Abstract
Background
Potato is a major staple food, and modification of its provitamin content is a possible means for alleviating nutritional deficiencies. beta-carotene is the main dietary precursor of vitamin A. Potato tubers contain low levels of carotenoids, composed mainly of the xanthophylls lutein, antheraxanthin, violaxanthin, and of xanthophyll esters. None of these carotenoids have provitamin A activity.
Results
We silenced the first dedicated step in the beta-epsilon- branch of carotenoid biosynthesis, lycopene epsilon cyclase (LCY-e), by introducing, via Agrobacterium-mediated transformation, an antisense fragment of this gene under the control of the patatin promoter. Real Time measurements confirmed the tuber-specific silencing of Lcy-e. Antisense tubers showed significant increases in beta-beta-carotenoid levels, with beta-carotene showing the maximum increase (up to 14-fold). Total carotenoids increased up to 2.5-fold. These changes were not accompanied by a decrease in lutein, suggesting that LCY-e is not rate-limiting for lutein accumulation. Tuber-specific changes in expression of several genes in the pathway were observed.
Conclusion
The data suggest that epsilon-cyclization of lycopene is a key regulatory step in potato tuber carotenogenesis. Upon tuber-specific silencing of the corresponding gene, beta-beta-carotenoid and total carotenoid levels are increased, and expression of several other genes in the pathway is modified.
Background
Potato is the fourth most important source of calories for mankind after the cereals wheat, rice and maize. Potato production worldwide stands at 293 million tons, of which 36% in developing countries, and covers more than 18 million hectares [1]. Therefore, the nutritional improvement of potato is of paramount importance to help eradicate nutritional deficiencies.
Vitamin A deficiency affects several hundred million people worldwide [2]. Vitamin A is ingested by humans in two forms: preformed vitamin A (retinol), found in foods of animal origin, and β-carotene (provitamin A), found in dark green vegetables like spinach, in palm oil, and in orange vegetables like yams or carrots.
The first dedicated step in the biosynthesis of carotenoids is the synthesis of phytoene, a colorless compound which is desaturated and isomerized into all-trans lycopene (Figure 1). Lycopene is the substrate for two competing cyclases: epsilon-cyclase, introducing a single epsilon-ring, and beta-cyclase introducing one or two beta-rings. Successive hydroxylation and epoxidation reactions result in the synthesis of lutein, on the branch starting at ε-cyclase, and β-β-ring xanthophylls on the branch starting at β-cyclase.
Figure 1.
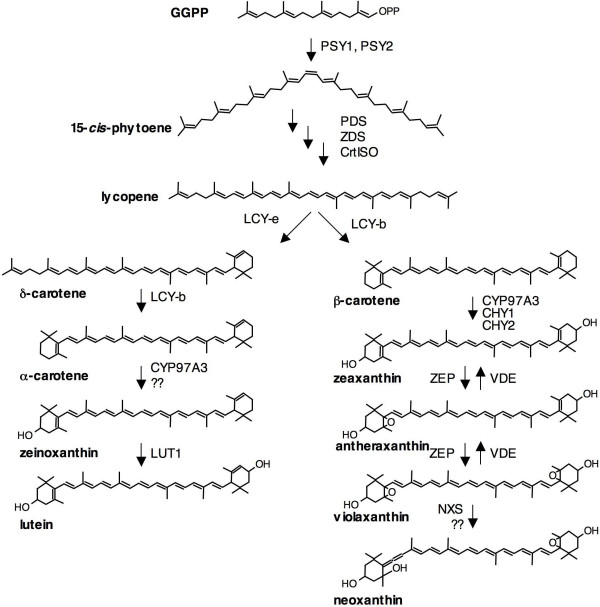
Carotenoid biosynthesis pathway. The enzymes involved in the various reactions are indicated.
Metabolic engineering of plant carotenoids has been achieved through different strategies (reviewed in [3,4]). These can be summarized in:
i) "push strategies", in which a gene encoding a rate-limiting step in the pathway is overexpressed. Examples of "push" strategies are the overexpression of the PSY gene, encoding phytoene synthase, in rice [5], canola [6], tomato [7] and potato [8], of later genes in the pathway, such as those encoding lycopene β-cyclases [9,10] or of "mini-pathways" composed of more than one gene, directing the formation of β-carotene in rice endosperm [11] or of zeaxanthin in tomato fruits [12]. A variant of "push" strategies consists in the introduction of non-plant genes to redirect metabolite flux and synthesize new compounds [13-15].
ii) "block" strategies, relying on the silencing of a biosynthetic step situated immediately downstream of the compound whose levels are to be increased. Examples of "block" strategies are the increase in lycopene in tomato fruits obtained through the silencing of lycopene β-cyclases [9,10], or the increase of zeaxanthin in potato tubers obtained through the silencing of zeaxanthin epoxidase [16]. In this paper, we present a variant "block" strategy, aimed at redirecting the metabolic flux into the β-β branch of the bifurcated carotenoid pathway by silencing the first step of the competing ε-β branch. We present the phenotypic analysis of transgenic potato plants in which the first biosynthetic step in the branch leading to lutein, lycopene ε-cyclase (Figure 1), has been inhibited through a tuber-specific antisense approach.
Results
Construction of transgenic plants
The first dedicated step in lutein biosynthesis is the ε-cyclization of lycopene by LCY-e (Figure 1). In cv. Desirée, lutein constitutes about 12% of tuber and 48% of leaf carotenoids, respectively. In Arabidopsis, lut2 mutants, which are deficient in ε-cyclization, are viable [17] but, under stress conditions, show reduced chlorophyll triplet quenching and, therefore, enhanced photooxidation [18]. In order not to interfere with leaf lutein biosynthesis, we decided to silence the lycopene ε-cyclase transcript (LCY-e) specifically in tubers. Several examples of organ-specific silencing are known in plants, such as the fruit-specific silencing of LYCOPENE β-CYCLASE (LCY-b) [9] or of the HP-2 gene [19] in tomato. In the present work, the patatin promoter [20] was used to target the expression of the silencing transcript in tubers. Appropriate fragments of the patatin B33 promoter [20] and of the LCY-e transcript were amplified by PCR, re-sequenced, and cloned in the pBI101 vector for Agrobacterium-mediated transformation [21] (see methods). Two plasmids were constructed: plasmid pB33:GUS contains the GUS reporter gene under the control of the B33 promoter, while plasmid pBI33:AS-e contains an antisense LCY-e cDNA fragment replacing GUS (See Figure 2 and Methods).
Figure 2.
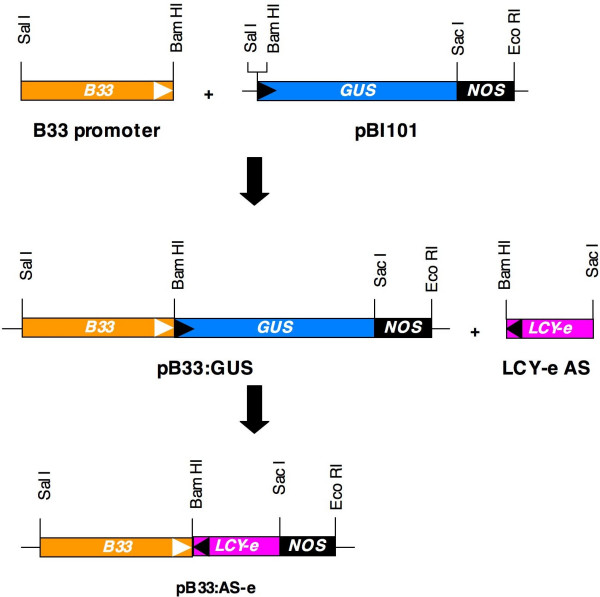
Construction of plasmids for Agrobacterium-mediated transformation. The patatin B33 promoter and a region of the LCY-e transcript were amplified, respectively, from genomic DNA and tuber cDNA using primers inserting appropriate restriction sites at the ends, resequenced, and cloned in the pBI101 plasmid [21]. White triangles indicate the direction of transcription, black triangles the direction of the coding strand. For details see Methods.
The two constructs were introduced in potato (cv Desirée) using published methods [22] and transgenic plants were selected on kanamycin. The presence of the transgene was confirmed via PCR (see Methods). Two independent transgenic lines for GUS and 6 for AS-e were selected for further characterization. For each clone, two plants were acclimated in the greenhouse for tuberization. At the end of the life cycle, tubers were harvested and the tuber production was evaluated. Neither GUS nor AS-e transgenic plants showed major alterations in tuber weight or number (data not shown).
Tuber and leaf carotenoid composition
The carotenoid composition of tubers from the Wt, one of the GUS lines and all the AS-e lines was analyzed through diode array HPLC (Table 1). The GUS line did not show significant changes in carotenoid content, with respect to wild-type Desirée. In contrast, four out of the six AS-e lines (lines 1 to 4) showed significant (>1.8-fold) increases in total tuber carotenoids, as well as changes in carotenoid composition. This increase was reflected in a minor increase in yellow pigmentation of the tubers (Figure 3). Consistent with the hypothesized silencing of LCY-e, the levels of β-β-carotenoids (β-carotene, zeaxanthin, antheraxanthin, violaxanthin and neoxanthin, see Figure 1) showed significant increases (up to 3-fold) in these lines. β-carotene was the single carotenoid compound showing the highest relative increases, up to 14-fold in line AS-e 4. Somewhat surprisingly, the levels of lutein remained fairly constant in most of the AS-e lines. The ratio of β-β- to β-ε-carotenoids increased from 5.4 in the wild-type, up to 16.3 in line AS-e 2.
Table 1.
HPLC analysis of tuber carotenoids (ng/g dry weight). Carotenoids were measured via diode array HPLC (see Methods) on a minimum of 4 different tubers from 2 different plants. Fold variation with respect to the wild-type is reported for each carotenoid compound and for each line.
| Line | Phytofluene | Lutein | β-carotene | Zeaxanthin | Antheraxanthin | Violaxanthin | Neoxanthin | Other Xanthophylls | Esters | Total Carotenoids |
| Wild-type | 43.07± 12.78 | 588.01± 108.14 | 3.17± 1.01 | 262.64± 108.08 | 1301.33± 479.34 | 1109.08± 420.06 | 541.02± 98.14 | 89.34± 13.02 | 734.35± 173.04 | 4672.02± 1414.62 |
| Gus 1 | 52.01± 5.87 | 531.46± 153.57 | 3.76± 1.32 | 223.02± 63.04 | 1091.14± 293.24 | 1157.69± 332.49 | 685.32± 204.95 | 81.34± 22.40 | 1270.39± 299.42 | 5096.15± 1376.25 |
| Fold Variation | 1.21 | 0.90 | 1.20 | 0.85 | 0.84 | 1.04 | 1.27 | 0.91 | 1.73 | 1.090 |
| AS-e 1 | 160.59± 8.03 | 725.46± 100.33 | 17.17± 3.59 | 626.28± 187.37 | 2173.42± 97.33 | 2788.91± 170.82 | 303.66± 83.88 | 110.94± 38.43 | 2015.11± 525.48 | 8921.56± 1218.27 |
| Fold Variation | 3.73 | 1.23 | 5.42 | 2.38 | 1.67 | 2.51 | 0.56 | 1.24 | 2.74 | 1.909 |
| AS-e 2 | 248.59± 62.92 | 506.12± 112.22 | 14.35± 3.14 | 669.39± 50.98 | 3045.66± 493.89 | 3320.07± 1061.82 | 1219.54± 20.21 | 158.88± 79.35 | 3089.75± 69.24 | 12272.36± 1961.77 |
| Fold Variation | 5.77 | 0.86 | 4.53 | 2.55 | 2.34 | 2.99 | 2.25 | 1.78 | 4.21 | 2.627 |
| AS-e 3 | 86.97± 6.34 | 866.91± 104.46 | 9.48± 1.84 | 465.48± 146.89 | 1703.35± 150.12 | 2917.70± 720.57 | 945.84± 318.98 | 125.80± 55.78 | 1555.41± 589.078 | 8677.12± 2094.07 |
| Fold Variation | 2.02 | 1.47 | 2.99 | 1.77 | 1.31 | 2.63 | 1.75 | 1.41 | 2.19 | 1.857 |
| AS-e 4 | 193.70± 1.23 | 1044.59± 144.40 | 43.56± 14.24 | 990.83± 197.51 | 2198.54± 559.12 | 2935.03± 797.13 | 612.56± 245.85 | 90.14± 24.62 | 1865.33± 1010.02 | 9974.29± 2994.11 |
| Fold Variation | 4.50 | 1.78 | 13.74 | 3.77 | 1.70 | 2.65 | 1.13 | 1.01 | 2.54 | 2.135 |
| AS-e 5 | 35.44± 5.22 | 510.42± 99.42 | 2.60± 0.92 | 324.83± 111.81 | 1266.71± 78.11 | 1124.44± 409.81 | 421.81± 173.67 | 98.73± 20.30 | 776.25± 87.11 | 4561.23± 986.38 |
| Fold Variation | 0.82 | 0.87 | 0.82 | 1.24 | 0.97 | 1.01 | 0.78 | 1.10 | 1.06 | 0.977 |
| AS-e 6 | 50.21± 8.87 | 469.53± 137.01 | 3.32± 2.01 | 338.81± 87.43 | 1369.53± 263.04 | 1378.81± 359.37 | 746.48± 221.87 | 76.81± 13.37 | 688.91± 118.44 | 5123.42± 1211.42 |
| Fold Variation | 1.17 | 0.80 | 1.05 | 1.29 | 1.05 | 1.24 | 1.38 | 0.86 | 0.99 | 1.097 |
Figure 3.
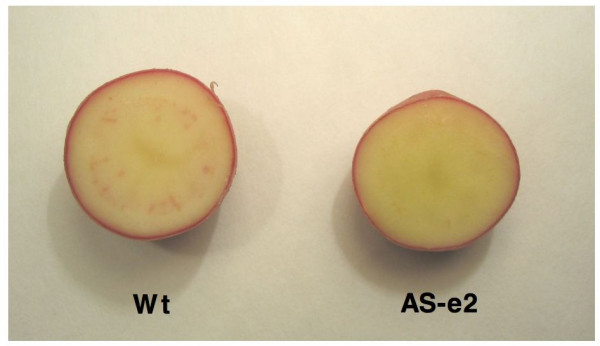
Pigmentation of Wt and transgenic tubers. Tubers were photographed immediately after harvesting.
A slight increase of the colorless biosynthetic intermediate, phytofluene, was observed in lines AS-e 1 to 4. The other early intermediates (phytoene, ζ- carotene) were below detection in all lines.
These changes in carotenoid content must be attributed to a specific effect of the introduced AS-e transgene, and not to non-specific events occurring during plant regeneration, such as somaclonal variation. These non-specific events account for a very limited variation in carotenoid levels, as shown by the data relative to the GUS line (Table 1) which shows only minor changes with respect to the wild-type.
Consistently with the tuber-specific nature of the promoter used for the silencing construct, no significant variations in carotenoid or chlorophyll content were observed in leaves of GUS or AS-e lines (data not shown).
Transgene expression in tubers and leaves
We performed fluorimetric GUS assays on the GUS lines and Real Time RT-PCR assays on four AS-e lines showing significant variations in tuber carotenoids (AS-e 1 to 4) and one showing wild-type carotenoid composition (AS-e 6). The results are shown in Figure 4. GUS activity was found in tissues of GUS transgenic plants, but not Wt plants, and was preferentially expressed in tubers. The AS-e silencing transcript was detected in the tubers of all the AS-e lines, except for AS-e 6. Lack of expression of the AS-e transcript in this line is not due to improper transgene integration, since the transgene is present and is not rearranged (data not shown). The silencing transcript was below the levels of detection of the Real Time assay in the leaves of all the AS-e lines (Figure 4). The levels of expression of the transgene in the tubers were rather low, ranging from 0.04-fold to 0.002-fold β-tubulin. Other than the lack of transgene expression in line AS-e 6, no evident correlation between the levels of transgene expression and carotenoid composition of tubers (Table 1) were observed.
Figure 4.
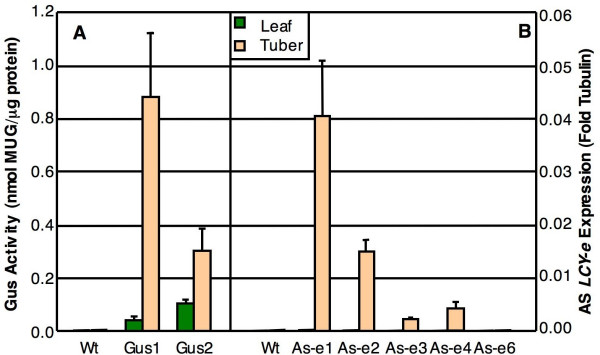
Transgene expression. (A): GUS activity was measured fluorimetrically. (B): AS-e tranSgene expression was measured via Real Time RT-PCR. For details see Methods.
Perturbations in endogenous carotenoid gene expression
Expression of some genes in the carotenoid pathway can be forward- or feedback-regulated by changes of the levels of pathway intermediates: for instance, the PDS transcript, encoding phytoene desaturase, is induced in leaves accumulating phytoene as a result of the ghost mutation or after treatment with the herbicide norflurazon [23,24], suggesting that expression of this gene may be driven by the concentration of the substrate of the reaction. In agreement with this hypothesis, tubers overexpressing a bacterial phytoene synthase show increased levels of the PDS transcript [8].
We measured the expression of carotenoid gene transcripts in the tubers, using Real Time quantitative RT-PCR. Alongside the Wt, we analyzed lines GUS1 and AS-e6, showing wild-type carotenoid levels, and lines AS-e 2, 3 and 4, showing significant changes in carotenoid composition. We analyzed transcripts for most genes in the carotenoid pathway (PSY1, PSY2, PDS, ZDS, CrtISO, LCY-b, LCY-e, CHY1, CHY2, LUT1, ZEP). The biosynthetic steps catalyzed by these genes are shown in Figure 1. The identity of the transcripts used for designing the oligonucleotides used for RT-PCR, is indicated in Methods. The tuber transcript levels, normalized first for the β-tubulin transcript and then for the Wt, are shown in Figure 5.
Figure 5.
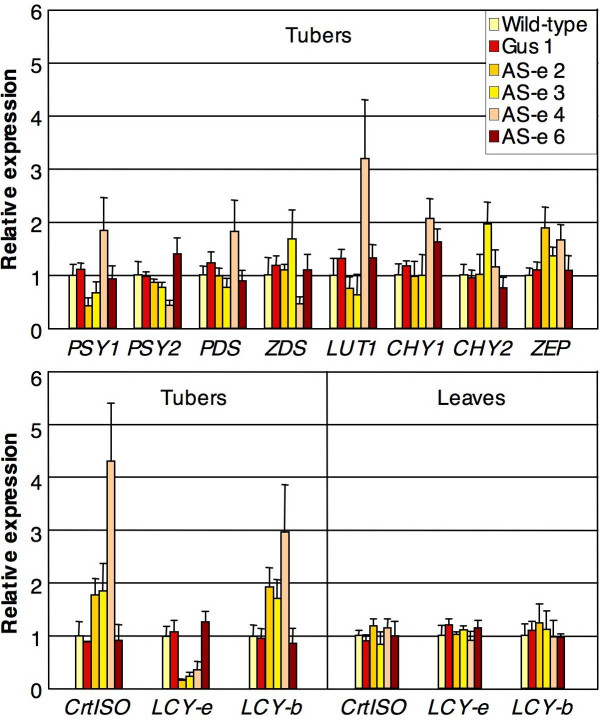
Endogenous carotenoid gene expression. Transcript levels were measured through Real Time RT-PCR and were first normalized for expression of the housekeeping β-tubulin gene, and then for the expression levels in the Wt. Data show the average and SE (error bars) of determinations from at least 4 different tubers (or leaves) from 2 different plants. For details see Methods.
The GUS1 line, as well as line AS-e6, showed only minor variations in gene expression with respect to the Wt line. This indicates that in vitro culture conditions, somaclonal effects due to regeneration procedures, or the presence of the silencing transgene by itself, do not cause any major variability in endogenous carotenoid gene expression. The endogenous LCY-e transcript shows consistent silencing in AS-e lines 2,3 and 4, but not in the other lines. Line AS-e 2, which is the one showing the highest increase in β-β-xanthophylls (Table 1), also shows the most efficient silencing of endogenous LCY-e.
Some clear trends in gene expression can be identified: CrtISO, LCY-b and ZEP show an increase in all the AS-e lines, with the exception of line AS-e 6, which has a wild-type carotenoid content. Other transcripts show more variable trends, being induced or repressed in only one or two of the AS-e lines.
Line AS-e4, which is the one showing the highest increase in total carotenoid levels (Table 1), stands out from the other lines analyzed, in that it shows a simultaneous induction of several transcripts: PSY1, PDS, CrtISO, LCY-b, LUT1, and CHY1.
The transcript levels of the three genes showing significant changes in tubers (LCY-e, Lcy-b and CrtISO) were measured also in leaves (Figure 5). In this tissue, they showed only minor oscillations. This confirms that, similarly to the alterations in carotenoid content, also the perturbations in endogenous carotenoid transcript levels were limited to tubers, the target tissue of AS-e transgene overexpression.
Discussion
Like for any other metabolic pathway, flux through the carotenoid pathway is thought to be controlled by rate-limiting enzymatic steps, whose modification through metabolic engineering should be able to change carotenoid composition. Metabolic flux analysis [25] should allow the identification of these rate-limiting steps, and predict the effects of their engineering. However, the experimental verification of these predictions often contradicts theory, mainly because the levels of expression of some of the genes, and thus of the corresponding enzymes, are sensitive to the levels of the intermediates and of the final products of the pathway.
In potato, overexpression of early genes of the carotenoid pathway is a promising strategy for increasing total carotenoids and β-carotene in the tuber. Overexpression of the bacterial CrtB gene, encoding phytoene synthase, leads to up to 7-fold increase in total carotenoids, mainly β-carotene and lutein [8]. This indicates that PSY is a rate-limiting step in wild-type tubers.
The role of LCY-e and LCY-b in directing the flux towards the synthesis of β-β and β-ε carotenoids can hardly be underestimated. Modulating the levels of these two competing cyclases is a strategy for obtaining tubers with a defined carotenoid composition. The results presented here indicate that the LCY-e transcript can be silenced in a tuber-specific fashion, resulting in a tuber-specific increase of β-β-carotenoids (up to 3-fold), and, more specifically, of β- carotene (up to 14-fold).
Real Time RT-PCR measurements showed that the silencing transgene, introduced under the control of the patatin promoter, was expressed in a tuber-specific fashion in all but one of the transgenic lines. The transgene was expressed at rather low levels, which is not surprising, since homology-driven post-transcriptional gene silencing is known to be a bidirectional phenomenon, affecting both the target gene and the silencing construct [26]. The lack of expression of the transgene in line AS-e 6 awaits an explanation, since the construct is stably integrated in the genome and it is not rearranged (data not shown). Whatever the case, this line, together with the GUS1 line, shows wild-type carotenoid composition, reinforcing the hypothesis that the alterations in carotenoid content observed in lines AS-e 1 to 4 are indeed the effect of expression of the AS-e transgene, and thus of the silencing of endogenous LCY-e. In fact, in all the lines expressing the AS-e transgene, the endogenous Lcy-e transcript was silenced, to various extents. The silencing was confined to tubers. This result suggests that, similarly to tomato fruits [9,19], a physical barrier must exist, preventing the spreading of the silencing signal outside of the tuber tissue.
The tuber-specific changes in LCY-e gene expression were accompanied by tuber-specific changes in carotenoid composition: first, total carotenoid levels increased significantly, albeit to variable extents; second, this increase involved, as expected, mostly carotenoids of the β-β- branch; third, β-carotene showed the highest relative increases (up to 14-fold) among all carotenoid compounds. Surprisingly, lutein showed little, if any, reduction in the silenced tubers. This observation suggests that LCY-e activity is not rate-limiting for tuber lutein levels. It is possible that, to decrease lutein, much higher levels of silencing of LCY-e are required, similar to the ones obtained with RNAi constructs [27]. Several additional enzymes, such as carotenoid hydroxylases [28,29] are involved in lutein synthesis. Furthermore, it is also possible that lutein levels are also controlled by hitherto undescribed carotenoid cleavage activities [30].
Endogenous carotenoid gene expression shows two distinct patterns of regulation in silenced tubers: three transcripts (CrtISO, LCY-b and ZEP) are significantly induced in all the silenced lines. This indicates that these transcripts are sensitive either to the levels of LCY-e expression, or to the levels of one or more carotenoid pathway intermediates.
In one of the lines, AS-e 4, showing the highest total carotenoid and β-carotene levels, a generalized induction of carotenoid transcripts is observed (CrtISO, LCY-b and ZEP, but also PSY1, PDS and the two paralogs showing preferential expression in tubers, i.e. LUT1 and CHY1). This result can be interpreted in two different ways:
a) the generalized induction of carotenoid gene transcripts could be an effect of the more extreme carotenoid level perturbation observed in this line with respect to the other ones
b) instead, the generalized induction could be the effect of a mutation, or of another somaclonal event, occurred during transformation or regeneration of this line, and having a generalized inductive effect on the carotenoid pathway. In this case, the induction of carotenoid gene expression, added to the silencing of LCY-e, would be the cause of the extreme variation in carotenoid content observed in this line.
No major changes in tuber productivity or leaf carotenoid composition were observed in any of the lines, suggesting that silencing of LCY-e is a viable strategy for changing tuber carotenoid composition without affecting agronomic performance. Field trials are however needed to verify the agronomic performance of these lines.
The presence of gene paralogs showing preferential expression in tubers constitutes an interesting parallel with tomato, a close relative of potato, where it has been suggested that duplication of the PSY and CHY genes has created two parallel pathways, one active in leaves and the second active in chromoplast-containing organs (fruits and flowers) [31]. In agreement with this hypothesis, mutation or silencing of PSY1 in tomato affects fruit and flower, but not leaf carotenoids [32,33]. It remains to be demonstrated what is the relative contribution of the different members of the PSY and CHY gene families to potato tuber carotenogenesis.
Conclusion
Using an antisense construct under the control of the tuber-specific patatin promoter, we obtained the tuber-specific silencing of the potato LCY-e transcript. The amount of β-β-carotenoids increased accordingly in a tuber-specific fashion, without a parallel decrease of the major tuber β-ε- carotenoid, lutein. This modification in carotenoid content is paralleled by modifications in endogenous carotenoid gene expression. The strategy described here, in combination with previously described ones, such as the overexpression of phytoene synthase and the antisense silencing of zeaxanthin epoxidase, constitutes a toolbox for the metabolic engineering of the carotenoid content of potato tubers.
Methods
Unless indicated differently, molecular biology methods are as described in [34]. The cloning strategy is outlined in Figure 2. Briefly, a 1.53 Kb patatin B33 promoter fragment [20] was amplified from potato (cv Berolina) genomic DNA using the primers B33-up and B33-dw (Table 2). These primers inserted, respectively, Sal I and Bam HI sites upstream and downstream of the promoter sequence. After intermediate cloning in the pBSK+ vector and re-sequencing, the fragment was inserted in the pBI101 vector [21] upstream of the GUS reporter gene, resulting in the plasmid pBI33:GUS.
Table 2.
Primers used. Sequences of the primers used for cloning of the promoter and gene fragments, for PCR screening of the putative transgenic plants, and for Real Time quantification of transcript levels. For further details, see Methods.
| Primer name | Sequence | Use |
| B33-149 Up | ACATATTTTACTTTTGACTTT | B33 cloning |
| B33-1950 Dw | GTAGTGTCACACTTACGC | B33 cloning |
| B33 Up | AGTCGACAATCATTGTTTTATTTTCTCTTTC | B33 cloning |
| B33 Dw | AGGATCCAAATTTTGTTGGTGCTTTGAG | B33 cloning |
| Lcy-e 322 Up | AGAGCTCGGCTGTGGTCCTGCTG | Lcy-e cloning |
| Lcy-e 1085 Dw | AGGATCCTGGCAAAGATCCACCAAC | Lcy-e cloning |
| Lcy-e 574 Up | TAGGATCATCATCATCAAG | Pcr screening |
| Nos-test2 | CGCGTATTAAATGTATAATTG | Pcr screening |
| Tub 865 Up | CTGTGAATTTAATCCCCTTG | Pcr screening |
| Tub 1260 Dw | TGCGCCTGAACATGGCAGT | Pcr screening |
| Tub Up | CAGACCTGAGGAAATTGGCTG | Real Time assay |
| Tub Dw | TTCTTGGCATCCCACATTTGT | Real Time assay |
| Ubi Up | ACAATGTCAAAGCCAAGATCCA | Real Time assay |
| Ubi Dw | CGGAGACGGAGCACGAGA | Real Time assay |
| AS-e Up | CCCCAATTTAGCAGACTCCG | Real Time assay |
| AS-e Dw | CAACAGAAATTATATGATAATCATCGCAA | Real Time assay |
| Psy1 Up | CGGTCTGCTATTGTTGCTACTCC | Real Time assay |
| Psy1 Dw | CAGGAACAGGTATGTCTGGCTTC | Real Time assay |
| Psy2 Up | AGCTTTAGATAGGTGGGAGGCA | Real Time assay |
| Psy2 Dw | CAAGTCCATACGCATTCCTTCAA | Real Time assay |
| PDS Up | AGAGACTTTGCATGCCGATTGT | Real Time assay |
| PDS Dw | AAAGCATCGCCCTCAACTGT | Real Time assay |
| ZDS Up | TTGCCATGTCAAAGGCCA | Real Time assay |
| ZDS Dw | ACAGGCACTCCGACCAATTT | Real Time assay |
| CrtIso Up | TTGGCAGCAGTAGGACGTAAAC | Real Time assay |
| CrtIso Dw | TCCCTTCCTTTTCATGTGGAA | Real Time assay |
| Lcy-e Up | GCCAAAATGGATGTGGCAG | Real Time assay |
| Lcy-e Dw | CAATGTTGCACCAGTAGGATCAG | Real Time assay |
| Lut1 Up | CGTTCTCCGCCCAAAAAAC | Real Time assay |
| Lut1 Dw | TTGGCCTAAAGTAAGTGACCTGG | Real Time assay |
| Lcy-b Up | AATGGGTGGTCCACTTCCAGTA | Real Time assay |
| Lcy-b Dw | GGATGGATGAACCATGCCAG | Real Time assay |
| Chy1 Up | CTTGGCCCAAAACCCACTT | Real Time assay |
| Chy1 Dw | CCTCAAATTGAGGTTTCAGCTTCT | Real Time assay |
| Chy2 Up | TTTTGCTGTCTCGAAGAAAGCC | Real Time assay |
| Chy2 Dw | AGCCAACAGGCAGCTAAACTCT | Real Time assay |
| ZEP Up | TCATGAATGCTGGCTGCATC | Real Time assay |
| ZEP Dw | TGCTGCAAAGTCATGCGG | Real Time assay |
A 0.75 Kb LCY-e cDNA fragment was amplified from potato (cv Desirée) tuber cDNA using the primers LCY-e 322 up and LCY-e 1085 dw (Table 2). These primers inserted, respectively, Sac I and Bam HI sites upstream and downstream of the cDNA fragment. After intermediate cloning in the pBSK+ vector and re-sequencing, the fragment was inserted, in antisense orientation, instead of the GUS gene in the pBI33:GUS vector, resulting in the plasmid pBI33: AS-e (Fig. 2).
Potato (cv Desirée) was transformed as previously described [22]. Plantlets growing on kanamycin were tested by PCR, using primers Lcy-e 574 Up and Nos-test 2. PCR-positive, rooted plantlets were adapted in greenhouse in pots (diameter: 25 cm) in a soil mixture composed of 1/3 sand and 2/3 of sterile soil (Terraplant 2, BASF). Photoperiod was set at 14 hours of light and 10 hours of dark, with temperature set at 24°C during the light period and at 16°C during the dark period. In the advanced phase of growth, the day temperature was kept around 20°C in order to promote tuberization. During tuberization, irrigation was reduced in order to prevent tuber decay.
Tubers from the lower 2/3 of the pot ("deep" tubers) were collected separately from superficial ones, washed in water, dried briefly at room temp, cut in pieces and frozen at -80°C. Tuber productivity for each line was estimated as the total number of tubers produced and as their total weight. All carotenoid and RT-PCR measurements were conducted on at least 4 different "deep" tubers per each line, to prevent possible alterations in carotenoid composition/gene expression resulting from light accidentally illuminating the superficial tubers.
Total RNA was isolated from frozen tissue and analyzed through Real Time RT-PCR using previously published methods [23,35]. Two independent RNA extractions and four cDNAs (two from each RNA) were used for the analyses; first strand cDNA was synthesized from 0.5 μg of RNA in 20 μl with oligo-dT(16) and Superscript II (Invitrogen), according to the manufacturer's instructions. QRT-PCR were performed using an ABI PRISM 7000 instrument and the SYBR Green Master Mix kit (cat. 4309155; Applera). Standard dilution curves were performed for each gene fragment and all data were normalized for the level of the β-TUBULIN transcript and for wild-type expression levels. Primers for Real Time experiments were designed using the Primer Express v2.0 software and validated with the Amplify v3.1 software. Sequences of the primers used for the various genes are shown in Table 2. The sequences used to design the primers were: β-TUBULIN: Z33382; PSY1: TC122598; PSY2: L23424 (from tomato); PDS: AY484445; ZDS: TC114158; CrtISO: TC117194; LCY-e: AF321537; LUT1: TC117729; LCY-b: X86452 (from tomato); CHY1: TC36005; CHY2: TC32024; ZEP: EST724320 (all numbers refer to GenBank accession numbers, with the exception of those starting with TC, which are Transcript Contig numbers from TIGR potato gene index release 10.0 [36]. In order to discriminate the introduced AS-e mRNA from the endogenous Lcy-e mRNA, the former was amplified using primers AS-e Up and AS-e Dw, while the latter was amplified using primers Lcy-e Up and Lcy-e Dw.
Fluorimetric GUS assays were performed as described previously [24].
For HPLC analysis, frozen tubers were lyophilized, peeled, ground to powder and pigments were extracted three times with 2 ml of acetone. In the first extraction, 200 μg tocopherol acetate per sample was added as an internal standard. Combined acetone extracts were dried, lipophilic compounds were resuspended in 2 ml of petroleum ether:diethyl ether (2:1, v/v) and 1 ml of distilled water. After centrifugation for 5 min at 3000 × g the organic phase was recovered and the aqueous phase was extracted for a second time as described above. The combined organic phases were dried and dissolved in 30 μl chloroform. 10 μl were subjected to quantitative analysis using HPLC with a C30 reversed-phase column (YMC Europe GmbH, Schermbeck, Germany) and a gradient system as described [37]. Carotenoids were identified by their absorption spectra, monitored using a photodiode array detector (PDA 2996; Waters, Eschborn, Germany). Normalization of the samples to the internal standard and the quantification of carotenoid amounts was performed as described previously [38].
Authors' contributions
GG, RT and PB planned and supervised the work. FM prepared the constructs for transformation, RT and VP performed the transformations and maintained the lines in vitro, DP and GD grew and sampled the plants, DP performed the GUS assays and GD the Real Time assays, RW performed the HPLC's.
Acknowledgments
Acknowledgements
Work supported by EC grant "ProVitA" to PB and GG. GD acknowledges the Univ. of L'Aquila for a doctoral fellowship and Prof. Laura Spano' (Univ. of L'Aquila) for supervision of his thesis work.
Contributor Information
Gianfranco Diretto, Email: gianfranco.diretto@casaccia.enea.it.
Raffaela Tavazza, Email: raffaela.tavazza@casaccia.enea.it.
Ralf Welsch, Email: welschra@web.de.
Daniele Pizzichini, Email: daniele.pizzichini@casaccia.enea.it.
Fabienne Mourgues, Email: fabienne.mourgues@trisaia.enea.it.
Velia Papacchioli, Email: velia.papacchioli@casaccia.enea.it.
Peter Beyer, Email: peter.beyer@biologie.uni-freiburg.de.
Giovanni Giuliano, Email: giuliano@casaccia.enea.it.
References
- http://www.cipotato.org/market/potatofacts/growprod.htm
- http://www.who.int/nutrition/topics/vad/en/
- Giuliano G, Aquilani R, Dharmapuri S. Metabolic engineering of plant carotenoids. Trends Plant Sci. 2000;5:406–409. doi: 10.1016/S1360-1385(00)01749-0. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res. 2004;43:228–265. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Burkhardt PK, Beyer P, Wunn J, Kloti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 1997;11:1071–1078. doi: 10.1046/j.1365-313X.1997.11051071.x. [DOI] [PubMed] [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci U S A. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducreux LJ, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J Exp Bot. 2005;56:81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]
- Rosati C, Aquilani R, Dharmapuri S, Pallara P, Marusic C, Tavazza R, Bouvier F, Camara B, Giuliano G. Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J. 2000;24:413–420. doi: 10.1046/j.1365-313x.2000.00880.x. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta -carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci U S A. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Dharmapuri S, Rosati C, Pallara P, Aquilani R, Bouvier F, Camara B, Giuliano G. Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett. 2002;519:30–34. doi: 10.1016/S0014-5793(02)02699-6. [DOI] [PubMed] [Google Scholar]
- Mann V, Harker M, Pecker I, Hirschberg J. Metabolic engineering of astaxanthin production in tobacco flowers. Nat Biotechnol. 2000;18:888–892. doi: 10.1038/78515. [DOI] [PubMed] [Google Scholar]
- Ralley L, Enfissi EM, Misawa N, Schuch W, Bramley PM, Fraser PD. Metabolic engineering of ketocarotenoid formation in higher plants. Plant J. 2004;39:477–486. doi: 10.1111/j.1365-313X.2004.02151.x. [DOI] [PubMed] [Google Scholar]
- Morris WL, Ducreux LJ, Fraser PD, Millam S, Taylor MA. Engineering ketocarotenoid biosynthesis in potato tubers. Metab Eng. 2006;8:253–263. doi: 10.1016/j.ymben.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Romer S, Lubeck J, Kauder F, Steiger S, Adomat C, Sandmann G. Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng. 2002;4:263–272. doi: 10.1006/mben.2002.0234. [DOI] [PubMed] [Google Scholar]
- Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Osto L, Lico C, Alric J, Giuliano G, Havaux M, Bassi R. Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. In preparation. [DOI] [PMC free article] [PubMed]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Bramley PM, Pennings HM, Bowler C. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald UWF, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson AR, Kavanagh TA, Bevan MW. GUS fusion: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazza R, Tavazza M, Ordas RJ, Ancora G, Benvenuto E. Genetic transformation of potato (Solanum tuberosum); an efficient method to obtain transgenic plants. Plant Science. 1988;59:175–181. doi: 10.1016/0168-9452(89)90135-0. [DOI] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona V, Aracri B, Kosturkova G, Bartley GE, Pitto L, Giorgetti L, Scolnik PA, Giuliano G. Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 1996;9:505–512. doi: 10.1046/j.1365-313X.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- Lange BM. Integrative analysis of metabolic networks: from peaks to flux models? Curr Opin Plant Biol. 2006;9:1–7. doi: 10.1016/j.pbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que QD, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs antisense constructs and single-copy vs complex T-DNA sequences. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA. Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet. 2003;4:29–38. doi: 10.1038/nrg982. [DOI] [PubMed] [Google Scholar]
- Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci U S A. 2004;101:402–407. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc Natl Acad Sci U S A. 2006;103:3474–3479. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, von Lintig J. Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 2003;8:145–149. doi: 10.1016/S1360-1385(03)00053-0. [DOI] [PubMed] [Google Scholar]
- Hirschberg J. Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol. 2001;4:210–218. doi: 10.1016/S1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- Bramley PM, Teulieres C, Blain I, Bird C, Schuch W. Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through the expression of antisense RNA to pTOM5. Plant J. 1992;2:343–349. doi: 10.1111/j.1365-313X.1992.00343.x. [DOI] [Google Scholar]
- Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual (Second Edition) Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Carbone F, Pizzichini D, Giuliano G, Rosati C, Perrotta G. Comparative profiling of tomato fruits and leaves evidences a complex modulation of global transcript profiles. Plant Sci. 2005;169:165–175. doi: 10.1016/j.plantsci.2005.03.011. [DOI] [Google Scholar]
- http://www.tigr.org/
- Hoa TT, Al-Babili S, Schaub P, Potrykus I, Beyer P. Golden Indica and Japonica rice lines amenable to deregulation. Plant Physiol. 2003;133:161–169. doi: 10.1104/pp.103.023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub P, Al-Babili S, Drake R, Beyer P. Why is golden rice golden (yellow) instead of red? Plant Physiol. 2005;138:441–450. doi: 10.1104/pp.104.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]


