Abstract
Mammals taste many compounds yet use a sensory palette consisting of only five basic taste modalities: sweet, bitter, sour, salty, and umami (the taste of monosodium glutamate)1,2. While this repertoire may appear modest, it provides animals with critical information about the nature and quality of food. Sour taste detection functions as an important sensory input to warn against the ingestion of acidic (e.g. spoiled or unripe) food sources1–3. We have used a combination of bioinformatics, genetic, and functional studies to identify PKD2L1, a polycystic kidney disease-like ion channel4, as a candidate mammalian sour taste sensor. In the tongue, PKD2L1 is expressed in a subset of taste receptor cells (TRCs) distinct from those responsible for sweet, bitter and umami taste. To examine the role of PKD2L1-expressing taste cells in vivo, we engineered mice with targeted genetic ablations of selected populations of TRCs. Animals lacking PKD2L1-cells are completely devoid of taste responses to sour stimuli. Notably, responses to all other tastants remained unaffected, proving that the segregation of taste qualities even extends to ionic stimuli. Our results now establish independent cellular substrates for four of the five basic taste modalities, and support a comprehensive labeled-line mode of taste coding at the periphery5–10. Interestingly, PKD2L1 is also expressed in specific neurons surrounding the central canal of the spinal cord. Here we demonstrate that these PKD2L1-expressing neurons send projections to the central canal, and selectively trigger action potentials in response to decreases in extracellular pH. We propose that these cells correspond to the long sought components of the cerebrospinal fluid chemosensory system11. Taken together, our results suggest a common basis for acid sensing in disparately different physiological settings.
A broad range of cell types, receptors and mechanisms have been proposed to mediate salt and acid sensing in TRCs1–3. These include the activation of ENaCs, ASICs, K2P channels, H+-gated calcium channels, as well as the involvement of Na+-H+-exchangers, TRPV pain receptors, and acid-inactivation of K+-channels1–3,12,13. Significantly, most of these proteins are broadly expressed in TRCs and other tissues. In contrast, we previously isolated and characterized the receptors for sweet, umami and bitter taste5–7,14–16, and showed that each of these three taste modalities is mediated by highly selective receptor proteins expressed in distinct and independent populations of taste receptor cells5–10. Therefore, we reasoned that salt and sour taste should also be mediated by highly selective dedicated cells, and consequently expected the receptor proteins to be very exclusive in their expression pattern.
To identify novel taste receptors, we developed a multi-step bioinformatics and expression screening strategy. First, since sensory receptors are expected to be membrane proteins, approximately 30,000 mouse open reading frames (ORFs) were scanned for the presence of at least one putative transmembrane segment. Second, because taste receptors are predicted to be very restricted in their expression pattern, ORFs encoding candidate transmembrane proteins were cross-searched against mouse EST databases to eliminate those broadly expressed. Next, to identify the subset specifically enriched in taste tissue, ORFs selected as encoding transcripts infrequently represented in EST databases (~880 candidates) were used in RT-PCR reactions templated with mRNA from TRCs versus control tongue epithelium. Finally, given that our goal was to discover membrane proteins selectively expressed in subsets of TRCs (and ideally not in sweet, bitter or umami sensing cells), we carried out detailed in situ hybridizations against taste papillae. Of 26 cDNAs used in situ studies, five were found to robustly and selectively label subsets of TRCs. Figure 1 shows that one of these candidates, PKD2L1 is expressed in TRCs of all taste papillae, including fungiform, circumvallate, foliate and palate taste buds.
Figure 1. PKD2L1 is expressed in a novel population of TRCs.
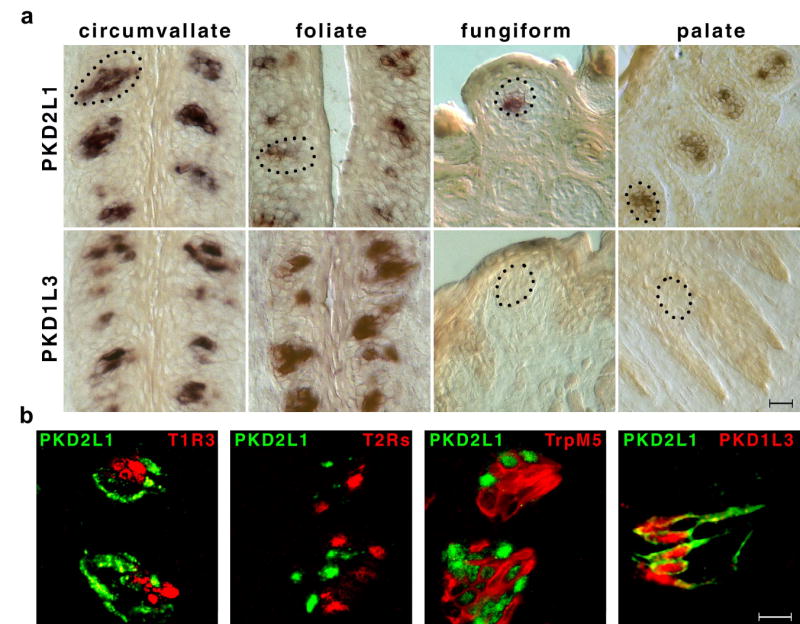
In situ hybridization (PKD2L1, PKD1L3, T1Rs, T2Rs and TRPM5) and double-label fluorescent immunohistochemistry (PKD2L1) were used to examine the overlap in cellular expression of taste receptors, TRPM5, PKD2L1 and PKD1L3. (a) In situ hybridization of PKD2L1 and PKD1L3 against circumvallate, foliate, fungiform and palate taste buds illustrating expression of PKD2L1 in subsets of TRCs of all taste buds, but lack of PKD1L3 in fungiform and palate TRCs. Dotted lines show the outline of sample taste buds. Scale bar represents 25 μm. (b) The first three panels show co-labeling with a PKD2L1 antisense RNA probe (PKD, green) and T1R3 (T1R, sweet and umami cells), a mixture of 20 T2Rs (bitter cells), and TRPM5 (sweet, umami and bitter cells), respectively. The last panel shows co-labeling with anti-PKD2L1 antibodies and an antisense PKD1L3 RNA probe. Note the absence of overlap between PKD2L1-expressing cells and those expressing sweet, umami or bitter receptors. However, PKD1L3 is always co-expressed with PKD2L1 in CV and foliate papillae. Scale bar represents 10 μm.
PKD2L1 encodes a polypeptide displaying significant amino acid sequence similarity to PKD24, a gene mutated in many cases of autosomal dominant polycystic kidney disease17,18. PKD2s are members of the TRP superfamily of ion channels19, and have been recently shown to function as non-selective cation channels when expressed in heterologous cells17,18,20. While the exact roles of PKDs remain unknown, they are believed to function as receptor/ion-channel complexes, often localized to ciliated compartments, and implicated in sensing extracellular signals (e.g. in renal epithelial cells17,18). We reasoned that if PKD2L1 has a specific role in taste it should be expressed in subpopulations of taste receptor cells with unique functional characteristics. To determine which type of TRCs express PKD2L1, we performed double labeling experiments with sweet, umami and bitter taste receptors (T1Rs and T2Rs), as well as TRPM5, the transduction channel of sweet, bitter and umami sensing cells. Our results (Figure 1) established that PKD2L1 is expressed in cells distinct from those mediating sweet, umami and bitter taste (see also 21).
Mammalian taste receptor cells project specialized apical microvilli to the taste pore, the site of interaction between tastants and taste receptor proteins. All known taste receptor proteins localize to, and function, in this TRC compartment 1,5–7,14,16,22. Therefore we would expect bona-fide candidate receptors to also be enriched in the taste pore. We generated antibodies to PKD2L1 and used them in immunofluorescence staining of tongue tissue sections. Examination of CV, foliate and fungiform papillae demonstrated that PKD2L1 protein is indeed enriched in the apical surface of taste receptor cells, with the antibodies robustly labeling the taste pore region (Supplementary Figure 1). These results implicate PKD2L1 as part of the taste sensing machinery.
Supplementary Figure 1.
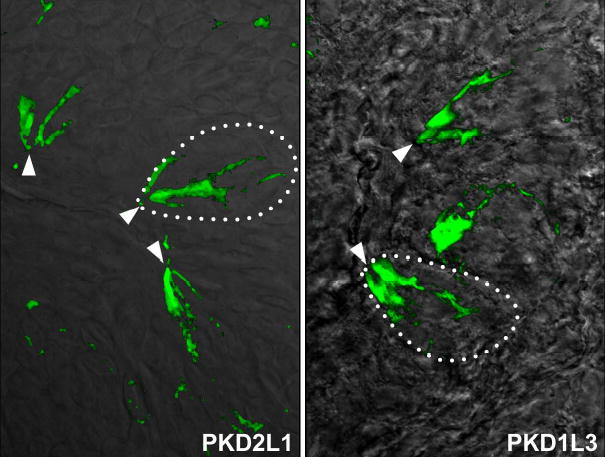
PKD2L1 and PKD1L3 are enriched in the taste pore
Immunofluorescent stainings of mouse taste buds with PKD2L1 (left panel) and with PKD1L3 (right panel) antibodies. The pictures show superposition of fluorescent antibody signals on DIC images of taste tissue. Dotted lines illustrate the outline of a taste bud, and arrows point to the taste pore region
PKD2 isoforms often require PKD1s for functional expression at the cell surface17,18,20. The mammalian genome contains 4 members of the PKD1 family: PKD1, PKD1L1, PKD1L2 and PKD1L317,18. We performed in situ hybridization studies with gene specific probes representing each family member, and determined that PKD1L3 is specifically co-expressed with PKD2L1 in CV and foliate TRCs (Figure 1, see also21). We also generated antibodies to PKD1L3 and demonstrated selective co-expression with PKD2L1 in non-TRPM5 expressing cells of the CV and foliate (Figure 1 and Supplementary Figure 1). Surprisingly, PKD1L3 transcript or protein is not detectable in fungiform or palate taste buds, suggesting that a different partner may be expressed in those TRCs.
To functionally dissect the role of PKD2L1-expressing cells in the tongue, we engineered mice where these cells were genetically ablated by targeted expression of attenuated diphtheria toxin23 (DTA). To validate this approach as a means of uncovering TRC function, we first generated mice where T1R2-regulatory sequences were used to target DTA expression24 (see Supplementary Figure 2). T1R2 is an essential subunit of the sweet receptor heterodimer (T1R2+3), and the selective ablation of these cells should generate animals with a specific loss of sweet taste6,9,10,16. To investigate the taste responses of the genetically modified mice, we recorded tastant-induced action potentials from nerves innervating taste receptor cells of the tongue; this physiological assay monitors the activity of the taste system at the periphery, and provides an accurate and reliable measure of taste receptor cell function. Indeed, animals expressing DTA in T1R2 cells have an extraordinary loss of sweet, but importantly retain umami, bitter, sour and salty tastes (Figure 2). These results further substantiate the exquisite segregation of taste modalities at the periphery, and demonstrate the utility of using DTA-mediated ablation of TRCs as a strategy for dissecting taste system function. Next, we engineered animals where the PKD2L1 gene was used to target Cre recombinase into PKD2L1-expressing cells (see Supplementary Figures 2 and 3). These mice were crossed to the conditional DTA lines24, and double-positive progeny were scrutinized both for the specificity and efficiency of killing, as well as the integrity of taste buds. We checked the expression of T1Rs, T2Rs, and TRPM5 8,25 in control and DTA-expressing animals, and found no significant differences in the number or distribution of T1R- or T2R-positive cells between wild type and ablated taste tissue (Supplementary Figure 2). In contrast, the DTA-targeted mice had a profound and practically complete loss of PKD2L1-expressing TRCs in the tongue (Supplementary Figure 2). Remarkably, genetic ablation of the PKD2L1-expressing cells produces animals with a devastating loss of sour taste (Figure 2). Responses to all acid tastants, including citric acid, HCl, tartaric acid and acetic acid are completely abolished, with no significant activity over a range of 5 orders magnitude of proton concentrations. However, responses to sweet, umami, bitter or salty tastants remain indistinguishable from wild type control animals. These results firmly establish PKD2L1-expressing cells as the sour taste sensors, and further substantiate a model of coding at the periphery in which individual taste modalities operate independently of each other.
Supplementary Figure 2.
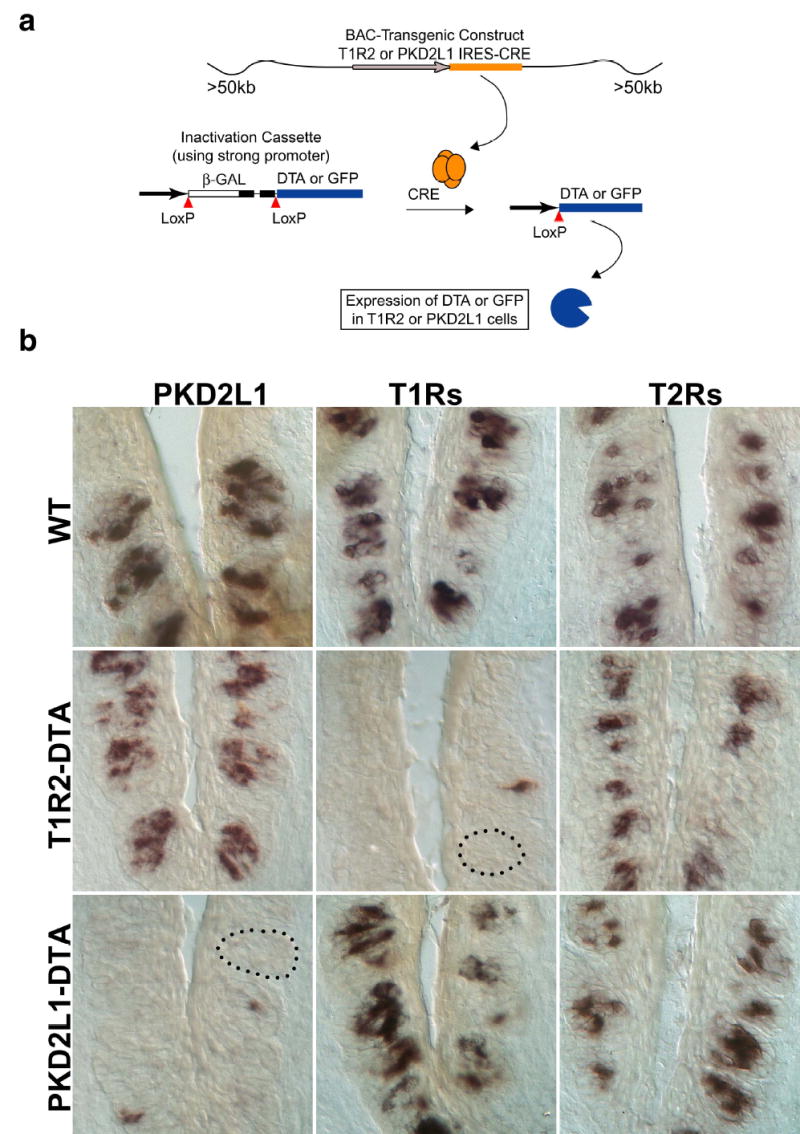
Loss of selective TRCs in DTA-expressing animals
Upper diagram illustrates the strategy used to target DTA or GFP to selective populations of TRCs. BAC constructs contained the entire T1R2 or PKD2L1 genes with the IRES-Cre added downstream of the termination codon, but upstream of polyA-addition signals. In both cases, the transgenic constructs included at least 50Kb of flanking sequences upstream and downstream of the target gene (see Methods). Fidelity of Cre and reporter expression in the correct cell types was confirmed by double labeling with a variety of TRC-specific gene probes. Lower panels show in situ hybridization experiments examining the presence of sweet (T1Rs), bitter (T2Rs) or PKD2L1-expressing cells in the two engineered lines. Targeting of DTA to T1R2- or PKD2L1-expressing cells eliminates over 95% of their respective TRC population. In situ hybridization probes were as in Figure 1.
Figure 2. PKD2L1-expressing TRCs are the mediators of sour taste.
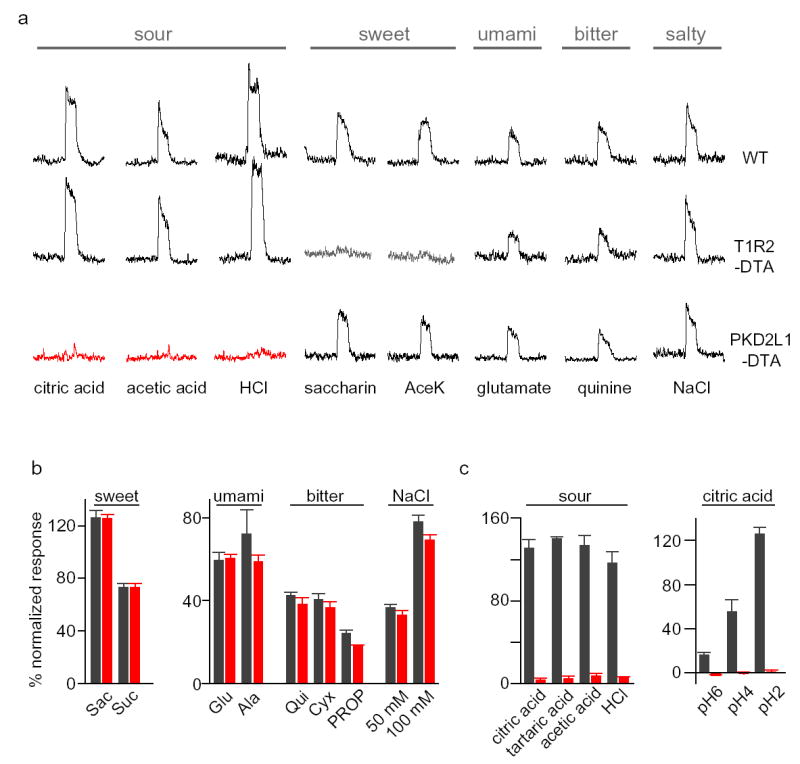
(a) Targeted expression of DTA to selective populations of TRCs produces animals with selective deficits in taste responses. Wild-type mice (WT) show robust neural responses to sour, sweet, umami (amino acid), bitter and salty tastants. However, ablation of sweet cells (T1R2-DTA) generates animals with a dramatic loss of sweet taste (middle panel). In contrast, ablation of PKD2L1-expressing cells eliminates responses to all acid stimuli (bottom panel). Importantly, responses to all other taste qualities remain unimpaired in the DTA-expressing animals. Shown are integrated chorda tympani responses (see Methods). (b) Average neural responses of animals lacking PKD2L1-expressing cells to an expanded panel of tastants; note normal responses to sweet, umami, bitter and salt stimuli. Wild type, gray bars; PKD2L1-DTA, red bars; the values are means ± s.e.m. (n=5) (c) Quantitation of acid responses of wild type and PKD2L1-DTA animals. The values are means ± s.e.m. (n=6). Only the differences in acid responses are significant between wild type and PKD2L1-DTA mice (P<0.00001).
Supplementary Figure 3.
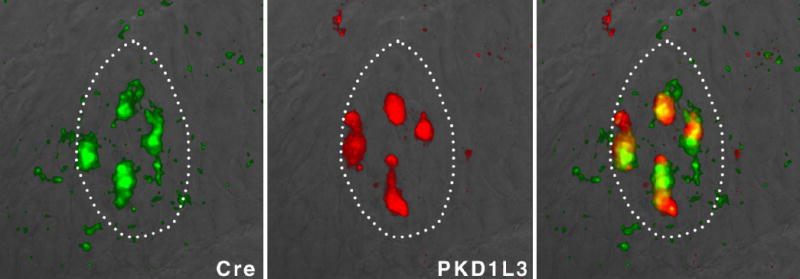
Targeting of Cre recombinase to PKD-expressing TRCs
In situ hybridization with double-labeled probes (Cre and PKD1L3) was used to examine the expression of Cre recombinase in PKD-expressing cells. Dotted lines illustrate the outline of a taste bud; note the cellular overlap in the hybridization signals. Similar results were obtained by crossing PKD2L1-Cre lines to GFP reporter lines30.
Acid sensing is important not only in the taste system, but also for monitoring the functional state of body fluids, including the internal milieu of the brain. This is particularly well-studied in the central and peripheral control of respiration, where pH sensing is the principal mechanism for monitoring CO2 levels in the blood and cerebrospinal fluid11,26,27 (CSF). Thus, we wondered whether PKD2L1 might be expressed in additional cell types, and if so whether such cells may also be involved in pH sensing in other physiological systems.
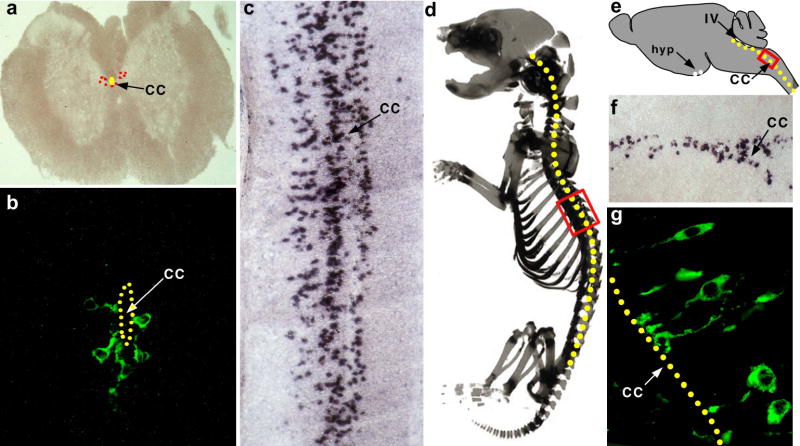
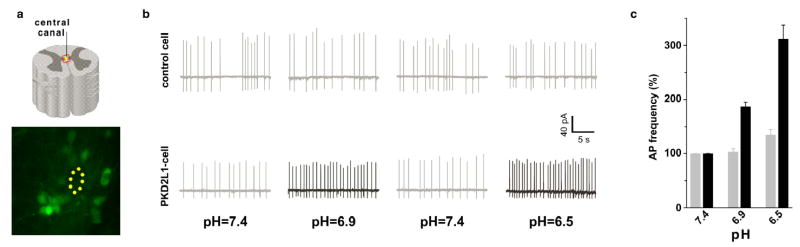
We carried out in situ hybridization and antibody staining experiments with PKD2L1 on a wide range of other tissues and identified a singular additional domain of expression: a discrete population of neurons surrounding the central canal of the spinal cord, through its entire length, from its origin in the brain stem to its end around the cauda equina (Figure 3). Notably, these neurons send processes into the central canal, suggesting they may function as chemoreceptors sensing the internal state of the CSF (Figure 3b,g, 11). Given their anatomical distribution and cellular morphology, we reasoned these cells might be part of the homeostatic circuitry responsible for monitoring and reporting the pH of the cerebrospinal fluid. This postulate predicts that these neurons should trigger action potentials in response to acid stimulation. Therefore, we engineered mice where a GFP reporter was targeted to PKD2L1-expressing cells, and performed patch clamp recordings from GFP-labeled cells in a spinal cord slice preparation28. A priori, we anticipated some notable differences in the behavior of these cells compared to TRCs; while the taste system is tuned to respond to acid stimulation in the range of multiple pH units (i.e. pH 2–5), we expected the CSF monitor cells to respond to pH changes within a range of a few tenths of deviation from pH 7.4. Indeed, Figure 4 shows that the PKD2L1-expressing neurons display exquisite sensitivity and selectivity to pH stimulation. Exposure to test solutions between pH 6.5 and 7.4 evoked a dramatic, dose dependent and reversible increase in action potential (AP) frequency (Figure 4 and data not shown). In contrast, the same acid stimuli have no significant impact on the response of control (e.g. unlabeled) cells, even after exposure to pH as low as 6.5 (lower pHs triggered irreversible damage to the slice preparation).
Figure 3. PKD2L1 is expressed in neurons contacting the central canal of the spinal cord.
(a) In situ hybridization with PKD2L1 specific probes to a coronal section of the spinal cord; signals are pseudocolored in red. (b) High magnification staining with anti-PKD2L1antibodies reveals a population of PKD2L1-expressing neurons surrounding the central canal of the spinal cord (cc; highlighted by yellow dots in all panels). (c) In situ hybridization with PKD2L1-specific probes on a sagital section of a P1 mouse. The PKD2L1-expressing cells are found throughout the entire length of the spinal cord. (d) Red box denotes the approximate area of the in situ shown in panel (c). (e–f) PKD2L1-expression extends through the brain stem and into the IV ventricle (IV). There is also a very small group of positive cells in the hypothalamus (hyp; data not shown). (g) Immunofluorescent stainings with anti-PKD2L1 antibodies. PKD2L1-expressing neurons project into the central canal; note robust expression of PKD2L1 receptors at the terminals.
Figure 4. PKD2L1-expressing neurons of the central canal fire action potentials in response to pH stimulation.
Spinal cord neurons were patched using a loose patch configuration28, tested for the presence of basal activity and recorded in the cell-attached configuration. (a) GFP-expressing (PKD2L1-positve cells) or unlabeled (control) cells were examined for pH responses. (b) Responses of a sample GFP-labeled or unlabeled neuron to test solutions under a perfusion regime consisting of pH 7.4, pH 6.9, pH 7.4 and pH 6.5; shown are AP traces in a window of ~25 sec. (c) Data were analyzed by examining records of ~4 minutes at each pH condition. Basal activity ranged between 1–5 Hz. Note the dramatic increases in pH-evoked firing frequency in GFP-labeled neurons versus unlabeled cells (P<0.001). A minimum of 8 GFP-labeled and 5 unlabelled cells were characterized for each stimulus. The values are means + s.e.m. normalized to basal activity at pH 7.4 (taken as 100%).
Most of the known CSF-contacting neurons in mammals project ciliated dendrites into the CSF, where they are proposed to sense fluid flow, pressure, pH or the composition of the CSF11. Our demonstration that PKD2L1-expressing cells of the spinal cord selectively fire in response to minor changes in proton concentration strongly suggests that they function as sentinels of cerebrospinal and ventricular pH. Collectively, these results uncover an entirely unexpected role for members of the PKD family of proteins, offer a new perspective into the potential significance of PKD2s in health and disease, and bring forth a surprising unity in the cellular basis of pH sensing in very different physiological systems. In the future, it will be of interest to develop an activity assay for PKD2L1 to establish the molecular mechanism of acid activation, to study the phenotype of PKD2L1 knockout animals, and determine whether PKD2L1 functionally associates or interacts with different partners in different cells types. In this regard, it would be worth exploring whether the differences in pH sensitivity between the tongue and spinal cord might be due to differences in PKD2L1-receptor complex composition.
The nature of the mammalian sour taste receptor and sour-sensing TRCs have been fertile ground for speculation over the years. A wide range of cell types, receptors, and even receptor-independent mechanisms, have been proposed to mediate acid detection in the tongue1–3. The results presented in this paper establish that sour taste, much like our previous findings for sweet, umami and bitter is mediated by a unique cell type, independent of all other taste qualities. In addition, our demonstration that sour-less mice have normal salt responses demonstrates that salt taste is also mediated by independent TRCs. Together, these results impose a considerable revision of the current views of taste representation at the periphery, and make a compelling case for a labeled line mode of coding across all five taste modalities and TRC types.
Methods
Molecular Cloning of PKD2L1
We used a strategy that combined bioinformatics and differential screening to isolate genes specifically expressed in taste receptor cells. Mouse genomic sequence information was obtained from Ensembl Mm.30 (http://www.ensembl.org). Approximately 30,000 predicted protein sequences were screened for the presence of at least one putative transmembrane segment, using both TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and f_TMHMM (San Diego Supercomputer center, http://www.sdsc.edu/pb/Group.html). The cDNA sequence for each candidate membrane protein was then extracted from NCBI (http://www.ncbi.nlm.nih.gov/blast/blastcgihelp.shtml#nucleotide_databases) and used to screen EST databases (http://www.ncbi.nlm.nih.gov/dbEST/index.html). Only EST hits with e-values of less than or equal to e−100 were considered in our analysis. A total of 884 genes expressed in 3 tissues or less were chosen for PCR reactions with cDNA prepared from taste papillae mRNA (CV and foliate) and from surrounding non-taste epithelial tissue (non-taste control). To ensure specificity of the PCR reactions, all primers sets included unique 3’UTR sequences (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi.). A total of 98 genes showed selective enrichment in taste versus non-taste tissue, and of these five were robustly expressed in subsets of TRCs. Full length clones were isolated from mouse taste cDNA libraries22.
In situ hybridization and immunostaining
In situ hybridization and immunostaining were as described previously8,22. Anti-peptide antibodies to PKD2L1 (KLKMLERKGELAPSPGMGE) and PKD1L3 (DFQEADNYCHAQRGRLAHT) were generated in rabbits and purified as described previously8.
Transgenic animals
Transgenic lines were produced by pronuclear injection of zygotes from FVB/N or CB6 (BALB/c × C57BL/6 hybrids) mice. The PKD2L1-IRES-Cre construct was generated in RP23-297K23 and the T1R2-IRES-Cre in RP23-348G10 (http://bacpac.chori.org/). Recombination was carried out exactly as described previously29. Z/EG reporter lines30 were obtained from Jackson Laboratories (Bar Harbor, Maine), and Rosa26-flox-lacZ-flox-DTA animals24 were a generous gift of Dr. Dieter Riethmacher.
Nerve Recordings
Lingual stimulation and recording procedures were performed as previously described7,9. All data analyses used the integrated response over a 25 s period immediately after the application of the stimulus. Tastants used for nerve recordings were: 10mM, 60mM acesulfameK (AceK); 10mM, 60mM sodium saccharin (saccharin); 300mM sucrose; 30mM mono potassium glutamate + 1mM inosine mono phosphate (Glu); 30mM L alanine + 1mM inosine mono phosphate (Ala); 10mM quinine hydrochloride (Qui); 100μM cycloheximide (Cyx); 10mM 6-n-propyl 2-thiouracil (PROP); 50mM, 100mM sodium chloride (NaCl); 10mM, 50mM citric acid; 10mM, 50mM tartaric acid; 50mM, 500mM acetic acid; pH 2 hydrochloric acid (HCl); 10mM citric acid pH 2, 4 and 6. The mean response to 60mM AceK was used to normalize responses to each experimental series in the wild type and PKD2L1-DTA animals (figure 2b-c).
Spinal cord slice recordings
Electrophysiological experiments were performed on P1-P4 mice as previously described28. All incubations included 10 μM CNQX and 50 μM APV. Spinal cord slices 250–300 μm thick were generated using a Vibratome® 3000 Plus at 0–4°C in a modified Ringers’ solution (0.5 mM CaCl2, 3.7 mM MgSO4). After at least a 1 h recovery period, slices were transferred to a recording chamber and perfused with oxygenated Ringers’ solution (pH 7.4) at room temperature. Cell-attached patch clamp recordings from GFP-labeled and unlabeled cells were performed using an EPC-10/2 amplifier and Patchmaster software (HEKA Electronik). Slices were stimulated with a solution containing 140 mM NaCl, 3 mM KCl, 1.3 mM MgSO4, 2.5 mM CaCl2, 10 mM glucose, 10 mM HEPES at various pH (7.4, 6.9, 6.5). 8 out of 8 GFP-labeled cells showed pH-dependent increases in AP frequency.
Acknowledgments
We thank Luxin Feng for help with expression studies, Ann Becker for generation of antibodies, D. Cowan for sequencing and Kristine Briedis for bioinformatics. We especially thank Ying Zhang for introducing us to the spinal cord slice preparation, her excellent technical guidance and generous help with equipment and animals. We thank members of the Zuker lab for valuable comments. This research was supported in part by a grant from the National Institute on Deafness and Other Communication Disorders to C.S.Z and the intramural research program of the NIH, NIDCR (N.J.P.R.). X.C. is a fellow of the H.F.S. program and D.T. is supported by an Emmy-Noether grant of the Deutsche Forschungsgemeinschaft. C.S.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information accompanies the paper.
References
- 1.Lindemann B. Receptors and transduction in taste. Nature. 2001;413:219–25. doi: 10.1038/35093032. [DOI] [PubMed] [Google Scholar]
- 2.Kinnamon SC, Margolskee RF. Mechanisms of taste transduction. Curr Opin Neurobiol. 1996;6:506–13. doi: 10.1016/s0959-4388(96)80057-2. [DOI] [PubMed] [Google Scholar]
- 3.DeSimone JA, Lyall V, Heck GL, Feldman GM. Acid detection by taste receptor cells. Respir Physiol. 2001;129:231–45. doi: 10.1016/s0034-5687(01)00293-6. [DOI] [PubMed] [Google Scholar]
- 4.Wu G, et al. Identification of PKD2L, a human PKD2-related gene: tissue-specific expression and mapping to chromosome 10q25. Genomics. 1998;54:564–8. doi: 10.1006/geno.1998.5618. [DOI] [PubMed] [Google Scholar]
- 5.Adler E, et al. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 6.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–90. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 7.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhao GQ, et al. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–66. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 10.Mueller KL, et al. The receptors and coding logic for bitter taste. Nature. 2005;434:225–9. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 11.Vigh B, et al. The system of cerebrospinal fluid-contacting neurons. Its supposed role in the nonsynaptic signal transmission of the brain. Histol Histopathol. 2004;19:607–28. doi: 10.14670/HH-19.607. [DOI] [PubMed] [Google Scholar]
- 12.Lyall V, et al. The mammalian amiloride-insensitive non-specific salt taste receptor is a vanilloid receptor-1 variant. J Physiol. 2004;558:147–59. doi: 10.1113/jphysiol.2004.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinnikova AK, et al. Na+-H+ exchange activity in taste receptor cells. J Neurophysiol. 2004;91:1297–313. doi: 10.1152/jn.00809.2003. [DOI] [PubMed] [Google Scholar]
- 14.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–4. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 15.Chandrashekar J, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–11. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 16.Li X, et al. Human receptors for sweet and umami taste. Proc Natl Acad Sci U S A. 2002;99:4692–6. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–37. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 18.Delmas P. Polycystins: polymodal receptor/ion-channel cellular sensors. Pflugers Arch. 2005;451:264–76. doi: 10.1007/s00424-005-1431-5. [DOI] [PubMed] [Google Scholar]
- 19.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–24. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 20.Murakami M, et al. Genomic organization and functional analysis of murine PKD2L1. J Biol Chem. 2005;280:5626–35. doi: 10.1074/jbc.M411496200. [DOI] [PubMed] [Google Scholar]
- 21.Lopezjimenez ND, et al. Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J Neurochem. 2006;98:68–77. doi: 10.1111/j.1471-4159.2006.03842.x. [DOI] [PubMed] [Google Scholar]
- 22.Hoon MA, et al. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–51. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- 23.Collier RJ. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39:54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brockschnieder D, et al. Cell depletion due to diphtheria toxin fragment A after Cremediated recombination. Mol Cell Biol. 2004;24:7636–42. doi: 10.1128/MCB.24.17.7636-7642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez CA, et al. A transient receptor potential channel expressed in taste receptor cells. Nat Neurosci. 2002;5:1169–76. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- 26.Lahiri S, Forster RE., 2nd CO2/H(+) sensing: peripheral and central chemoreception. Int J Biochem Cell Biol. 2003;35:1413–35. doi: 10.1016/s1357-2725(03)00050-5. [DOI] [PubMed] [Google Scholar]
- 27.Richerson GB, Wang W, Hodges MR, Dohle CI, Diez-Sampedro A. Homing in on the specific phenotype(s) of central respiratory chemoreceptors. Exp Physiol. 2005;90:259–66. 266–9. doi: 10.1113/expphysiol.2005.029843. [DOI] [PubMed] [Google Scholar]
- 28.Gosgnach S, et al. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–9. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- 29.Lee EC, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 30.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cremediated excision. Genesis. 2000;28:147–55. [PubMed] [Google Scholar]