Abstract
Cell ploidy in the ovarian follicle and corpus luteum was investigated by DNA in situ hybridization to a reiterated, chromosome 3 transgene in mice that were hemizygous for the transgene. This approach was first validated by analysis of mouse kidney, pancreas and liver control tissues, which contain different frequencies of polyploid nuclei. Polyploid nuclei (with multiple hybridization signals) were seen in histological sections of both ovarian follicles and corpora lutea. The frequency of polyploid nuclei in follicles showed no consistent relationship with age (between 6 weeks and 10 months) but polyploid nuclei were significantly more abundant in corpora lutea than follicles (6.3% vs. 2.5%). This implies that production of polyploid cells is more closely associated with differentiation of ovarian follicles into corpora lutea than with the age of the female. Polyploidy tended to be more frequent in corpora lutea of mice that had mated even if they did not become pregnant. This study has highlighted the presence of polyploid cells in the mouse ovarian follicle and corpus luteum and has identified mating as a possible trigger for polyploidy in the corpus luteum. Further work is required to determine the physiological role of polyploid ovarian cells in reproduction.
Keywords: corpus luteum, DNA in situ hybridization, liver, ovarian follicle, pancreas, polyploidy
Introduction
Many adult mammalian tissues contain polyploid cells alongside diploid cells (mixoploidy). Brodsky & Uryvaeva (1985) reviewed the evidence for cells with polyploid nuclei in many tissues, including the liver, exorbital gland (lacrimal gland), urinary bladder, uterine decidua, cardiac muscle and cerebellar Purkinje cells. Megakaryocytes are all polyploid (Odell et al. 1969) and the placenta contains both polyploid and polytene nuclei (Zybina & Grishchenko, 1970; Zybina & Zybina, 1996; Klisch et al. 1999). There is also an age-related increase in the frequency of polyploid nuclei in the mouse pancreas (Webb et al. 1982) and liver (Alfert & Geschwind, 1958; Inamdar, 1958; Epstein, 1967; Evans, 1976; Brodsky & Uryvaeva, 1977). Several authors have suggested that the extent of polyploidy in the liver is a function of liver growth or differentiation rather than age per se and that polyploidy may be advantageous in differentiated tissues that need a high biosynthetic capability (Epstein, 1967; Wheatley, 1972; Brodsky & Uryvaeva, 1985). However, it is usually difficult to distinguish between the effects of age and differentiation and it is not understood what causes polyploid cells to arise.
There is evidence that polyploid cells also occur in ovarian follicles and corpora lutea. These are transient structures that undergo growth, differentiation and degeneration throughout adult life so different stages coexist in the same ovary. Oocytes are surrounded by a single layer of flattened epithelial cells in the fetus to produce primordial follicles. During follicular growth the follicle granulosa cells become cuboidal and increase in number to produce a multilayered follicle. Fluid accumulates in spaces between the epithelial cells and later these fluid-filled vesicles coalesce to form an antral cavity. At this stage, follicle cells are classified as either mural granulosa cells (which line the inside of the follicle wall) or cumulus cells (which project into the antral cavity and surround the oocyte). At ovulation the oocyte and surrounding cumulus cells are expelled into the oviduct and the remainder of the follicle collapses and differentiates into the corpus luteum. In many species the theca cells (which form the follicle wall) also contribute to the corpus luteum but in the mouse it seems likely that only the mural granulosa follicle cells contribute (Tong et al. 1998). The mural granulosa cells differentiate into larger flattened granulosa–lutein cells of the corpus luteum, which synthesize progesterone required to maintain the pregnancy. Most granulosa–lutein cells are terminally differentiated and cease division (Robker & Richards, 1998) but a subpopulation continues to proliferate during the early and mid luteal phases, at least in some species (Gaytan et al. 1998; Young et al. 2000). The corpus luteum is maintained throughout pregnancy but, if pregnancy does not ensue, infiltrating fibroblasts cause the corpus luteum to regress to scar tissue after a few days.
Stangel et al. (1970) reported the occurrence of polyploid cells in human corpora lutea in late pregnancy, and Coulson (1979) detected polyploid cells in small follicles in ovaries of pigs and cows but not in larger follicles. In the mouse, Telfer and Gosden found no polyploid nuclei in mural granulosa cells of antral follicles using microdensitometry (cited in Telfer et al. 1988) but Keighren & West (1993) identified a low frequency of polyploid cells in ovarian follicles and corpora lutea by DNA in situ hybridization.
The aim of the present study was to determine whether the frequency of polyploid cells varied among stages during the sequential differentiation of pre-antral follicles to antral follicles, and then to terminally differentiated corpora lutea. Since this sequence occurs throughout adulthood it uncouples the effects of age vs. follicle growth and differentiation, and provides a unique opportunity to test which has the greater influence on the frequency of polyploidy. Also, since both follicles and corpora lutea exist in pregnant and non-pregnant females we were able to compare the frequency of polyploidy in these two different physiological conditions. To detect polyploid nuclei we used DNA in situ hybridization to a chromosome-specific transgene, which is a sensitive method that allows visual identification of low levels of polyploid nuclei in histological sections. The method was validated by determining the proportion of polyploid nuclei in several control mouse tissues, for which polyploidy has been investigated by other methods.
Materials and methods
Mice
The mice were all hemizygous for the transgene TgN(Hbb-b1)83Clo (here abbreviated to Tg) comprising approximately 1000 tandemly repeated copies of a mouse β-globin plasmid inserted near the telomere of chromosome 3 (Lo, 1986; Lo et al. 1987). Males, homozygous for the β-globin transgene (Tg+/+), were crossed to wild-type (C57BL × CBA/Ca)F1 females, with no transgene (Tg–/–), to produce hemizygous (Tg+/–) offspring. Unless stated, pregnancies were obtained by natural matings and were timed from the day of the vaginal plug which was designated embryonic day 0.5 (E0.5). Those that were superovulated were injected with 5 iu pregnant mares’ serum gonadotrophin (PMSG) at approximately 12:00 h, followed by 5 iu human chorionic gonadotrophin (hCG) 48 h later. Mating was confirmed by the presence of a vaginal plug the following morning and pregnancy was confirmed by the presence of implantation sites or the recovery of pre-implantation embryos.
Tissue preparation
Mice were killed by cervical dislocation and tissues immediately dissected and fixed. Most samples were fixed in acetic ethanol (3 ethanol : 1 acetic acid, v/v) and either embedded in paraffin wax for histology or used to prepare cell spreads. For histological sections of liver, samples were fixed in 4% paraformaldehyde because, for this tissue, it proved superior to acetic ethanol fixation for DNA in situ hybridization. Ovaries were analysed only by histological sections to facilitate staging the follicles and scoring follicles located deep within the ovary. For spreads of pancreas, liver and kidney, small pieces of tissue were placed in multiwell plates and the wells were half filled with hypotonic solution (1 : 3 mixture of 1.93% citrate and 0.56% KCl), minced with scissors and left for 20–30 min. The hypotonic solution was replaced with freshly prepared acetic ethanol fixative for 2 h followed by 60% acetic acid for 5 min. Tissue was disrupted with a Pasteur pipette, cells spread on a hot microscope slide (from a 50 °C hot plate), excess acid removed and slides allowed to air dry. Slides were stored in a cool dry box until needed.
DNA in situ hybridization
The hybridization signal was detected as a brown spot in the nucleus by diaminobenzidine staining for a peroxidase-conjugated antibody bound to the digoxygenin-labelled DNA probe (Keighren & West, 1993). Slides were counter-stained with haematoxylin and eosin and examined by bright-field microscopy and the number of hybridization signals was recorded for each nucleus scored.
In order to quantify the frequency of polyploid nuclei, all the nuclei in one field of view (×63 objective) were scored according to the number of hybridization signals, using a 10 × 10 index square eyepiece graticule (Graticules Ltd, Tonbridge, Kent, UK). Nuclei were only classified as having two hybridization signals if the signals were of similar size (not significantly smaller than those in nuclei with a single signal) and clearly separated by a space so they were not touching. Two hybridization signals very close together were classified as a single split signal (split spot) and may represent chromosomes in G2 of the cell cycle. The numbers of hybridization signals were scored as 0, 1, 2 or > 2. Transgenic mice were hemizygous (one copy of the reiterated transgene per diploid genome), so diploid nuclei were identified as those with 1 hybridization signal and polyploid nuclei had > 1 signal. Apart from the liver, few nuclei had > 2 signals so the results are presented as percentage of nuclei with > 1 signal. Since all nuclei should have at least one chromosome with the target β-globin transgenic marker (positive nuclei), the percentage of nuclei with no signal represents the percentage of false negatives (e.g. sections that did not include the part of the nucleus with the target sequence). The percentage of putatively polyploid nuclei was estimated as the percentage of positive nuclei that had more than one signal. To ensure analysis of sections near the middle of the follicle, they were only analysed and classified if part of the oocyte was present. Mural granulosa cells and cumulus cells (where present) were scored but theca cells were excluded. Ovarian follicles were classified according to the number of layers of granulosa cells (Mandl & Zuckerman, 1951).
Statistical analysis
Mann–Whitney U-tests were performed on an Apple Macintosh computer using the statistical package ‘StatView 5.0’ (SAS Institute Inc.).
Results
Identification of polyploid nuclei in control tissues by DNA in situ hybridization
All nuclei were hemizygous for the target DNA sequences (β-globin transgene), so the diploid genome is represented by a single hybridization signal (brown spot in the nucleus). Nuclei with multiple hybridization signals were classified as putative polyploid nuclei. To validate the use of in situ hybridization on histological sections to identify polyploid nuclei, the technique was first tested on mouse kidney (which served as a negative tissue control), and on pancreas and liver (which served as positive tissue controls). Cell spreads and 7-µm histological sections were prepared from mice at three ages and coded prior to analysis. In addition, tissue sections and spreads were prepared from non-transgenic (C57BL × CBA)F1 mice (negative control mice). In situ hybridization was less efficient for the liver, and two samples were excluded from the analysis because too few nuclei had a hybridization signal. However, in most experimental samples, 80–90% of nuclei had at least one hybridization signal and tissues from non-transgenic negative control mice showed negligible levels of false positive hybridizations.
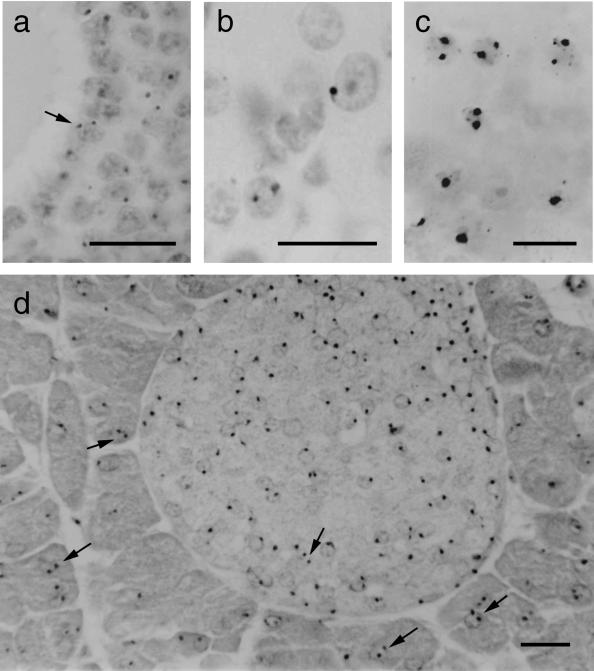
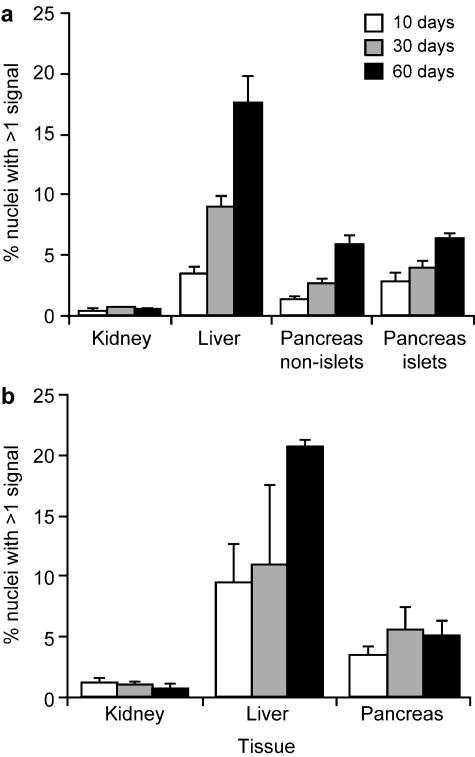
Putatively polyploid nuclei with multiple hybridization signals are illustrated in Fig. 1 and the frequencies of polyploid nuclei in the kidney, liver and pancreas are shown in Fig. 2. Few (< 1%) kidney nuclei had multiple hybridization signals in either the histological sections or the cell spreads but, as expected, these occurred at much higher levels in liver sections and spreads. An intermediate frequency was found in both the pancreas spreads and sections. In histological sections these putatively polyploid nuclei were seen among both the endocrine cells of the islet of Langerhans and the non-islet, exocrine cells (Fig. 1d). The overall percentages of nuclei with multiple hybridization signals was similar for cell spreads and tissue sections (Fig. 2a,b).
Fig. 1.
Nuclei with two hybridization signals (some are arrowed) in (a) a section of ovarian follicle, (b) a section of ovarian corpus luteum, (c) a cell spread from pancreas of 60-day-old mouse and (d) a section of pancreas of 60-day-old mouse showing an islet of Langerhans (small, paler stained cells). Some positive nuclei appear negative in the photographs because the hybridization signals are out of the plane of focus. Scale bar = 20 µm.
Fig. 2.
Histograms showing the percentages of putatively polyploid nuclei (with multiple hybridization signals) in kidney, pancreas and liver from mice (males and females pooled) at different ages in (a) histological sections and (b) cell spreads. Pancreases were analysed from six mice at each age and kidneys were analysed from three mice at each age. Livers were analysed from three mice at each age, except that only two samples were analysed at 10 days by histological sections and at 30 days by cell spreads.
Mann–Whitney U-tests, on pooled results for different ages and sexes, showed that the percentage of nuclei with multiple hybridization signals was significantly higher in the pancreas cell spreads than the kidney spreads (P = 0.0003), and for histological sections it was significantly higher in both the pancreas non-islet cells (P < 0.0001) and pancreas islet cells (P = 0.0002) than kidney sections. Analysis of the pancreas showed that the in situ hybridization method is capable of identifying relatively low levels of polyploid nuclei. Figure 2(a) shows the expected age-related increase in frequency of polyploid nuclei in liver and pancreas sections.
Polyploidy in ovarian follicles and corpora lutea
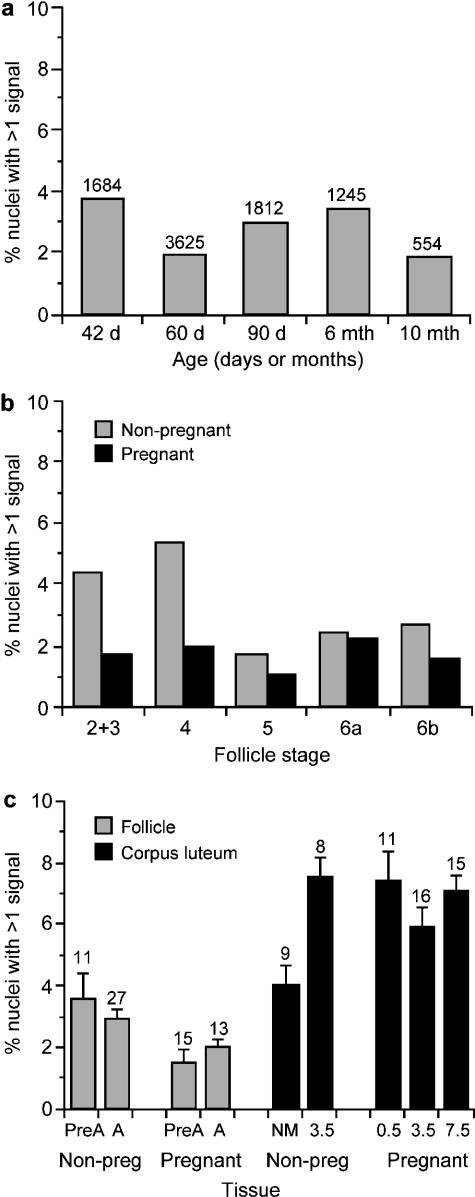
Putative polyploid nuclei were found in 5-µm sections of ovarian follicles and corpora lutea (Fig. 1a,b) and almost all of these were putative tetraploid nuclei (two hybridization signals). For the ovarian follicles, there was no consistent trend in the frequency of polyploid nuclei with age for 17 non-pregnant mice between 6 weeks and 10 months (Fig. 3a). For these non-pregnant mice the smallest follicle classes (stages 2–4) had the highest proportion of polyploid nuclei (14/324 and 19/361, respectively; Fig. 3b) but this trend was not seen in the six pregnant mice analysed. When the follicles were grouped as pre-antral (stages 2–5) vs. antral (stages 6a and 6b) there was no significant difference in the frequency of polyploid nuclei for either pregnant or non-pregnant mice (Fig. 3c).
Fig. 3.
Histogram showing the mean percentages of putatively polyploid nuclei (with multiple hybridization signals) in mouse ovaries. (a) Ovarian follicles from non-pregnant females of different ages (the number of nuclei with hybridization signals analysed are shown above the bars). (b) Ovarian follicles at different stages (stages 6a and 6b are antral follicles; others are pre-antral follicles). A total of 8920 nuclei with hybridization signals were analysed from non-pregnant mice (ranging from 125 for stage 5 follicles to 5917 for stage 6a); 6236 positive nuclei were scored from non-pregnant mice (ranging from 299 for stages 2 and 3 to 2896 for stage 6a). (c) Ovarian follicles and corpora lutea from pregnant and non-pregnant mice. (The mean and standard error are shown for percentages calculated separately for each follicle or corpus luteum and the numbers of follicles and corpora lutea analysed are shown above the bars.) Abbreviations: Non-preg, non-pregnant; PreA, pre-antral follicles; A, antral follicles; NM, female not housed with male; 0.5, 3.5 and 7.5 indicate number of days post-coitum. No embryos were found in three mice, 3 days after finding vaginal plugs (3.5 non-pregnant group).
For comparison with the results for the follicles, polyploidy was analysed in corpora lutea from five groups of mice (three females in each group), including pregnant and non-pregnant females (Fig. 3c). The percentage of polyploid nuclei was calculated separately for each follicle and corpus luteum to enable calculation of standard errors for each group and statistical comparison between groups by Mann–Whitney U-tests. Figure 3(c) shows that the overall percentage of putatively polyploid nuclei was higher in corpora lutea (6.32 ± 0.35%) than in ovarian follicles (2.45 ± 0.23%) and that this difference was statistically significant (P < 0.0001).
It was notable that the frequency of polyploid nuclei in the corpora lutea was lower in females that were not housed with males (3.9%) than in all three groups of pregnant females (range 5.8–7.3%). This difference was significant for the pregnant females examined at E0.5 (P = 0.030) and E7.5 (P = 0.0066), but not for those at E3.5 (P = 0.079). The frequency of polyploid corpora lutea cells in females that were not housed with males was also significantly lower than in females that mated with males (vaginal plug present) but from which no embryos were recovered at E3.5 (3.9% vs. 7.5%; P = 0.0071).
Influence of mating on the frequency of polyploidy in the corpus luteum
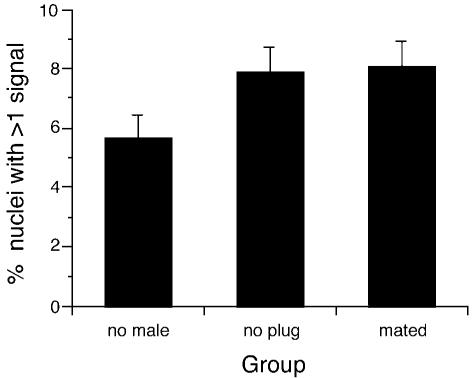
The results shown in Fig. 3(c) indicate that the differentiation of the follicle to a corpus luteum after ovulation is accompanied by an increase in the frequency of polyploid nuclei. The results also raise the possibility that this increase depends on mating (or housing overnight with a male) rather than pregnancy itself. To distinguish between these effects, the frequency of polyploid nuclei was analysed in three groups of superovulated females, two of which were housed overnight with sterile, vasectomized males and killed the following afternoon (28 h after hCG). In each group approximately 3000 nuclei were scored from 11 to 12 corpora lutea from 4–6 mice. The three groups comprised six females not paired with vasectomized males, four females that were housed overnight with vasectomized males but failed to produce a vaginal plug, so may not have mated, and four females that were housed overnight with vasectomized males and produced a vaginal plug as evidence of mating. Figure 4 shows that the frequency of polyploidy was lower in the group of superovulated females that had not been housed with males (5.6%) than the two groups of females that had been housed overnight with vasectomized males (7.9% and 8.1%, respectively). Although these differences did not reach statistical significance (P = 0.065 and P = 0.083, respectively, by Mann–Whitney U-tests), the trends were consistent with the earlier evidence that mating (or the presence of a male) increases the frequency of polyploidy in the corpus lutea even without pregnancy. Moreover, the frequency of polyploidy seen in the females housed with, or mated by, vasectomized males (Fig. 4) was comparable to that in the pregnant females shown in Fig. 3(c) (5.8–7.3%).
Fig. 4.
Histogram showing the mean percentages of putatively polyploid nuclei (with multiple hybridization signals) in mouse corpora lutea in three groups of superovulated females. The mean and standard error are shown for percentages calculated separately for each corpus luteum. Females in the ‘no male’ group were not housed with males. Females in the ‘no plug’ group were housed overnight with vasectomized males but did not have vaginal plugs the following morning so there was no evidence of mating. Females in the ‘mated’ group were housed overnight with vasectomized males and had vaginal plugs the next morning (evidence of mating).
Discussion
The occurrence of hemizygous Tg+/– nuclei with multiple hybridization signals is most simply interpreted as polyploidy, particularly in tissues where polyploidy has been inferred by other methods. Since we used in situ hybridization to a transgene on chromosome 3, we could not formally distinguish polyploid nuclei from nuclei made polysomic for chromosome 3 (e.g. by somatic non-disjunction) or diploid nuclei made homozygous for the transgene by somatic recombination. However, such events are likely to be rare and sporadic (Grüneberg, 1966; Fisher et al. 1986; Panthier & Condamine, 1991). Two signals could also occur in G2 stage interphase nuclei if the signals from sister chromatids became separated after DNA replication, but our scoring criteria for multiple hybridization signals were chosen to ensure that hybridization sites on sister chromatids were scored as split signals (split spots) rather than two separate hybridization signals (Masumoto et al. 1989; Hopman et al. 1990; Lawrence et al. 1990). Possible technical artefacts that could affect the frequency of multiple hybridization signals include undetected overlapping nuclei in histological sections and differences in survival of small and large nuclei during the preparation of cell spreads. The overall similarity between the results for the histological sections and cell spreads for the pancreas, liver and kidney suggest that such technical artefacts, which are specific to one method of preparation, do not significantly distort the analysis. We therefore conclude that technical artefacts are negligible and most nuclei with multiple hybridization signals were polyploid.
The results for the control tissues agree with previous findings obtained by other methods in showing a higher frequency of polyploid nuclei in the liver (Epstein, 1967; Brodsky & Uryvaeva, 1977, 1985) than in the pancreas (Larsen & Nielsen, 1978; Webb et al. 1982; Ornitz et al. 1987; Levine et al. 1991). They also showed the expected age-related increases in polyploidy in both organs. This indicated that the DNA in situ hybridization method was suitable for the analysis of polyploidy in histological sections of the mouse ovary. This method provides obvious visual cues for the identification of polyploid nuclei and is useful for detecting low frequencies of polyploid nuclei in specific regions of a complex tissue or organ.
Analysis of histological sections of mouse ovaries demonstrated that polyploid nuclei exist in mouse ovarian follicles and corpora lutea. There was no increase in frequency of polyploid follicle cells with age and no significant difference between pre-antral and antral follicles. However, there was some suggestion that, in non-pregnant mice, the frequency of polyploid nuclei was highest in the smallest classes of pre-antral follicles. This was not seen in the pregnant mice but is consistent with observations on pig and cow follicles (Coulson, 1979). One possibility is that it could reflect initial accumulation of polyploid cells that are diluted out during the later phase of rapid follicular growth. Corpora lutea generally had a significantly higher frequency of polyploid nuclei than follicles, which suggests that the increased frequency is more likely to be related to differentiation of follicle cells into luteal cells than to age or follicle growth.
The frequency of polyploid nuclei in corpora lutea was higher in pregnant females than females that had not been with males. This elevated frequency occurred in pregnant females as early as E0.5 and was also seen in non-pregnant females that had mated to vasectomized males or to fertile males but failed to become pregnant (no embryos found). Overall, these results suggest that mating is more likely than pregnancy to be the main stimulus responsible for the increase in polyploidy in corpora lutea. It is known that functional changes are induced in the mouse corpus luteum by vaginal stimulation at mating but that these changes do not necessarily require the production of a vaginal plug (McGill et al. 1968; McGill & Coughlin, 1970; McGill, 1970). This raises the possibility that an increase in the frequency of polyploid cells accompanies the changes induced in the mouse corpus luteum by the stimulus of mating. Although polyploid cells also exist in human corpora lutea, their frequency does not increase until later in pregnancy (Stangel et al. 1970). In the human, a subpopulation of proliferating granulosa–lutein cells persists into the mid-luteal phase (Gaytan et al. 1998) but comparable studies have not been reported for the mouse. Further work on the mouse corpus luteum is needed to clarify the relationship between mating and the frequency of polyploidy, and the relationship between cell proliferation and polyploidy.
Polyploid cells accumulate in several adult mouse tissues including the liver, pancreas and corpus luteum but the reason for this remains unclear (also see Martin et al. 2002). This contrasts with the situation in mixoploid pre-implantation embryos, where both naturally occurring and experimentally induced tetraploid cells are depleted between the early and late blastocyst stages (Everett & West, 1998; Viuff et al. 2002). If polyploid cells in early embryos are at a proliferative or selective disadvantage and are not replenished they would become diluted or lost. Any proliferative disadvantage of polyploid cells would be less marked in adult tissues that have completed their growth phase and so have a lower mitotic index. The accumulation of polyploid cells in some adult tissues suggests multiple diploid-to-polyploid conversions occur and this may reflect an association of terminal differentiation with acytokinetic mitosis in some cells (Epstein, 1967; Brodsky & Uryvaeva, 1977, 1985; Klisch et al. 1999). Possible beneficial properties of polyploidy include protection against accumulation of recessive mutations or enhanced biosynthetic capability. Polyploid cells often occur in tissues with specialized biosynthetic secretary or storage functions (Brodsky & Uryvaeva, 1977, 1985; Webb et al. 1982), as is the case with the liver, pancreas and corpus luteum. However, the frequency of polyploidy in the liver varies widely among different mammalian species (Brodsky & Uryvaeva, 1985) which makes it difficult to generalize about possible functional advantages of polyploidy.
This study has demonstrated the occurrence of polyploid cells in the mouse ovarian follicle and corpus luteum and highlighted their accumulation in the mouse corpus luteum after it is formed from the antral follicle. Here, differentiation is not age-dependent and our results imply that the accumulation of polyploid cells is more clearly associated with differentiation of the corpora luteum than age. Our results also suggest that mating may trigger the accumulation of polyploid cells in the corpus luteum. Further studies will be required to investigate the trigger mechanism and identify whether polyploid cells have a physiological role in the ovarian follicle or corpus luteum.
Acknowledgments
We thank Jean Flockhart for expert technical help, Denis Doogan, Maureen Ross, Jim Macdonald, Ted Pinner and Tom McFetters for specialist assistance, John Donaldson for performing some of the in situ hybridizations on kidney, liver and pancreas samples, and Gillian MacKay for comments on the manuscript. The work was funded in part by The Wellcome Trust (grant 046359 awarded to J.D.W. to support M.A.K. and vacation scholarships for L.P.M. and A.S.H.) and the Moray Endowment Fund of The University of Edinburgh. We are grateful to Professor John Dale for additional financial support.
References
- Alfert M, Geschwind I. The development of polysomaty in the rat liver. Exp. Cell Res. 1958;15:230–232. doi: 10.1016/0014-4827(58)90079-x. [DOI] [PubMed] [Google Scholar]
- Brodsky VY, Uryvaeva IV. Cell polyploidy: its relation to tissue growth and function. Int. Rev. Cytol. 1977;50:275–332. doi: 10.1016/s0074-7696(08)60100-x. [DOI] [PubMed] [Google Scholar]
- Brodsky VY, Uryvaeva IV. Genome Multiplication in Growth and Development. Biology of Polyploid and Polytene Cells. Cambridge: Cambridge University Press; 1985. [Google Scholar]
- Coulson PB. Characterization of polyploidy in ovarian granulosa cells. In: Midgley AR, Sadler WA, editors. Ovarian Follicular Development and Function. New York: Raven Press; 1979. pp. 385–394. [Google Scholar]
- Epstein CJ. Cell size, nuclear content and the development of polyploidy in the mammalian liver. Proc. Natl. Acad. Sci. USA. 1967;57:327–334. doi: 10.1073/pnas.57.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans IH. Polyploidization in the rat liver: the role of binucleate cells. Cytobios. 1976;16:115–124. [PubMed] [Google Scholar]
- Everett CA, West JD. Evidence for selection against tetraploid cells in tetraploid ↔ diploid mouse chimaeras before the late blastocyst stage. Genet. Res. 1998;72:225–228. doi: 10.1017/s0016672398003498. [DOI] [PubMed] [Google Scholar]
- Fisher G, Stephenson DA, West JD. Investigation of the potential for mitotic recombination in the mouse. Mut. Res. 1986;164:381–388. doi: 10.1016/0165-1161(86)90031-2. [DOI] [PubMed] [Google Scholar]
- Gaytan F, Morales C, Garcia-Pard L, Reymundo C, Bellido C, Sanchez-Criado JE. Macrophages, cell proliferation, and cell death in the human menstrual corpus luteum. Biol. Reprod. 1998;59:417–425. doi: 10.1095/biolreprod59.2.417. [DOI] [PubMed] [Google Scholar]
- Grüneberg H. The case for somatic crossing over in the mouse. Genet. Res. 1966;7:58–75. doi: 10.1017/s0016672300009472. [DOI] [PubMed] [Google Scholar]
- Hopman AHN, Ramaekers FCS, Vooijs GP. Interphase cytogenetics of solid tumours. In: Polack JM, McGee JOD, editors. In Situ Hybridization Principles and Practice. Oxford: Oxford Science Publications; 1990. pp. 165–186. [Google Scholar]
- Inamdar N. Development of polyploidy in mouse liver. J. Morphol. 1958;103:65–90. [Google Scholar]
- Keighren M, West JD. Analysis of cell ploidy in histological sections of mouse tissues by DNA–DNA in situ hybridization with digoxygenin labelled probes. Histochem. J. 1993;25:30–44. doi: 10.1007/BF00161042. [DOI] [PubMed] [Google Scholar]
- Klisch K, Hecht W, Pfarrer C, Schuler G, Hoffmann B, Leiser R. DNA content and ploidy level of bovine placentomal trophoblast giant cells. Placenta. 1999;20:451–458. doi: 10.1053/plac.1999.0402. [DOI] [PubMed] [Google Scholar]
- Larsen JK, Nielsen O. Flowcytometric evaluation of the DNA distribution in isolated pancreatic islets from normal and diabetic mice. J. Histochem. Cytochem. 1978;27:410–412. doi: 10.1177/27.1.374603. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, McNeil JA. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science. 1990;249:928–932. doi: 10.1126/science.2203143. [DOI] [PubMed] [Google Scholar]
- Levine DS, Sanchez CA, Rabinovitch PS, Reid BJ. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc. Natl. Acad. Sci. USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C. Localization of low abundance DNA sequences in tissue sections by in situ hybridization. J. Cell Sci. 1986;81:143–162. doi: 10.1242/jcs.81.1.143. [DOI] [PubMed] [Google Scholar]
- Lo CW, Coulling M, Kirby C. Tracking of mouse cell lineage using microinjected DNA sequences: analysis using genomic Southern blotting and tissue-section in situ hybridizations. Differentiation. 1987;35:37–44. doi: 10.1111/j.1432-0436.1987.tb00149.x. [DOI] [PubMed] [Google Scholar]
- Mandl AM, Zuckerman S. Numbers of normal and atretic oocytes in unilaterally spayed rats. J. Endocrinol. 1951;7:112–119. doi: 10.1677/joe.0.0070112. [DOI] [PubMed] [Google Scholar]
- Martin NC, McCullough CT, Bush PG, Sharp L, Hall AC, Harrison DJ. Functional analysis of mouse hepatocytes differing in DNA content: volume, receptor expression, and effect of IFN-gamma. J. Cell Physiol. 2002;191:138–144. doi: 10.1002/jcp.10057. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugimoto K, Okazaki T. Alphoid satellite DNA is tightly associated with centromere antigens in human chromosomes throughout the cell cycle. Exp. Cell Res. 1989;181:181–196. doi: 10.1016/0014-4827(89)90192-4. [DOI] [PubMed] [Google Scholar]
- McGill TE, Corwin DM, Harrison DT. Copulatory plug does not induce luteal activity in the mouse Mus musculus. J. Reprod. Fert. 1968;15:149–151. doi: 10.1530/jrf.0.0150149. [DOI] [PubMed] [Google Scholar]
- McGill TE. Induction of luteal activity in female house mice. Horm. Behav. 1970;1:211–222. [Google Scholar]
- McGill TE, Coughlin RC. Ejaculatory reflex and luteal activity induction in Mus musculus. J. Reprod. Fert. 1970;21:215–220. doi: 10.1530/jrf.0.0210215. [DOI] [PubMed] [Google Scholar]
- Odell TT, Burch EA, Jackson CW, Friday TJ. Megakaryocytopoiesis in mice. Cell Tissue Kinet. 1969;2:363–367. [Google Scholar]
- Ornitz DM, Hammer RE, Messing A, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987;238:188–193. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- Panthier JJ, Condamine H. Mitotic recombination in mammals. Bioessays. 1991;13:351–356. doi: 10.1002/bies.950130709. [DOI] [PubMed] [Google Scholar]
- Robker RL, Richards JS. Hormonal control of the cell cycle in ovarian cells: proliferation versus differentiation. Biol. Reprod. 1998;59:476–482. doi: 10.1095/biolreprod59.3.476. [DOI] [PubMed] [Google Scholar]
- Stangel JJ, Richart RM, Okagaki T, Cottral G. Nuclear DNA content of luteinized cells of the human ovary. Am. J. Obstet. Gyn. 1970;108:543–549. doi: 10.1016/0002-9378(70)90229-2. [DOI] [PubMed] [Google Scholar]
- Telfer E, Ansell JD, Taylor H, Gosden RG. The number of clonal precursors of the follicular epithelium in the mouse ovary. J. Reprod. Fert. 1988;84:105–110. doi: 10.1530/jrf.0.0840105. [DOI] [PubMed] [Google Scholar]
- Tong W, Kiyokawa H, Soos TJ, Park MS, Soares VC, Manova K, et al. The absence of p27Kip1, an inhibitor of G1 cyclin-dependent kinases, uncouples differentiation and growth arrest during the granulosa → luteal transition. Cell Growth Differ. 1998;9:787–794. [PubMed] [Google Scholar]
- Viuff D, Palsgaard A, Rickords L, Lawson LG, Greve T, Schmidt M, et al. Bovine embryos contain a higher proportion of polyploid cells in the trophectoderm than in the embryonic disc. Mol. Reprod. Dev. 2002;62:483–488. doi: 10.1002/mrd.90004. [DOI] [PubMed] [Google Scholar]
- Webb SR, Dore BA, Grogan WM. Cell cycle analysis of the postnatal mouse pancreas. Biol. Neonate. 1982;42:73–78. doi: 10.1159/000241578. [DOI] [PubMed] [Google Scholar]
- Wheatley DN. Binucleation in mammalian liver. Exp. Cell Res. 1972;74:455–465. doi: 10.1016/0014-4827(72)90401-6. [DOI] [PubMed] [Google Scholar]
- Young FM, Rodger FE, Illingworth PJ, Fraser HM. Cell proliferation and vascular morphology in the marmoset corpus luteum. Hum. Reprod. 2000;15:557–566. doi: 10.1093/humrep/15.3.557. [DOI] [PubMed] [Google Scholar]
- Zybina EV, Grishchenko TA. Polyploid trophoblast cells in different parts of the rat placenta. Tsitologiya. 1970;14:284–290. [Google Scholar]
- Zybina EV, Zybina TG. Polytene chromosomes in mammalian cells. Int. Rev. Cytol. 1996;165:53–119. doi: 10.1016/s0074-7696(08)62220-2. [DOI] [PubMed] [Google Scholar]