Abstract
The organization of the actin cytoskeleton in prefusion aligning myoblasts is likely to be important for their shape and interaction. We investigated actin filament organization and polarity by transmission electron microscopy (TEM) in these cells. About 84% of the filaments counted were either found in a subplasmalemma sheet up to 0.5 µm thick that was aligned with the long axis of the cell, or in protrusions. The remaining filaments were found in the cytoplasm, where they were randomly orientated and not organized into bundles. The polarity of the subplasmalemma filaments changed progressively from one end of the cell to the other. At the ends of the cells and in protrusions, the majority of filaments were organized such that their barbed ends faced the tip of the protrusion. We did not find any actin filament bundles or stress fibres in these cells. Time-lapse phase microscopy demonstrated that aligned cells were still actively migrating at the time of our TEM observations, and their direction of movement was restricted to the long axis of the cell group. The ability of these cells to locomote actively in the absence of actin filament bundles suggests that in these cells the subplasmalemma actin sheet contributes not only to cell shape but also to cell locomotion.
Keywords: cell locomotion, cytoskeleton, muscle differentiation, S1-decoration, TEM, video microscopy
Introduction
During embryonic development, multinucleated skeletal muscle fibres are formed by the fusion of mononucleated myoblasts, leading to the formation of mature skeletal muscle fibres (Miller, 1992). The cellular and molecular processes of myoblast fusion and fibre formation can be studied in isolation in tissue culture. In the early stages of muscle differentiation in vitro, myoblasts proliferate, recognize each other, align, adhere and eventually fuse to form multinucleated myotubes (reviewed in Wakelam, 1985). The signal that promotes fusion is not clear, but Ca2+ entry is essential (Shainberg et al. 1969; Bernheim & Bader, 2002).
Myoblast fusion is tightly controlled spatially, in that myoblasts almost always fuse into long linear myotubes rather than spheroid structures. This is partly explained by the elongation of myoblasts that occurs when they align and fuse, and partly by restricting myoblasts from fusing at regions of the cell other than at the ends. Evidence for this was provided by experiments in which myoblasts fused on laminin tracks of varying width (Clark et al. 1997), where it was found that although the number of myoblasts increased with track width, the diameter of myotubes formed remained constant. However, both side-to-side and end-to-end interactions are important in fusion, because if lateral movement is inhibited, fusion is significantly reduced (Clark et al. 2002). Furthermore, time-lapse imaging of fusing myoblasts in culture has demonstrated end-to-end fusion (Lu et al. 2001; Musa et al. 2003).
Elongation of myoblasts in aligned groups is likely to be a result of the re-organization of the actin and microtubular cytoskeleton that occurs in aligned prefusion myoblasts as demonstrated by light microscopy (Tassin et al. 1985; Wells et al. 1997; Lu et al. 2001; Musa et al. 2003). We and others have also shown that microtubules become organized linearly, parallel with the long axis of the cell (Tassin et al. 1985; Lu et al. 2001; Musa et al. 2003). We previously described, using confocal microscopy, that actin stress fibre-like bundles are found in migrating myoblasts, but not in aligned groups, where the actin filaments are found mainly at the periphery of the aligned cells (Wells et al. 1997). Furthermore, we showed that non-muscle myosin II and myosin Iα associate with actin at the plasma membrane, and could contribute to the elongated shape of myoblasts (Wells et al. 1997). However, fusing myoblasts are still able to locomote (Musa et al. 2003), and thus the re-organized actin cytoskeleton in aligned myoblasts is also likely to contribute to this process.
Combined with these changes in the organization of the actin and microtubule cytoskeletons, aligned, fusing myoblasts tend to become less well attached to the substratum, and cell–cell interactions become more important. This feature can partly be attributed to changes in integrin expression (Steffensen et al. 1992; Blaschuk et al. 1997) and increased activity of extracellular matrix proteases such as calpain and metalloproteinases (Yagami-Hiromasa et al. 1995). Surface proteins known to be important for fusion in Drosophila (Bour et al. 2000; Ruiz-Gomez et al. 2000; Taylor, 2000) are also likely to be important in the mammalian system, although mammalian homologues have not yet been clearly identified for some of these.
Clearly, myoblast locomotion, cell–cell interaction and cell shape are important for the fusion of myoblasts into multinucleated myotubes. To investigate how the actin cytoskeleton contributes to shape and locomotion in aligned prefusion myoblasts, we have determined its ultrastructural organization in these cells. The polarity of actin filaments in other cell types has been determined previously by decoration of the filaments with myosin subfragment-1 using whole mounts (Small et al. 1978), sections (Ishikawa et al. 1969; Begg et al. 1978), scanning electron microscopy (Isobe & Shimada, 1986; Svitkina et al. 1995) and, more recently, in sections combined with a detailed analysis of the resultant transmission electron microscopy (TEM) images (Cramer et al. 1997). We have used this latter approach here to analyse the organization and polarity of actin filaments in consecutive sections through aligned prefusion myoblasts. We further correlated this organization with the locomotion of differentiating myoblasts in aligned groups using time-lapse video microscopy.
Materials and methods
Growth and differentiation of H2kb-tsA58 myogenic cells
Conditionally immortal H2kb-tsA58 myogenic clones were isolated from the immortal mouse and grown as described previously (Morgan et al. 1994; Clark et al. 2002). Myoblasts were proliferated in growth media: DMEM supplemented with 20% fetal calf serum (FCS), 2% chick embryo extract, 100 µg mL−1 penicillin/streptomycin, and 20 units mL−1 of murine recombinant γ-interferon (IFNγ, Gibco) at 33 °C. For electron microscopy (EM), cells were seeded onto 12.7-mm-diameter Aclar coverslips (Agar scientific), precoated with 1 mg mL−1 matrigel (BD Biosciences) at a density of approximately 5 × 104 cells cm−2 and cultured for 1–2 days at 33 °C to allow aligned groups to form. The cells were then cultured under differentiating conditions (DMEM supplemented with 5% FCS, 1% chick embryo extract, 100 µg mL−1 penicillin/streptomycin at 37 °C) for 24 h at 37 °C before preparing them for EM. Under these conditions, in our experience, the maximum rate of fusion occurs between 24 and 72 h after the switch to differentiation conditions.
Preparation of cells for TEM
To label actin filaments in cells with S1, cells on Aclar coverslips were permeabilized with 0.1 mg mL−1 Saponin in MOPS-buffered Ringer's solution for 10–15 s, rinsed gently 2–3 times in MOPS-buffered Ringer's solution containing 1 µg mL−1 phalloidin (Sigma Chemical Co.) and incubated with 6 mg mL−1 myosin subfragment 1 (S1) in MOPS-buffered Ringer's solution for 15–30 min at room temperature. The S1 used was prepared by digesting purified rabbit skeletal muscle myosin II with papain in the presence of Mg2+ (Margossian et al. 1975). As a control for the effects of permeabilization on cell morphology, the same procedure was performed using a second set of cells but omitting saponin from the MOPS-buffered Ringer's solution. Following incubation with S1, the solution was aspirated from the coverslips and the cells were fixed immediately, without prior rinsing, in MOPS-buffered Ringer's solution containing 3% glutaraldehyde, 0.2% tannic acid and 1 µg mL−1 phalloidin. Cells were thoroughly rinsed in MOPS-buffered Ringer's solution to remove residual tannic acid, which precipitates upon contact with osmium tetroxide. Next, they were post-fixed on ice for 10 min in 1% osmium tetroxide (made up from a 2% stock in 10 mm MgCl2 and 100 mm KPO4, pH 6.1), then thoroughly rinsed in MOPS-buffered Ringer's solution to remove all traces of phosphate buffer, which produces crystalline artefacts upon contact with uranyl acetate in the TEM. Finally, they were block stained in 2% aqueous uranyl acetate for 15 min at 4 °C, and subsequently washed with distilled water.
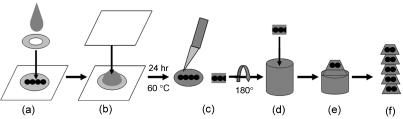
Cells were dehydrated in an ethanol gradient consisting of 5-min incubations in 35% followed by 50% ethanol at 4 °C, then 50% followed by 80, 95 and 100% ethanol at −20 °C; and finally 100% ethanol at room temperature. Embedding used propylene oxide as an intermediate solvent. Coverslips were washed in propylene oxide and infiltrated in propylene oxide/Epon Araldite (2 : 1) solution for 30 min; propylene oxide/Epon Araldite (1 : 2) solution for a further 30 min and ultimately left to infiltrate for at least 1 h in Epon Araldite (Agar Scientific). An Aclar ‘sandwich’ was made at the embedding step, which, after polymerization at 60 °C, allowed all the Aclar to be peeled away, leaving a thin disc of Araldite containing the cells (Fig. 1).
Fig. 1.
Embedding a monolayer of cells. (a) Cells, fixed and prepared for electron microscopy on their circular coverslips, were placed on squares of Aclar, and coated with a drop of Araldite. An Aclar washer was placed on top. (b) A second square of Aclar was pressed onto the upper surface of the washer to sandwich the Araldite-coated coverslip and washer. The Araldite was then polymerized at 60 °C for 24 h. (c) After polymerization the Aclar squares, washer and coverslip were peeled away and a hardened Araldite disc with the cells embedded remained. Cells were selected based on their morphology under the light microscope and the part of the disc containing them was excised using a scalpel. (d) These cells were then flipped over 180° so that their ventral surface was uppermost and mounted onto an Araldite stub using rapid cure Araldite and a specimen mounting apparatus. (e) The block was shaped and (f) 60–90-nm silver/gold sections were cut using a diamond knife and ultramicrotome.
The Araldite disc was then attached to a microscope slide and a region containing cells of the desired morphology was identified in the light microscope and excised with a scalpel. To enable exact parallel sectioning of the monolayer of cells, in a ventro-dorsal direction, the dorsal surface of the excised region was glued onto a 2-cm Araldite stub using rapid cure Araldite. This was positioned in a mounting apparatus, a modification of a previous design (Woodcock & Bell, 1967), in which spring-loaded pins forced the outer face of the Araldite stub, ventral cell surface upwards, against the flat surface of a glass slide to ensure that the excised region was mounted perpendicular to the axis of the stub. This was essential for enabling accurate parallel orientation of cells with the knife edge during sectioning and that sections containing cells were obtained in the first few sections cut.
Cells were sectioned parallel to the substrate from ventro-dorsally at 60- to 90-nm intervals using a diamond knife (Diatome, Switzerland) and ultramicrotome set at a cutting speed of 0.8 mm s−1. The microtome used could not cut sections thinner than this. All sections were retained in order on distilled water in the boat of the knife. Complete, undamaged sections were picked up in the meniscus of a slot grid in constant orientation and allowed to dry on a Pioloform-coated ‘domino rack’ (Rowley & Moran, 1975). Pioloform was selected over Formvar because of its higher mechanical and thermal stability and its lower material bulk, which results in decreased electron scatter along with insignificant intrinsic structure (Stockem, 1970).
Sections were stained on grids in 2% potassium permanganate for 10 min, washed briefly in a dilute bleaching solution (PALS bleach: three drops of 0.5% sodium sulphate and 0.5% oxalic acid, diluted in 10 mL distilled water), and then transferred onto drops of saturated aqueous uranyl acetate for 1 h. They were subsequently washed in distilled water and placed onto drops of Sato lead (Sato, 1968) (1 g lead nitrate, 1 g lead citrate, 1 g lead acetate, 2 g tri-sodium citrate dissolved in 82 mL distilled water and then mixed with 18 mL 4% NaOH freshly prepared from pellets) for 10 min before a final wash in 0.02% NaOH. Care was taken to avoid tearing the pioloform film over the slots of the grids by drying and storing them vertically in a rack.
TEM sections were observed using a JEOL 1200EX operated at 80 kV and a magnification of 4000× to observe whole cell morphology. To identify filament polarity, images of selected areas of interest were photographed at a nominal magnification of 12 000 or 15 000×. EM negatives were scanned at 20-µm pixel size and calibrated using Fourier transforms of the 14.4-nm paramyosin repeat. Digitized images were viewed in Adobe Photoshop 7.0 (Adobe Systems Inc., Mountain View, CA, USA).
EM quantitation
The orientation and distribution of actin filaments was determined from electron micrographs. Aligned cells were selected that appeared to have normal morphology compared with non-permeabilized cells. The S1-decorated filaments could be clearly identified, and they showed the distinctive arrowhead appearance in the electron micrographs. Detailed analysis of actin filament polarity was performed on a set of three aligned cells, although brief analysis of other aligned cell groups showed similar results. Actin filaments were either found in a zone close to the plasma membrane (subplasmalemma), scattered deeper in the cytoplasm or in protrusions. Filaments in all of these regions were analysed. The width of the subplasmalemma actin network was measured using an eye-piece graticule.
To classify the orientation of filaments in the subplasmalemmal zone, filaments that were orientated at an angle of ±30° parallel to the plasma membrane were classified as pointing ‘towards’ or ‘away from’ one end of the cell, depending on their arrowhead orientation. Those filaments falling outside of this range were termed ‘other’. A similar classification strategy was employed for cytoplasmic filaments. For protrusions, filament orientation was classified as either pointing towards or away from the tips of the protrusions.
Filament polarity was measured from ventral sections (three sections from the ventral 0.75 µm of the cell), middle sections (three sections from the middle 0.75 µm of the cell) and dorsal sections (three sections from the final 0.75 µm of the cell) through each of these cells. Measurements made ‘blind’ (without knowing from which cell or section they came, without knowing the orientation of the image relative to the cell and without knowing the orientation of the cell) produced similar results to measurements made using a sighted method.
The polarity of actin filaments in the subplasmalemma zone was analysed in two ways for each of the three cells in the aligned group. First, the filament polarity was scored within a 1-µm × 1-µm area every 5 µm over the entire length of the cell, from the arrowhead decoration pattern. Decorated filaments were scored in selected areas in each section of each cell from 12 000× or 15 000× images viewed through a 7× anastigmatic lupe on a light box. Approximately 50% of filaments were unscorable in the dense regions of actin filaments owing to the super positioning of actin filaments, or the close proximity of filaments to one another, which made arrowhead complexes difficult to distinguish. This is mainly due to the section thickness used, with in turn was a limitation of the microtome available. Second, actin filaments in a 5-µm region at the middle of the cell were scored at shorter (0.1 µm) intervals by a transect running at right angles to the plasmalemma.
Time-lapse video microscopy
Cells were plated into 25-cm2 plastic culture flasks (Iwaki, Japan) at a density of 2 × 104 cells cm−2, in growth medium, and allowed to settle for at least 4 h prior to filming at 37 °C. The flask was sealed, and placed on a Nikon Eclipse TE300 inverted microscope contained in an incubator (Solent Scientific) at 37 °C. An area of cells was selected and phase contrast images were captured at 5-min intervals for 48 h using Kinetic Acquisition Manager Advance 6 (Kinetic Imaging, Medical Solutions plc). Movement of individual cells within aligned groups of cells was tracked using Kinetic Imaging Tracking software (Medical Solutions plc) by determining the x and y coordinates of the nucleus of each cell in each frame. Cell movements were assessed and statistically evaluated as described previously (Clark et al. 2002).
Results
Morphology of aligned myoblasts
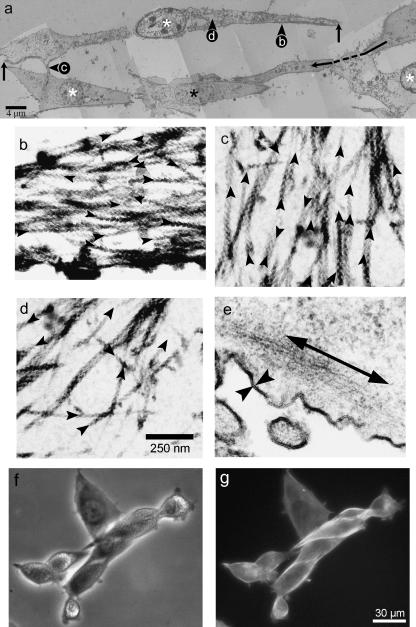
Preservation of the overall morphology of fixed, saponin-treated cells was good as compared with control (non-permeabilized) cells in electron micrographs (Fig. 2). Furthermore, we were able to find aligned groups of cells in EM sections that looked similar to those that had been fixed and stained for the light microscope as we reported previously (Wells et al. 1997), and as shown here (Fig. 2f,g). The cells in these groups have a typical elongated morphology that is easily identifiable. Electron micrographs showed that the aligned cells were elongated, between 45 µm and 60 µm long, only slightly wider than the width of the nucleus, and 2.7–4.0 µm thick (calculated from the number of TEM sections between the first ventral and last dorsal section, which was 45 on average). Elongated aligned cells possessed protrusions that were often in contact with the cell membranes or the protrusions of surrounding cells (Fig. 2a). Owing to the mild action of the low-concentration saponin used as a permeabilization agent, some cells within groups were not permeabilized (Fig. 2a, black asterisks), and their morphology could be directly compared with permeabilized cells.
Fig. 2.
Electron micrographs of aligned myoblasts, permeabilized with saponin and decorated with S1. (a) A low-power montage of electron micrographs depicting a ventral section through several saponin permeabilized cells. (b–d) Enlargements of the corresponding areas within the montage; (e) from a control cell not shown (scale bar 250 nm). In (a) long vertical arrows highlight the regions between which the subplasmalemma filaments were measured. Long dashed arrow links two parts of a single cell separated by a protrusive structure extending from another cell, highlighting the ‘bridging’ interactions of neighbouring cells. The montage shows that some cells were mildly permeabilized (white asterisks), whilst others were not permeabilized (black asterisk) and acted as controls for morphology (scale bar 4 µm). Panel (b) shows an area of S1-labelled subplasmalemma filaments. The plasma membrane is located at the bottom of the image. Panel (c) shows S1-labelled actin filaments within the protrusive end of a cell. Panel (d) shows S1-labelled actin filaments within the cytoplasm. Panel (e) shows the subplasmalemma region of a non-permeabilized control cell. Added arrowheads in (b), (c) and (d) emphasize the orientatation of S1-labelled actin filaments. In (e) opposing arrowheads indicate the intact plasma membrane, and the long arrow highlights the location of undecorated subplasmalemma actin filaments. Panel (f) shows a phase contrast image taken of myoblasts at the same stage of development as that for the EM above, fixed using 4% paraformaldehyde and viewed in the light microscope, and (g) shows the fluorescent image of the same cells stained for filamentous actin with rhodamine phalloidin.
Organization of actin filaments in aligned myoblasts
Three classes of actin filament were identified in cells: subplasmalemma filaments, filaments within protrusions and cytoplasmic filaments (Fig. 2b–d). Out of a total of 8229 filaments counted, 69% (5718) of the actin filaments were organized into a well-defined zone lying beneath the plasma membrane (subplasmalemma filaments; Fig. 2b), 14% (1157) were found in protrusions (Fig. 2c) and the remainder (1354) were found in the rest of the cytoplasm (Fig. 2d): subplasmalemma filaments appeared in every section observed (with the exception of the most ventral and most dorsal sections) and thus formed a continuous sheet of longitudinally orientated filaments inside the walls of the cell, rather than bundles. The thickness of the sheet was similar for dorsal (0.32 ± 0.05 µm), middle (0.40 ± 0.07 µm) and ventral (0.39 ± 0.05 µm) sections (all values are given as mean ± SD, where n (the number of measurements taken) = 54). The majority of filaments were orientated within ±30° of a parallel dictated by the plasma membrane; we observed no instances of filaments in this region orientated at right angles to the plasma membrane. In protrusions, there was some variation in length, width and thickness and at least 30 filaments were counted in any one section through them. The remaining 38% of filaments counted were found in the cytoplasm (Fig. 2d). These filaments were not organized into longitudinal or transverse bundles.
Polarity of subplasmalemma actin filaments
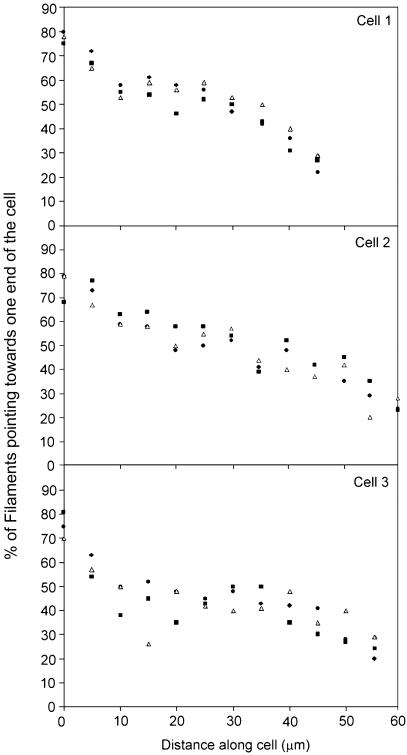
Decorated actin filaments showed the expected strong arrowhead polarity. We were only able to determine the polarity unambiguously for about 50% of the observed filaments due to their overlap with others, where actin filament density was high (Fig. 2b). In all three cells examined in detail, and in all of the sections (ventral, middle and dorsal) the average polarity of the actin filaments gradually changed from one end of the cell to the other, such that at each end of the cell the majority of actin filaments had their barbed ends pointing towards the cell apex (Fig. 3). In the middle of each cell, approximately 50% of filaments are orientated with the barbed ends towards one end of the cell, and the remaining 50% are orientated with their barbed ends pointing towards the other end of the cell. Thus the filaments in the subplasmalemma sheet display the graded polarity detected previously in filament bundles in fibroblasts (Cramer et al. 1997).
Fig. 3.
Graphs showing the graded polarity of actin filaments in the subplasmalemma zone for the ventral sections of three cells within an aligned group. The three symbols represent data taken from each of the three ventral sections. In these sections the subplasmalemma sheet runs parallel to the plasma membrane and spans the entire length of the cell.
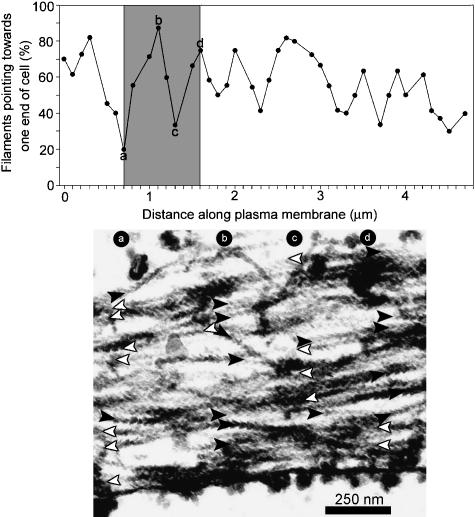
Subplasmalemma actin filaments have been reported to have an alternating polarity in chick heart fibroblasts (Cramer et al. 1997). In that report, distinct changes in polarity could be observed by eye, and the periodicity of this switch in polarity was 0.8 µm on average. However, this alternating polarity was not obvious in our sections. Therefore, we sampled the polarity of the filaments at regular, short (0.1 µm) intervals within a randomly selected 5-µm stretch in the central third of the cell, for all three cells. The results were the same for ventral, middle and dorsal sections, and the data for a ventral section are shown here (Fig. 4).
Fig. 4.
Short-range random polarity of subplasmalemma actin filaments in a ventral section of an aligned myoblast. Transects were drawn at 90° to the membrane every 0.1 µm over a 5-µm stretch at the central region of the cell. Polarity was scored where filaments intersected these lines. To avoid bias, transects were scored in a random sequence. Points a–d are shown in the micrograph. The total number of filaments scored = 406. Although the filament polarity appears to shift from about 80% that point towards one end of cell (left-hand side of micrograph) to 20% in an alternating fashion, in fact these observations can be explained by a random organization of filaments with mixed polarity. The polarity of the filaments is indicated by the filled and empty arrows; filled arrows represent filaments pointing towards one end of the cell, and the empty arrows represent filaments pointing towards the other end.
The polarity of the subplasmalemma actin filaments fluctuated, appearing to show abrupt changes in filament polarity as expected if the polarity was alternating. However, by a statistical analysis of these data, we find that they conform to a random organization of filaments with opposite polarities. We showed this by scoring whether the percentage polarity value rose or fell between successive transects, and then analysing the pattern of these +/– scores taken in pairs. For a random arrangement, ++, + –, – + and – – should be equally frequent, whereas ++ and – – will predominate when an alternating polarity is present and the sampling interval is shorter than the filament length, as here. Chi-squared testing of the null hypothesis of a random arrangement showed it was a good description of the data. For example, for one of the ventral sections for one of the cells (data shown in Fig. 4), the test gave 0.3 > P > 0.2, which means we can accept the null hypothesis. Similar results were obtained for the remaining sections. Therefore, there is no evidence for a more complex, alternating arrangement.
In non-permeabilized cells, a zone of undecorated actin filaments can be observed just beneath the plasma membrane, running parallel to it (long arrow, Fig. 2e). The similarity of the overall organization of the actin cytoskeleton in permeabilized and non-permeabilized cells suggests that the underlying actin cytoskeleton is not significantly perturbed by permeabilization and S1 decoration.
Actin polarity within protrusions
Actin filaments in protrusions were orientated with the majority of filaments running in arrays parallel with the long axis of the protrusion (Small, 1988; Lewis & Bridgman, 1992; Small et al. 2002). In aligned myoblasts, 73% of all filaments classified within protrusions (n = 1157) were orientated with their barbed ends facing the tip of the protrusion. This preferred orientatation was found at all levels within a protrusion, in all protrusions observed, including each end of cells that were also classified as protrusions. This arrangement of filament polarity was maintained in these regions irrespective of whether they were in direct interaction with other cells, or where two protrusive structures extending from different cells were in contact, or where protrusions were not involved in any cell contact. The filaments within any given protrusion were observed to terminate in direct contact with the plasmalemma in the arrangement described.
Actin polarity within the cytoplasm
Examination of the polarity of the actin filaments in the cytoplasm in aligned cells in areas remote from or close to the nucleus showed that the orientation of filaments was random (Fig. 2d), as demonstrated by an increase in the percentage of filaments falling into the ‘other’ category compared with the subplasmalemma zone. In aligned myoblasts, 38% of all filaments scored in the cytoplasm (n = 1335) pointed away from the nucleus, 37% pointed towards the nucleus and 25% were classified as ‘other’. Comparison of filaments in front of and behind the nucleus showed no difference in their organization. There were no significant differences between ventral, middle and dorsal regions of the cells. Cytoplasmic filaments sometimes appeared short, possibly because they left the plane of the section.
Movement of aligned myoblasts
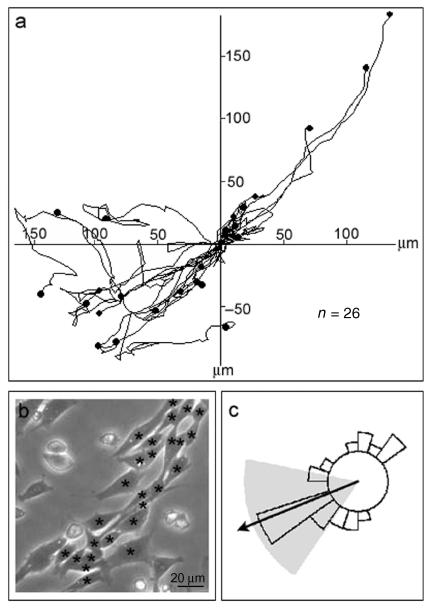
The actin filament organization observed above in aligned myoblasts following 24 h under differentiation conditions suggested to us that myoblast movement should be restricted linearly. Therefore, by time-lapse phase microscopy, we analysed myoblast locomotion under the same culture conditions that we had used prior to fixing the cells for EM. Analysis of the locomotion of myoblasts in aligned groups showed that their direction of migration is restricted to the long axis of the linear aggregates (Fig. 5). Cells tracked over an 80-min period showed that the direction of cell movement was restricted to between 281.6° and 213.8° (95% confidence interval; P < 0.006). The long axis of the aligned group (approximately 225°, Fig. 5) is found within this range, as shown by the arrow in the circular histogram (Fig. 5c). In contrast, cells not in aligned groups displayed a ‘random walk’ (data not shown) and showed no significant clustering of directions, as reported previously (Clark et al. 2002). Furthermore, when cells are recruited to the aligned group, the direction of their migration changes from ‘random walk’ and becomes restricted in the direction of the long axis of the aggregate. During this movement, cells are able to slide past each other as well as move over or beneath neighbouring cells. Cells in the process of this type of movement appear to have been captured in TEM sections (Fig. 2a, partially dotted arrow). The average velocity of locomotion of cells tracked was 1.2 ± 0.4 µm per min (mean ± SD, n = 26). This is similar to the value measured for isolated myoblasts reported previously (Peckham et al. 2001).
Fig. 5.
Analysis of cell locomotion within aligned groups. The cells contributing to the aligned group shown in (b) and highlighted with black asterisks (n = 26) were quantified and analysed as described in the Materials and methods section. In each case, the cell tracks are shifted to a common point of origin as shown in the trajectory diagram (a) and a horizon was chosen as the largest distance reached by 95% of the cells. The direction in which each cell first crossed the horizon is shown in the circular histogram (c). There is a significant preference for one direction (shown by the arrow) and the 95% confidence interval of the mean is shown by the shaded sector. The long axis of the aligned group of cells runs in this sector.
Discussion
We have found that there are three distinct types of actin filament organization in aligned prefusion myoblasts. Most of the actin filaments run parallel to the plasmalemma in a zone close to the plasmalemma and in protrusions. The polarity of subplasmalemma filaments gradually changes from one end of the cell to the other, such that at each end of the cell, the barbed ends of these filaments face the plasmalemma. In the central region of the cell, where approximately 50% of the filaments point towards one end of the cell, and 50% point to the other, the polarity of the filaments is random. The polarity of actin filaments in protrusions is such that most of these filaments are orientated with their barbed ends pointing towards the tips of the protrusions. The remainder of the actin filaments are found in the cytoplasm, where they are randomly arranged and not bundled. Time-lapse microscopy showed that the aligned cells are still actively moving, and that their movement is restricted in the direction of the long axis of the aligned group.
The overall organization of actin filaments in aligned myoblasts is similar to that which we found previously using immunofluoresence (Wells et al. 1997). In that study, we found that most of the actin filaments were located close to the plasmalemma, and were co-localized with a number of myosins (myosin Iα, non-muscle myosin IIA and non-muscle myosin IIB) in aligned cells. The majority of aligned cells at this stage of differentiation have not yet begun to express skeletal actin and myosin isoforms, and are not post-mitotic. The numbers of cells still dividing at the end of the first 24 h under differentiation conditions is roughly 30% (Morgan et al. 1994). We have also shown that very few of the cells in cultures at this stage have begun to express skeletal myosin (Wells et al. 1997). Therefore, we expect that the actin at the plasmalemma also mostly consists of non-skeletal actin isoforms, and does not contain skeletal actin that is starting to form early muscle sarcomeres at the start of myofibrillogenesis.
The organization of actin filaments in protrusions in aligned myoblasts is similar to that found previously for lamellipodia and filopodia (Edds, 1977; Hoglund et al. 1980; Karlsson et al. 1984; Small, 1988; Rinnerthaler et al. 1991; Small et al. 1995; Cramer et al. 1997). The protrusions in aligned myoblasts contain unipolar bundles of actin filaments in which the barbed ends face outwards towards the tip. The protrusions in aligning myoblasts that make contact with neighbouring cells in the aggregate are likely to be involved in cell recognition and alignment, and may help to trigger changes in the actin cytoskeleton described here when cells make contact and form aligned groups.
The organization and polarity of the actin filaments in the cell body of aligned myoblasts shows some differences to that reported for fibroblasts and PtK2 cells (Cramer et al. 1997). Aligned myoblasts do not contain ventral actin filament bundles with a graded polarity or short overlapping actin filament bundles with alternating polarity on the inner surface of the plasma membrane as reported for fibroblasts (Cramer et al. 1997). However, they do contain a continuous sheet of actin filaments that lies just beneath the plasmalemma in which the polarity changes gradually from one end of the cell to the other. Although our data on polarity initially suggested an alternating arrangement for the subplasmalemma actin filaments in the central region of the cell, a fine-grained analysis indicated that this was merely the fluctuation expected from a random arrangement. Thus the peaks and troughs seen when sampling at 1-µm intervals were filled with further peaks and troughs when the sample interval was reduced to 0.1-µm intervals.
Furthermore, we did not find any stress fibres in aligned myoblasts, as found in PtK2 cells (Cramer et al. 1997). Stress fibres, which are different to graded polarity bundles, display an alternating polarity, contain much shorter filaments (Cramer et al. 1997) and are usually associated with focal adhesions. The lack of stress fibres in aligned myoblasts could account for their weakened attachment to the substrate, which may be linked to the known changes in integrin expression during early muscle differentiation (Steffensen et al. 1992; Blaschuk et al. 1997). In agreement with this, we have also found that focal adhesions are restricted to the ends of aligned cells (M.P., unpublished observation).
The actin filament organization observed in aligned myoblasts is consistent with the idea that their locomotion is driven by actin polymerization in protrusions and by myosin force generation in graded polarity actin filaments that are aligned in the direction of movement. Cell locomotion is thought to be partly driven by the polymerization of actin filaments at the barbed ends to form new protrusions (Mitchison & Cramer, 1996). In aligned myoblasts, new protrusions can form at each end of the cell, as the actin filaments here are orientated with their barbed ends facing outwards. Graded polarity actin bundles, found in the ventral region of motile chick heart fibroblasts, have been suggested to play a role in the generation of motile force for cell body motility (Cramer et al. 1997). Despite the different spatial organization, the graded polarity actin subplasmalemma sheet could act in a similar way to generate cell body motility in aligned myoblasts. Our earlier observations showed that non-muscle myosin II is concentrated in this region, as expected (Wells et al. 1997).
Furthermore, the organization of subplasmalemma actin filaments in aligned myoblasts suggests that it is highly unlikely that they will form lateral protrusions and move in a direction other than along the long axis of the aligned group. In the central region of the cell, all the filaments run parallel to the plasma membrane, suggesting that it is unlikely that they will be able to form new protrusions laterally. In agreement with this, we found that locomotion of aligned myoblasts tended to be restricted to the direction of the long axis of the aligned group.
As well as contributing to cell locomotion, the subplasmalemma actin filaments are also likely to contribute to cortical tension in these cells. Although in the central region of the cell, the actin filaments have a mixed random rather than an alternating polarity, this organization would still be expected to enable interaction with non-muscle myosin II to produce a contractile force, and therefore cortical tension. This cortical tension would help to generate the elongated shape of these cells. However, it is worth pointing out that this tension could also generate a pressure gradient, which would tend to force fluid flow towards weaker regions of the cell cortex to enable the formation of protrusions (Cunningham, 1995; Stossel et al. 1999) and contribute towards cell locomotion.
This leaves the question of the function of the cytoplasmic actin filaments. They are not organized into graded actin polarity bundles, or stress fibre-like bundles, and so are unlikely to be involved in cell locomotion or adhesion. They are not close to the cell membrane, and so are unlikely to contribute to cortical tension. Their polymerization is unlikely to drive protrusions. Moreover, the density of these filaments is similar in dorsal, ventral and middle sections. One possibility is that their main function is in intracellular movements, i.e. vesicle trafficking, and/or in the re-arrangement of structures such as the Golgi and the endoplasmic reticulum that are known to occur in cells as they fuse (Lu et al. 2001).
In conclusion, our examination of the organization and polarity of actin filaments in aligned myoblasts suggests that these cells do not strongly attach to the substratum, as they have very few if any stress fibres; have a direction of movement that is likely to be limited to either end of the cell, due to the orientation of actin filaments within these regions; have centrally located subplasmalemma actin filaments orientated in a manner that does not favour lateral movements but is likely, with the aid of co-localized myosins, to be involved in the production of membrane tension that would help to generate the bipolar shapes of these cells, and contribute to their locomotion.
Acknowledgments
We thank the Anatomical Society of Great Britain and Northern Ireland for a studentship to N.S. We are also grateful to Mary Reedy, Louise Cramer and Margaret Siebert for their advice on methods used, Adrian Hick for assistance with microtomy and special thanks to Ian Swailes for construction and modification of the specimen mounting apparatus.
References
- Begg DA, Rodewald R, Rebhun LI. The visualization of actin filament polarity in thin sections. Evidence for the uniform polarity of membrane-associated filaments. J. Cell Biol. 1978;79:846–852. doi: 10.1083/jcb.79.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim L, Bader CR. Human myoblast differentiation: Ca(2+) channels are activated by K(+) channels. News Physiol. Sci. 2002;17:22–26. [PubMed] [Google Scholar]
- Blaschuk KL, Guerin C, Holland PC. Myoblast alpha v beta3 integrin levels are controlled by transcriptional regulation of expression of the beta3 subunit and down-regulation of beta3 subunit expression is required for skeletal muscle cell differentiation. Dev. Biol. 1997;184:266–277. doi: 10.1006/dbio.1997.8527. [DOI] [PubMed] [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Clark P, Coles D, Peckham M. Preferential adhesion to and survival on patterned laminin organizes myogenesis in vitro. Exp. Cell. Res. 1997;230:275–283. doi: 10.1006/excr.1996.3429. [DOI] [PubMed] [Google Scholar]
- Clark P, Dunn GA, Knibbs A, Peckham M. Alignment of myoblasts on ultrafine gratings inhibits fusion in vitro. Int. J. Biochem. Cell Biol. 2002;34:816–825. doi: 10.1016/s1357-2725(01)00180-7. [DOI] [PubMed] [Google Scholar]
- Cramer LP, Siebert M, Mitchison TJ. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 1997;136:1287–1305. doi: 10.1083/jcb.136.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC. Actin polymerization and intracellular solvent flow in cell surface blebbing. J. Cell Biol. 1995;129:1589–1599. doi: 10.1083/jcb.129.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edds KT. Dynamic aspects of filopodial formation by reorganization of microfilaments. J. Cell Biol. 1977;73:479–491. doi: 10.1083/jcb.73.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund AS, Karlsson R, Arro E, Fredriksson BA, Lindberg U. Visualization of the peripheral weave of microfilaments in glia cells. J. Muscle Res. Cell Motil. 1980;1:127–146. doi: 10.1007/BF00711795. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Bischoff R, Holtzer H. Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J. Cell Biol. 1969;43:312–328. [PMC free article] [PubMed] [Google Scholar]
- Isobe Y, Shimada Y. Organization of filaments underneath the plasma membrane of developing chicken skeletal muscle cells in vitro revealed by the freeze-dry and rotary replica method. Cell Tissue Res. 1986;244:47–56. doi: 10.1007/BF00218380. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Lassing I, Hoglund AS, Lindberg U. The organization of microfilaments in spreading platelets: a comparison with fibroblasts and glial cells. J. Cell Physiol. 1984;121:96–113. doi: 10.1002/jcp.1041210113. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Joseph D, Bugnard E, Zaal KJ, Ralston E. Golgi complex reorganization during muscle differentiation: visualization in living cells and mechanism. Mol. Biol. Cell. 2001;12:795–808. doi: 10.1091/mbc.12.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margossian SS, Lowey S, Barshop B. Effect of DTNB light chain on the interaction of vertebrate skeletal myosin with actin. Nature. 1975;258:163–166. doi: 10.1038/258163a0. [DOI] [PubMed] [Google Scholar]
- Miller JB. Myoblast diversity in skeletal myogenesis: how much and to what end? Cell. 1992;69:1–3. doi: 10.1016/0092-8674(92)90111-o. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Morgan JE, Beauchamp JR, Pagel CN, et al. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: a model system for the derivation of tissue-specific and mutation-specific cell lines. Dev. Biol. 1994;162:486–498. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- Musa H, Orton C, Morrison EE, Peckham M. Microtubule assembly in cultured myoblasts and myotubes following nocodazole induced microtubule depolymerisation. J. Muscle Res. Cell Motil. 2003;24:301–308. [PMC free article] [PubMed] [Google Scholar]
- Peckham M, Miller G, Wells C, Zicha D, Dunn GA. Specific changes to the mechanism of cell locomotion induced by overexpression of beta-actin. J. Cell Sci. 2001;114:1367–1377. doi: 10.1242/jcs.114.7.1367. [DOI] [PubMed] [Google Scholar]
- Rinnerthaler G, Herzog M, Klappacher M, Kunka H, Small JV. Leading edge movement and ultrastructure in mouse macrophages. J. Struct. Biol. 1991;106:1–16. doi: 10.1016/1047-8477(91)90058-5. [DOI] [PubMed] [Google Scholar]
- Rowley JC, 3rd, Moran DT. A simple procedure for mounting wrinkle-free sections on formvar-coated slot grids. Ultramicroscopy. 1975;1:151–155. doi: 10.1016/s0304-3991(75)80018-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Sato T. A modified method for lead staining of thin sections. J. Electron Microsc. (Tokyo) 1968;17:158–159. [PubMed] [Google Scholar]
- Shainberg A, Yagil G, Yaffe D. Control of myogenesis in vitro by Ca2+ concentration in nutritional medium. Exp. Cell Res. 1969;58:163–167. doi: 10.1016/0014-4827(69)90127-x. [DOI] [PubMed] [Google Scholar]
- Small JV, Isenberg G, Celis JE. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. doi: 10.1038/272638a0. [DOI] [PubMed] [Google Scholar]
- Small JV. The actin cytoskeleton. Electron Microsc. Rev. 1988;1:155–174. doi: 10.1016/s0892-0354(98)90010-7. [DOI] [PubMed] [Google Scholar]
- Small JV, Herzog M, Anderson K. Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- Steffensen B, Magnuson VL, Potempa CL, Chen D, Klebe RJ. Alpha 5 integrin subunit expression changes during myogenesis. Biochim. Biophys. Acta. 1992;1137:95–100. doi: 10.1016/0167-4889(92)90105-k. [DOI] [PubMed] [Google Scholar]
- Stockem W. [Utilization of pioloform F in producing electron microscopic carrier films] Mikroskopie. 1970;26:185–189. [PubMed] [Google Scholar]
- Stossel TP, Hartwig JH, Janmey PA, Kwiatkowski DJ. Cell crawling two decades after Abercrombie. Biochem. Soc. Symp. 1999;65:267–280. [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J. Struct. Biol. 1995;115:290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MV. Muscle development: molecules of myoblast fusion. Curr. Biol. 2000;10:R646–R648. doi: 10.1016/s0960-9822(00)00664-3. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ. The fusion of myoblasts. Biochem. J. 1985;228:1–12. doi: 10.1042/bj2280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C, Coles D, Entwistle A, Peckham M. Myogenic cells express multiple myosin isoforms. J. Muscle Res. Cell Motil. 1997;18:501–515. doi: 10.1023/a:1018607100730. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Bell PR. A method for mounting 4mu resin sections routinely for ultra-thin sectioning. J. R. Microsc. Soc. 1967;87:485–487. doi: 10.1111/j.1365-2818.1967.tb04527.x. [DOI] [PubMed] [Google Scholar]
- Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]