNA-His/ND5 | 60 |
| 6963–6983 | ND5/ND1 | 21 |
| 7929 | ND1/tRNA-Gly | 1 |
| 7992–7998 | tRNA-Gly/tRNA-Asp | 7 |
| 8063–8105 | tRNA-Asp/tRNA-Tyr | 43 |
| 8166 | tRNA-Tyr/ND4 | 1 |
| 9529–9533 | ND4-tRNA-Pro | 5 |
| 9601–9621 | tRNA-Pro/tRNA-Thr | 21 |
| 9682–8690 | tRNA-Thr/tRNA-Ala | 9 |
| 9753–10366 | tRNA-Ala/Atp8 | 614 |
| 10527–10730 | Atp8/tRNA-Ile | 204 |
| 10796–10800 | tRNA-Ile/tRNA-LeuI | 5 |
| 10932–10933 | tRNA-Leu/tRNA-Val | 2 |
| 11062 | tRNA-Arg/tRNA-Cys | 1 |
| 11124–11129 | tRNA-Cys/tRNA-Asn | 6 |
| 11094–11311 | tRNA-Asn/Nd4l | 218 |
| 11612–11672 | Nd4l/Nd3 | 61 |
| 12030–12055 | ND3/tRNA-SerI | 26 |
| 13707–13924 | tRNA-Gln/COIII | 218 |
| 14753–14878 | COIII/ND6 | 126 |
| 15482–15722 | ND6/tRNAMetII | 241 |
| 15855–15875 | tRNA-Trp/tRNA-SerII | 21 |
| 15931–16000 | tRNA-SerII/Cytb | 70 |
| 18197–18244 | ND2/COI | 48 |
| Overlapping | ||
| 10864 | tRNA-LeuI/tRNA-LeuII | 1 |
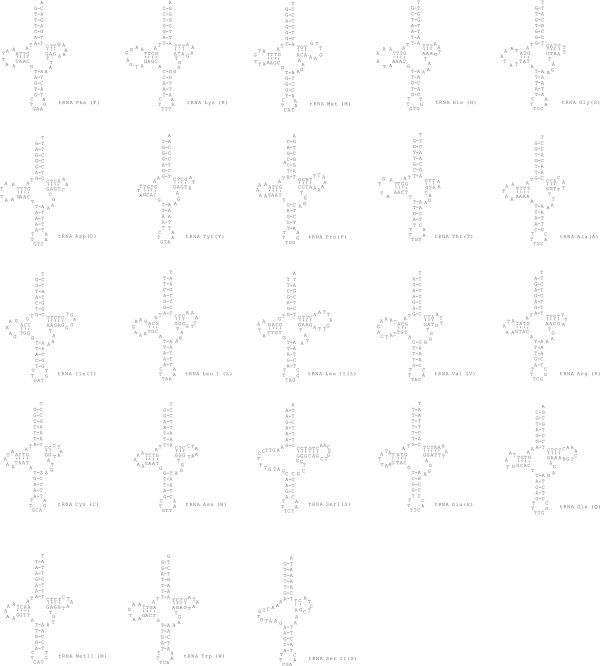
Figure 4.
Cloverleaf structures of the 23 tRNA genes in the mitochondrial genome of Hiatella arctica.
The Hiatella mt-genome contains all 37 mitochondrial genes including atp8 and a second copy of the trn-Met (Fig. 4). A total of 3.985 amino acids are encoded. As in Acanthocardia, the most frequent codons are TTT (Phe; n = 359) and TTA (Leu; n = 284). A or T are present in 2.873 third codon positions (72.09 %). Seven of the 13 protein coding genes start with ATA, the other six genes with ATG. The codon ATT terminates seven, and the codon ATG four protein coding genes (Tab 4). Truncated stop codons (T) are inferred for the atp8 and the coxII genes.
Table 4.
Organisation of the mitochondrial genome of Hiatella arctica
| Gene | Position | Strand | Start | Stop |
| COI | 1–1653 | + | ATG | TAG |
| Atp6 | 1820–2602 | + | ATG | TAA |
| tRNA-Phe | 2610–2673 | + | ||
| tRNA-Lys | 2679–2743 | + | ||
| tRNA-Met | 2745–2808 | + | ||
| COII | 2810–3893 | + | ATA | T incomplete |
| 12S rRNA | 4271–5171 | + | ||
| tRNA-His | 5172–5234 | + | ||
| ND5 | 5295–6962 | + | ATA | TAA |
| ND1 | 6984–7928 | + | ATA | TAA |
| tRNA-Gly | 7930–7991 | + | ||
| tRNA-Asp | 7999–8062 | + | ||
| tRNA-Tyr | 8106–8165 | + | ||
| ND4 | 8167–9528 | + | ATA | TAA |
| tRNA-Pro | 9534–9600 | + | ||
| tRNA-Thr | 9622–9681 | + | ||
| tRNA-Ala | 9691–9752 | + | ||
| Atp8 | 10367–10526 | + | ATG | T incomplete |
| tRNA-Ile | 10731–10795 | + | ||
| tRNA-LeuI | 10801–10864 | + | ||
| tRNA-LeuII | 10864–10931 | + | ||
| tRNA-Val | 10934–10998 | + | ||
| tRNA-Arg | 10999–11061 | + | ||
| tRNA-Cys | 11063–11123 | + | ||
| tRNA-Asn | 11130–11193 | + | ||
| ND4l | 11312–11611 | + | ATG | TAG |
| ND3 | 11673–12029 | + | ATA | TAG |
| tRNA-SerI | 12056–12125 | + | ||
| tRNA-Glu | 12126–12191 | + | ||
| 16S rRNA | 12192–13638 | + | ||
| tRNA-Gln | 13639–13706 | + | ||
| COIII | 13925–14752 | + | ATA | TAA |
| ND6 | 14879–15481 | + | ATG | TAG |
| tRNA-MetII | 15723–15788 | + | ||
| tRNA-Trp | 15789–15854 | + | ||
| tRNA-SerII | 15876–15930 | + | ||
| Cytb | 16001–17155 | + | ATA | TAA |
| ND2 | 17156–18196 | + | ATG | TAA |
Phylogenetic analysis of nucleotide and protein coding sequences
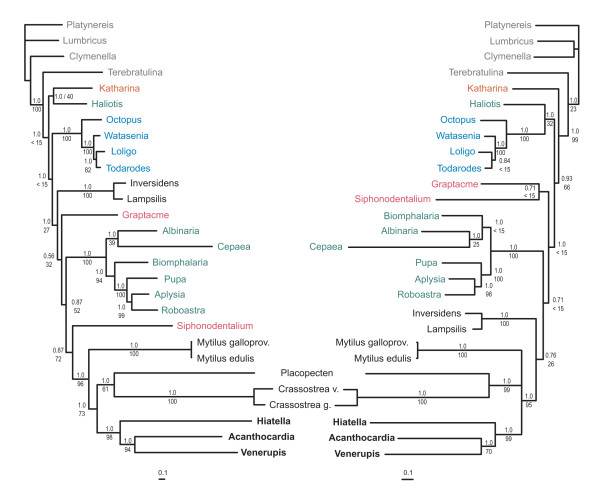
The concatenated amino acid alignment of 28 species (Tab 5) consists of 5.004 positions of which 3.085 are parsimony-informative. The corresponding nucleotide alignment including the rrnL sequences has 16.862 positions in total, 11.854 without 3rd codon positions, of which 7.130 are parsimony-informative. The Bayesian analyses resulted in almost fully resolved trees (Fig. 5) with total marginal -lnL of 156.458,59 for the amino acid data and 209.800,11 for the nucleotide data (arithmetic means). Most branches have posterior probabilities of 1.0. The deeper nodes tend to be less supported. The parsimony analyses of both data sets yielded a single most parsimonious tree each (amino acid data: tree length 31.163, consistency index 0.5819, rescaled consistency index 0.2719; nucleotide data: tree length 49.859, consistency index 0.3440, rescaled consistency index 0.1366; trees not shown). All Bayesian and parsimony analyses recover the three heterodont species as a robust monophylum. Acanthocardia is the sister taxon of Venerupis with high to moderate support (fig 5). The Pteriomorpha are resolved as monophyletic from the nucleotide data in the parsimony analysis only. In most analyses the two unionid species are separated from the remaining bivalves and placed in a more basal position in the tree. Only the Bayesian tree of the nucleotide data resolves monophyletic Bivalvia, although with low support (posterior probability 0.76). This is also the only tree showing monophyletic Scaphopoda and Pulmonata. The unstable position of the vetigastropod Haliotis, near the base of the molluscan clade renders the Gastropoda diphyletic in all analyses. Cephalopoda are always robustly supported, and only the parsimony analysis of the amino acid data fails to resolve Mollusca as monophyletic. The exclusion of highly variable alignment positions using GBLOCKS had no effect on the topologies of the trees and brought only minimal changes in branch support.
Table 5.
List of taxa used in the phylogenetic analysis
| Taxon | Classification | GenBank Accession number |
| Outgroup | ||
| Terebratulina retusa | Brachiopoda | AJ245743 |
| Lumbricus terrestris | Annelida, Clitellata | U24570 |
| Platynereis dumerilii | Annelida, Polychaeta | AF178678 |
| Clymenella torquata | Annelida, Polychaeta | AY741661 |
| Polyplacphora | ||
| Katharina tunicata | Mollusca, Polyplacophora, Neocoleida | U09810 |
| Gastropoda | ||
| Haliotis rubra | Mollusca; Gastropoda, Orthogastropoda, Vetigastropoda | AY588938 |
| Aplysia californica | Mollusca; Gastropoda, Orthogastropoda, Apogastropoda | AY569552 |
| Pupa strigosa | Mollusca; Gastropoda, Orthogastropoda, Apogastropoda | AB028237 |
| Roboastra europaea | Mollusca; Gastropoda, Orthogastropoda, Apogastropoda | AY083457 |
| Biomphalaria glabrata | Mollusca; Gastropoda, Pulmonata, Basammatophora | AY380531 |
| Albinaria caerulea | Mollusca; Gastropoda, Pulmonata, Stylommatophora | X83390 |
| Cepaea nemoralis | Mollusca; Gastropoda, Pulmonata, Stylommatophora | U223045 |
| Scaphopoda | ||
| Graptacme eborea | Mollusca, Scaphopoda, Dentaliida | AY484748 |
| Siphonodentalium lobatum | Mollusca, Scaphopoda, Gadilida | AY342055 |
| Cephalopoda | ||
| Octopus vulgaris | Mollusca, Cephalopoda, Coleoidea, Neocoleoidea, Octopodiformes | AB158363 |
| Loligo bleekeri | Mollusca, Cephalopoda, Coleoidea, Neocoleoidea, Decapodiformes | AB029616 |
| Todarodes pacificus | Mollusca, Cephalopoda, Coleoidea, Neocoleoidea, Decapodiformes | AB158364 |
| Watasenia scintillians | Mollusca, Cephalopoda, Coleoidea, Neocoleoidea, Decapodiformes | AB086202 |
| Bivalvia | ||
| Placopecten magellanicus | Mollusca, Bivalvia, Pteriomorphia, Pectinoida | DQ088274 |
| Mytilus galloprovincialis | Mollusca, Bivalvia, Pteriomorphia, Mytiloida | AY497292 |
| Mytilus edulis | Mollusca, Bivalvia, Pteriomorphia, Mytiloida | AY484747 |
| Crassostrea gigas | Mollusca, Bivalvia, Pteriomorphia, Ostreoida | AF177226 |
| Crassostrea virginica | Mollusca, Bivalvia, Pteriomorphia, Ostreoida | AY905542 |
| Lampsilis ornata | Mollusca, Bivalvia, Palaeoheterodonta, Unionida | AY365191 |
| Inversidens japanensis | Mollusca, Bivalvia, Palaeoheterodonta, Unionida | AB055625 |
| Venerupis phillipinarium | Mollusca, Bivalvia, Heterochonchia, Veneroida | AB065375 |
| Acanthocardia tuberculata | Mollusca, Bivalvia, Heterochonchia, Veneroida | DQ_632743 this study |
| Hiatella arctica | Mollusca, Bivalvia, Heterochonchia, Myoida | DQ_632742 this study |
Figure 5.
Phylogenetic analyses. Bayesian trees of the amino acid sequences of all protein coding genes (left) and nucleotide sequences of the protein coding genes and the rrnL gene (right). Bivalve species are in black font (Heterodonta in bold), Gastropoda in green, Cephalopoda in blue, Scaphopoda in magenta, Polyplacophora in red and the outgroup taxa in grey font. Posterior probabilities (above) and parsimony bootstrap values (below) are given for each branch.
Discussion
Base frequencies, codon usage and amino acid frequencies in the mt genome of Acanthocardia tuberculata compare well with that of other bivalves. For instance, the A+T content of 59.6 % is similar to that Inversidens (57.2), Lampsilis (62.3), Crassostrea gigas (63.4%) and Crassostrea virginica (62.8%), Placopecten (55.7) and Mytilus (61.8%), but it is lower than that of Venerupis (69.7%) and Hiatella arctica (66.4).
The functional and selective significance of the duplicated regions in Acanthocardia is unclear. Tandem repeats are also present in other bivalve mitochondrial genomes: Venerupis, e.g., has four tandem repeats of 203 bp between the nad2 and the nad4l genes [9]; Placopecten has seven repeats of a 79 bp motive between trn- Asn and trn-Glu and two repeats of 1.435 bp between nad6 and trn-Met [18]. More unusual is the duplication in Hiatella starting 12 bases upstream of the 3' end of the nad6 gene. Although the copies are 79 % identical the second repeat has no open reading frame. It is likely that this non-functional copy of the coding part accumulated substitutions more rapidly due to relaxed selection and, thus, lost the reading frame. Nearly identical duplications of complete genes occur in mt-genomes of the cephalopods Watasenia and Todarodes [19,20].
Acanthocardia and Hiatella mt genomes encode 23 transfer RNA genes which can be folded in a typical secondary structure. Both genomes have an additional tRNA for Methionine. A second Methionine tRNAs is present in the bivalves Mytilus edulis [11], Mytilus galloprovincialis [7], Crassostrea virginica [21], Placopecten magellanicus [18] and Venerupis phillipianarum [9]. Overlaps of tRNA genes as observed in Acanthocardia and Hiatella are a common feature in mt-genomes [1].
The atp6 gene of Acanthocardia lacks a Methione or Isoleucine at the putative 5' end and a complete stop codon. The first ATN codon is 48 bp downstream of the putative point of start as inferred from the alignment of the molluscan atp6 genes. The assumed start codon is GTG as in the nad 2 gene of polyplacophore Katharina tunicata [12]. Truncated stop codons like in the Acanthocardia atp6 and the Hiatella co II and the atp8 genes require the inference of the ends of the genes from the alignment with other species. The completion of truncated stop codons by polyadenylation after transcript processing was described by Ojala [22].
Hiatella arctica is the first marine bivalve reported to have a complete atp8 gene consisting of 53 amino acids. The alignment of this atp8 gene (Fig. 6) shows a Methionine at the start and a truncated stop codon T. We also identified a putative atp8 gene in the mt-genome of Venerupis, between the genes rrnL and nad4 at positions 5.974 to 6.088. Although this region was annotated as part of the rrnL by the authors [9], it represents an open reading frame encoding for only 37 amino acid positions. It starts with Leucine instead of Methionine, but ends with a complete stop codon. The more conserved 5' region of the gene resembles other molluscan atp8 genes in amino acid sequence (Fig. 6) and in the hydrophilicity profile. The positively charged 3' region of the gene, which is known to vary greatly in length and composition [23,24], is reduced to a few residues in Venerupis. This is confirmed by the alignment of the amino acid sequence corresponding to the conserved atp8 profiles in other metazoans [25]. It remains open, however, whether this gene is functional. Dreyer and Steiner [15] reported a comparably short atp8 gene for the scaphopod Siphonodentalium lobatum. Serb and Lydeard [26] discuss a non functional version of the atp8 gene in the freshwater mussel Inversidens, and Milbary and Gaffney [21] describe a potential remnant of the atp8 gene in the eastern oyster Crassostrea virginica.
Figure 6.
Alignment of the atp8 genes of Hiatella arctica, Venerupis philippinarum and Katharina tunicata.
Many metazoan mt genomes have neighbouring atp6 and atp8 genes on the same strand. This arrangement is likely to be selected for, if the uncleaved transcripts are co-translated [25,2]. Several taxa lacking this gene arrangement in the mt genome, e.g. Plathyhelminthes, Nematoda, Annelida, Sipunculida, the brachipods Laqueus [27] and Terebratalia [28], and, among Mollusca, Bivalvia and Scaphopoda. Of these genomes, Plathyhelminthes, Nematoda except for Trichinella, and the pteriomorph bivalves lack atp8 altogether. The disparate distribution of this feature clearly indicates that the loss of the atp6 – atp8 coupling and the loss of atp8 occurred several times independently in metazoan evolution. This is corroborated by finding truncated atp8 genes separated from the atp6 gene in the nematode Trichinella and in the scaphopod Siphonodentalium. It is possible that this situation represents an evolutionary stepping stone from the fully functional atp6 – atp8 coupling, via decoupled but complete genes like in annelids and the scaphopod Graptacme, and the complete loss of atp8.
The location of all mt-genes on the same strand, as in Acanthocardia and Hiatella, is uncommon among Metazoa, but is reported for several taxa [28] including all published marine bivalves. Only in the unioid freshwater bivalves Lampsilis ornata and Inversidens japanensis genes are located on both strands. Under the Heteroconchia concept postulating a sister group relationship of Unionida and Heterodonta, the "all-on-one-strand" situation either evolved independently in Heterodonta and Pteriomorph or was lost in the Unionida.
Comparing the gene arrangements of Acanthocardia and Venerupis no identities are apparent, if the tRNA genes are included. The tRNAs are more variable because the secondary structure allows them to translocate more frequently [12]. Even after excluding the tRNAs from the comparison the two mt-genomes show few identical gene junctions. These are limited to the block containing the Cytb – rrnL – nad4 – atp6 genes in Acanthocardia, although this is interrupted by the putative atp8 gene in Venerupis. This gene order may be inherited from the common ancestor of Acanthocardia and Venerupis, with the apomorphic loss of atp8 in Acanthocardia. The mt-genome of Hiatella appears almost completely rearranged. Only the neighbourhood of the nad2 and cox I genes is present in other molluscs like Katharina, Haliotis and Octopus and may represent a plesiomorphic trait.
The mitochondrial genome sequence data confirm previous results [16,17] on the monophyly of Cardiidae and Veneridae relative to the Hiatellidae. Their common branch and the heterodont clade are robustly supported in all analyses. Similarly, the clade uniting Heterodonta and Pteriomorpha is well supported, although to the exclusion of the unionid branch. This is in accordance with the topology of Giribet and Distel [16] but contrasts that of Waller [29] and Steiner and Hammer [30] supporting the Heteroconchia clade (Unionida + Heterodonta). The Bivalvia clade is resolved by the Bayesian analysis of the nucleotide data only. This may indicate the higher potential of recovering correct topologies by this method or the superiority of nucleotide substitution models over amino acid substitution models or a combination of these factors. Note that the Bayesian nucleotide analysis also succeeds in resolving the pteriomorph, scaphopod, and pulmonate branches.
What could cause the unexpected position of the Unionida rendering the Heteroconchia diphyletic? In both amino acid and nucleotide-based trees the unionid species have conspicuously shorter branches compared to the other bivalves. Although the present data set is not large enough for statistical assessment, such obvious differences in substitution rates may cause phylogenetic analyses to find incorrect trees, as previously documented for Bivalvia [30]. In addition to lower substitution rates, different substitution patterns in the unionids may confound phylogenetic analyses. All bivalve mt-genomes have the genes encoded on the same strand, except for those of the unionids where three to four genes are encoded on the opposite strand. Due to the asymmetric replication process the strands show different substitution skews. Hassanin et al.[31] showed that skew differences may influence phylogenetic analyses.
The mitochondrial gene order in the Bivalvia is too divergent and the present taxon set too small to make use of this character set for phylogenetic inference at this point. In addition, the substitution models for phylogenetic inference presently do not take strand specific patterns into account. Similarly, gene rearrangement models are limited to one type of rearrangement only, either translocation or inversion. However, with a growing set of mt-genomes – their nucleotide and gene sequences – we are likely to enhance our understanding of patterns and modes of nucleotide substitutions and gene rearrangements. This will help to improve phylogenetic reconstructions by refining the models for these evolutionary processes. Improved taxon sampling and refined phylogenetic inference models are likely to resolve more open questions of bivalve phylogeny and evolution than with previously used markers.
Methods
Material, DNA isolation, PCR, sequencing
Hiatella arctica (Linné, 1767) and Acanthocardia tuberculata (Linné, 1758) were collected in the Adriatic Sea (Rovinj, Croatia) and frozen in liquid nitrogen. Total DNA was isolated with the DNeasy Tissue Kit (Qiagen).
Partial cox I, rrnL and rrnS genes were amplified (Acanthocardia rrnL and cox I, Hiatella rrnS and rrnL) using the primers HCO2198/LCO 1490, 16Sfmwg/16Srmwg and 12Sai/12Sbi (Table 6). The PCR was done on a Primus 96 advanced Gradient (Peqlab) in a 30 μl reaction containing 1,5 mM Mg Cl2, each dNTP at 250 μM, each primer at 0,5 μM, 0,6 units Taq polymerase (Biotaq Red, Bioline) and the supplied buffer at 1× concentration. The PCR cycle conditions were: Initial denaturation step of 2 min at 94°C, 35 cycles of 30 sec denaturation at 94°C, 40 sec annealing at 48°C and 2 min (HCO2198/16Sbr) or 45 sec primer extension at 72°C followed by a final primer extension step at 72°C for 7 min. PCR products were purified with the E.Z.N.A Cycle-Pure Kit (Peqlab, Germany). PCR products were sequenced automatically with the amplification primers on an ABI-capillary-sequencer at Eurofins-Medigenomix GmbH (Martinsried, Germany). The sequences were used to design sets of long range primers (12SrRNA, CO1, 16SrRNA Primer see table 3) to amplify the whole mitochondrial Genome in three fragments with the TaKaRa LA Taq (Takara) on a Primus 96 advanced Gradient (Peqlab). The 50 μl reactions contained 5 μl of the supplied buffer 3 (including 22.5 mM MgCl2), 25 pmol of each primer, 500 μM dNTPs, 5.25 units of Taq DNA polymerase, 0.3 μl Nonidet P40 and about 100 ng of total DNA. The PCR conditions for both fragments are: initial denaturation at 92°C for 2 min, 30 cycles of 15 sec denaturation at 92°C, 35 sec annealing at 63°C and 10 min primer extension at 72°C followed by a final primer extension step at 72°C for 10 min. The products were sequenced by primer walking.
Table 6.
Amplification primers used in this study
| Species | Primer | Sequence |
| Both species | HCO 2198a | TAAACTTCAGGGTGACCAAAAAATCA |
| LCO 1490a | GGTCAACAAATCATAAAGATATTGG | |
| 16S Fmwgb | CTCGCCTTTTAWCAAAAACAT | |
| 16S Rmwgb | ACGCCGGTCTKAACTCAG | |
| 12Saic | AAACTAGGATTAGATACCCTATTAT | |
| 12Sbic | AAGAGCGACGGGCGATGTGT | |
| Acanthocardia tuberculata | Ac16Flong | GAAGCTTAAACAGTGGGACTGTTCGTCC |
| Ac16Rlong | CCTATAAGACAGCTATTCCATCGTCAAC | |
| AcHCOrev | CGGTGAATTCATAAGATTCCAATGCTACC | |
| AcLCOrev | GAGCATGGTTATGTAGGCATGAAGATG | |
| Hiatella arctica | Hi12Flong | ACCTTCAATAGCTGATCTCTACCCCAGG |
| Hi12Rlong | GCATCATATCCTGTAGGGGAACTTGGCC | |
| Hi16Flong | AAGCTACCACGGGATAACAGCGTG | |
| Hi16rlong | CACGTCAACCCCTTCTTCCTAGACTTC |
aFolmer et al. [47]
bmodified after Palumbi et al. [48]
cSimon et al. [49]
Data analysis
Protein coding genes were analysed by the Open Reading Frame Finder [32] using the invertebrate mitochondrial code. Protein and rRNA genes were identified by their similarity to published gene sequences by BLAST searches [33]. The tRNA genes are usually too little conserved for BLAST hits. Some of them were identified by tRNA-scan SE Search Server [34] and DOGMA [35], others could only be reckognised by manually folding intergenic sequences to cloverleaf structures with anticodons. Codon usage analysis was performed by CodonW version 1.3 [36]. The whole sequence was tested for potentially tandem repeats by TANDEM REPEAT FINDER, Version 4.0 [37]. The hydrophobicity profiles for the atp8 genes are generated using the general method of Kyte and Doolittle [38] with BIOEDIT version 7.0.5 [39].
Phylogenetic analysis
Deduced amino acid sequences were aligned with CLUSTAL X 1.83 [40] at default settings followed by manual correction. The nucleotide alignment was based on the amino acid alignment. All protein coding and the rrnL gene sequences of 24 molluscs (10 bivalves, 4 cephalopods, 2 scaphopods, 7 gastropods and 1 polyplacophore [tab 6]) were concatenated in a single Nexus file. Three annelids and one brachiopod served as outgroups. Separate analyses were run with all positions and with hypervariable positions excluded with GBLOCKS 0.91 [41]. We used PAUP* 4.0b10 [42] for equally weighted parsimony analyses with the heuristic search option and 50 random addition sequences with TBR branch swapping. Bootstrap support was assessed by 10.000 (amino acid data) or 20.000 replicates (nucleotide data) with three random addition sequences each. Bayesian inference was performed with MRBAYES 3.1 [43,44] on the Schrödinger II cluster of the Univ. Vienna computing facility under the Mtrev+Γ+I substitution model for the amino acid data set. The AIC criterion implemented in MODELTEST 3.06 [45] returned the GTR+Γ+I model as most appropriate for the nucleotide data set. We used separate and unlinked partitions for each gene and 2 × 4 chains of 5 × 105 generations, sampling every 100th tree. Burnin estimation by lnL and convergence diagnostics were used as implemented in the software. We excluded 3rd codon positions from the nucleotide analyses to reduce phylogenetic noise due to substitution saturation. Trees were visualized with TREEVIEW 1.6.6 [46].
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
HD carried out the genome sequencing and annotation, contributed to the phylogenetic analyses and drafted parts of the manuscript. GS designed the study, collected the animals, carried out the phylogenetic analyses, drafted parts of the manuscript and is responsible for the final editing.
Acknowledgments
Acknowledgements
This study was supported by the Austrian Science Fund (FWF) project no. P16954.
Contributor Information
Hermann Dreyer, Email: Hermann.Dreyer@univie.ac.at.
Gerhard Steiner, Email: Gerhard.Steiner@univie.ac.at.
References
- Wolstenholme DR. Animal mitochondrial DNA: Structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- Boore JL, Staton JL. The mitochondrial genome of the sipunculid Phascolopsis gouldii supports its association with Annelida rather than Mollusca. Mol Biol Evol. 2002;19:127–137. doi: 10.1093/oxfordjournals.molbev.a004065. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DT, Saavedra C, Stanwood RR, Ball AO, Zouros E. Male and female mitochondrial DNA lineages in the Blue Mussel Mytilus. Mol Biol Evol. 1995;12:735–747. doi: 10.1093/oxfordjournals.molbev.a040252. [DOI] [PubMed] [Google Scholar]
- Hoeh WR, Stewart DT, Sutherland BE, Zouros E. Multiple origins of gender-associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia) Evolution. 1996;50:2276–2286. doi: 10.2307/2410697. [DOI] [PubMed] [Google Scholar]
- Liu HP, Mitton JB, Wu SK. Paternal mitochondrial DNA differentiation far exceeds maternal mitochondrial DNA and allozyme differentiation in the freshwater mussel, Anodonta grandis grandis. Evolution. 1996;50:952–957. doi: 10.2307/2410870. [DOI] [PubMed] [Google Scholar]
- Mizi A, Zouros E, Moschonas N, Rodakis GC. The complete maternal and paternal mitochondrial genomes of the mediterranean mussel Mytilus galloprovincialis : Implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol. 2005;22:952–967. doi: 10.1093/molbev/msi079. [DOI] [PubMed] [Google Scholar]
- Breton S, Burger G, Stewart DT, Blier PU. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.) Genetics. 2006;172:1107–1119. doi: 10.1534/genetics.105.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki M, Ueshima R. Gender-associated mtDNA of Tapes philippinarum http://www.ncbi.nlm.nih.gov/ (unpublished, Genbank [AB065374], [AB065375])
- Okazaki M, Ueshima R. Evolutionary diversity between the gender-associate mitochondrial DNA genomes of freshwater mussels http://www.ncbi.nlm.nih.gov/ (unpublished, Genbank [AB055624], [AB0055625])
- Hoffmann RJ, Boore JL, Brown WM. A novel mitochondrial genome organization for the Blue Mussel, Mytilus edulis. Genetics. 1992;131:397–412. doi: 10.1093/genetics/131.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. The complete DNA sequence of the mitochondrial genome of the black chiton Katharina tunicata. Genetics. 1994;138:423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki N, Ueshima R, Terrett JA, Yokobori SI, Kaifu M, Segawa R, Kobayashi T, Numachi KI, Ueda T, Nishikawa K, Watanabe K, Thomas RH. Evolution of pulmonate gastropod mitochondrial genomes: Comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implications of unusual tRNA secondary structures. Genetics. 1997;145:749–758. doi: 10.1093/genetics/145.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Medina M, Rosenberg LA. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol Biol Evol. 2004;21:1492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. The complete sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca) Mol Phylogenet Evol. 2004;31:605–617. doi: 10.1016/j.ympev.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Giribet G, Distel DL. Bivalve phylogeny and molecular data. In: Lydeard C, Lindberg DR, editor. Molecular Systematics and Phylogeography of Mollusks. Washington and London: Smithsonian Books; 2003. pp. 45–90. [Google Scholar]
- Dreyer H, Steiner G, Harper EM. Molecular phylogeny of Anomalodesmata (Mollusca: Bivalvia) inferred from 18S rRNA sequences. Zool J Linnean Soc. 2003;139:229–246. doi: 10.1046/j.1096-3642.2003.00065.x. [DOI] [Google Scholar]
- La Roche J, Snyder M, Cook DI, Fuller K, Zouros E. Molecular characterization of a repeat element causing large-scale size variation in the mitochondrial DNA of the sea scallop Placopecten magellanicus. Mol Biol Evol. 1990;7:45–64. doi: 10.1093/oxfordjournals.molbev.a040586. [DOI] [PubMed] [Google Scholar]
- Akasaki T, Nikaido M, Tsuchiya K, Segawa S, Hasegawa M, Okada N. Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Mol Phylogenet Evol. 2006;38:648–658. doi: 10.1016/j.ympev.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Fukuda N, Nakamura M, Aoyama T, Oshima T. Long-term conservation of six duplicated structural genes in cephalopod mitochondrial genomes. Mol Biol Evol. 2004;21:2034–2046. doi: 10.1093/molbev/msh227. [DOI] [PubMed] [Google Scholar]
- Milbury CA, Gaffney PM. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar Biotechnol. 2005;7:697–712. doi: 10.1007/s10126-005-0004-0. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi J. tRNA punctation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acid Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Complete mtDNA of Ciona intestinalis Reveals Extensive Gene Rearrangement and the Presence of an atp8 and an Extra trnM Gene in Ascidians. J Mol Evol. 2004;58:376–389. doi: 10.1007/s00239-003-2559-6. [DOI] [PubMed] [Google Scholar]
- Watkins RF, Beckenbach AT. Partial Sequence of a sponge mitochondrial genome reveals sequence similarity to cnidaria in cytochrome oxidase subunit II and the large ribosomal RNA subunit. J Mol Evol. 1999;48:542–554. doi: 10.1007/PL00006497. [DOI] [PubMed] [Google Scholar]
- Serb JM, Lydeard C. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): An eximination of the evolution and phylogenetic utility of mitochondrial genome organisation in Bivalvia (Mollusca) Mol Biol Evol. 2003;20:1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- Noguchi Y, Endo K, Tajima F, Ueshima R. The mitochondrial genome of the brachiopod Laqueus rubellus. Genetics. 2000;155:245–259. doi: 10.1093/genetics/155.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfenbein KG, Brown WM, Boore JL. The Complete Mitochondrial Genome of the Articulate Brachiopod Terebratalia transversa. Mol Biol Evol. 2001;18:1734–1744. doi: 10.1093/oxfordjournals.molbev.a003961. [DOI] [PubMed] [Google Scholar]
- Waller TR. Origin of the molluscan class Bivalvia and a phylogeny of major groups. In: Johnston PA Haggart, editor. Bivalves: An eon of Evolution – Paleobiological Studies honoring Norman D Newell. University of Calgary Press; 1998. pp. 1–47. [Google Scholar]
- Steiner G, Hammer S. Molecular phylogeny of Bivalvia (Mollusca) inferred from 18S rDNA sequences with particular reference to the Pteriomorphia. In: Harper EM, Taylor JD, Crame JA, editor. The Evolutionary Biology of the Bivalvia. Geological Society of London Special Publications 77; 2000. pp. 11–29. [Google Scholar]
- Hassanin A, Léger N, Deutsch J. Evidence for Multiple Reversals of Asymmetric Mutational Constraints during the Evolution of the Mitochondrial Genome of Metazoa, and Consequences for Phylogenetic Inferences. Systemat Biol. 2005;54:277–298. doi: 10.1080/10635150590947843. [DOI] [PubMed] [Google Scholar]
- NCBI Open Reading Frame Finder http://www.ncbi.nlm.nih.gov/gorf/gorf.html
- The Basic Local Alignment Search Tool (BLAST) http://www.ncbi.nlm.nih.gov/BLAST/ [DOI] [PubMed]
- tRNAscan-SE Search Server http://www.genetics.wustl.edu/eddy/tRNAscan-SE/
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- codonw: Correspondence Analysis of Codon Usage http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–142. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Sym Ser. 1999;41:95–98. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods), version 4beta10. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis-coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh R, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol and Biotec. 1994;3:294–299. [PubMed] [Google Scholar]
- Palumbi SR. Nucleic Acids II: The Polymerase Chain Reaction. In: Hillis, DM, Moritz C, Mable BK, editor. Molecular Systematics. 2. Sunderland, MA: Sinauer; 1996. pp. 205–247. [Google Scholar]
- Simon C, Frati F, Beckenback A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primers. Annals Entomol Soc Am. 1994;87:651–701. [Google Scholar]