Abstract
Computational resources are now using the tissue names of the major model organisms so that tissue-associated data can be archived in and retrieved from databases on the basis of developing and adult anatomy. For this to be done, the set of tissues in that organism (its anatome) has to be organized in a way that is computer-comprehensible. Indeed, such formalization is a necessary part of what is becoming known as systems biology, in which explanations of high-level biological phenomena are not only sought in terms of lower-level events, but are articulated within a computational framework. Lists of tissue names alone, however, turn out to be inadequate for this formalization because tissue organization is essentially hierarchical and thus cannot easily be put into tables, the natural format of relational databases. The solution now adopted is to organize the anatomy of each organism as a hierarchy of tissue names and linking relationships (e.g. the tibia is PART OF the leg, the tibia IS-A bone) within what are known as ontologies. In these, a unique ID is assigned to each tissue and this can be used within, for example, gene-expression databases to link data to tissue organization, and also used to query other data sources (interoperability), while inferences about the anatomy can be made within the ontology on the basis of the relationships. There are now about 15 such anatomical ontologies, many of which are linked to organism databases; these ontologies are now publicly available at the Open Biological Ontologies website (http://obo.sourceforge.net) from where they can be freely downloaded and viewed using standard tools. This review considers how anatomy is formalized within ontologies, together with the problems that have had to be solved for this to be done. It is suggested that the appropriate term for the analysis, computer formulation and use of the anatome is anatomics.
Keywords: anatomics, C. elegans anatomy, databases, Drosophila anatomy, gene expression database, human anatomy, mereology, mouse anatomy, ontology, systems biology
Introduction
Over the past few years, considerable effort has been devoted to articulating the anatomy of humans and other organizms in a way that is computer-comprehensible and so can mesh with other computational resources such as gene-expression databases. Such formalized anatomies are available for a range of model organisms (Table 1) that include the mouse, Drosophila,* Caenorhabditis elegans* and the zebrafish* as well as various plants, and these are linked to the core databases for these organisms (in this review, an asterisk indicates a website that is listed in Table 2). There are also versions of human adult and developmental anatomies although there is, as yet, no central resource for human data. In addition, there is a hierarchy for subcellular location within the Gene Ontology or GO* that is used for linking genetic data with knowledge about gene products (Harris et al. 2004, and see below).
Table 1.
Anatomy ontologies and associated resources. All are available athttp://obo.sourceforge.net
| Animal anatomy ontologies |
| Caenorhabditis gross anatomy |
| Drosophila gross anatomy |
| Human anatomy and development |
| Human developmental anatomy, abstract version |
| Human developmental anatomy, timed version |
| Medaka fish anatomy and development |
| Mouse adult anatomy |
| Mouse developmental anatomy |
| Zebrafish anatomy |
| Plant, fungal, etc., anatomy ontologies |
| Arabidopsis gross anatomy |
| Dictyostelium anatomy |
| Cereal plant gross anatomy |
| Fungal gross anatomy |
| Maize gross anatomy |
| Cell anatomy-related ontologies |
| Gene ontology cell component |
| Cell type |
Table 2.
Websites (marked * in text)
The purpose of this article is to consider from an anatomical viewpoint what is involved in articulating our knowledge of tissue organization for human and other species so that it can be made computer comprehensible. [For the purposes of this article, a tissue is defined more broadly than is usual: it is an identifiable anatomical component that may or may not be named and may or may not have subsidiary tissues – it is to be equated with a concept or term in an ontology (see later).] This exercise turns out to be very different from writing normal text descriptions of anatomy, and for two obvious reasons. First, computer-readable and human-readable syntaxes are distinct, as anyone who has looked at a computer program will know. Second, although the reader always brings considerable knowledge to any text, the ignorance of a computer is deep, wide and complete. The exercise of producing a computer-comprehensible anatomy thus requires thinking anatomy through from first principles, embracing the strengths of computer speak (e.g. logic and rigour), while accepting its weaknesses (e.g. no sense of subtlety or semantics). Thus, while some aspects of anatomy lend themselves to formalization, others are just too hard to handle within a text representation and can only be expressed visually, if at all. An example of the former might be the parts of the mouse heart and one of the latter might be the natural variation in the course (and hence the neighbours) of a major nerve in humans: it is easy to discuss exceptions and variation using normal prose, but remains impossible to do in any way that is useful computationally. A similar, as yet unsolved, problem now under active investigation is how to formalize mutant anatomy in a way that is appropriate for all organisms (Bard & Rhee, 2004; Gkoutos et al. 2004).
This paper starts by discussing how anatomy can be articulated in ways that can be used computationally and moves on to consider their formal representations and the tools that are used for this. There are then summaries of the core vertebrate anatomy and related ontologies and the uses to which they are currently being put. At the end, some broader implications are considered.
Definitions
It is both convenient and sensible to start by defining two new terms that summarize the approach taken here for formulating anatomies for computational biology. They have been chosen to mesh with the equivalent terms for handling basic biological building blocks such as genes and proteins.
The anatome. This word is used to represent the complete set of tissues and organs associated with an organism. As with the genome, proteome and metabolome of an organism, the meaning in a computational context is clear in principle, but hard to be precise about in practice. Like the proteome (the set of proteins expressed by an organism), the actual set of tissues and organs present in an organism will depend on its developmental stage, but, unlike any of the other ‘-omes’, it is hard to be precise about the actual number of components in an anatome. The cardiovascular component of the anatome, for example, holds the heart and the sets of arteries, veins and capillaries and the anatome should thus include each artery, vein and capillary, whether named or not. This is clearly impossible, partly because it is hard to include tissues that are not named and partly because complex organisms have some developmental flexibility; in addition, the fine detail of the anatome is unique for each individual. If we allow for individual variation due to single nucleotide polymorphism (SNPs), the same is of course true for the proteome and genome.
The intrinsic complexity of the anatome means that, as it is difficult to incorporate variation in a standard anatomy, we have to accept that the standard anatome for an organism represents an idealization in terms of the choices of minor tissues that are included. This choice is relatively straightforward for an adult organism in that one can, in practice, only include named tissues, but can be more difficult when one considers the individual stages in developing organisms: there is, for example, variation in the development of mouse embryos in the same litter while a tissue may, say, only become apparent towards the end of a developmental stage. Furthermore, the practical requirements imposed on the anatome mean that only a subset may be needed for a particular context. For handling microarray data, for example, the anatomical resolution required will be limited by the level of clean dissection that can be achieved and this may be relatively low (Parkinson et al. 2004). If, however, one wishes to assign gene-expression data as gleaned from sectioned material, then a much higher level of detail will be required (e.g.Ringwald et al. 2001). In practice, the subset of the full anatome required for a particular purpose will be determined by that purpose and, for any organism with a rich and variable anatomy (e.g. the mouse), the full anatome will be an idealization containing all of the major (named) and as many of the minor (un-named) tissues (e.g. the undifferentiated mesenchyme associated with the developing mouse hindgut) as is practical.
Anatomics. This is the formalization of our knowledge of the anatome in ways that are readily comprehensible to computer-based resources. Here, one should note that anatomics is different from proteomics and genomics because, while the basics of our knowledge of these two domains can be described by flat, annotated tables and hence included in standard relational databases (e.g. Uni-Prot* and GenBank*), a simple list of tissues, even when annotated, does not reflect even the basics of our knowledge of anatomy. While there are far fewer tissues in an organism than there are genes, the relationships between the tissues are most naturally seen as spatial, developmental, functional and partitive, and hence unlike those required for listing genes or proteins (but not, of course, for discussing their functional relationships), and the task of anatomics is to produce a hierarchical formalism that incorporates both the tissues and the complexity of their relationships with one another.
Before discussing how these formal hierarchies are made, it is necessary to consider which anatomical relationships they need to incorporate, and obvious examples include part of, develops from, is-a[is a member of the class of], connects with, and next to. One might also wish to represent functional relationships such as stimulates (e.g. neuronaly or by signalling), responds to, and represses, as well as histological relationship: such as includes cell type. It soon becomes apparent that producing a complete formalism for the whole anatomy of an organism is not dissimilar to writing a major textbook; in practice, it is harder because a textbook can detail what is known and either mention or ignore what is not known without misleading the reader. Formalisms are different as they have to be explicit about the knowledge that they contain and omissions can lead to errors of inference. These difficulties are discussed in detail below. At this stage, it is enough to point out that the essential formalism for handling the anatome is based on ontologies rather than standard relational databases.
Ontologies
The basics
It is not easy to represent knowledge hierarchies in relational databases for two key reasons. First, although hierarchies can in principle be included in relational databases, searching such databases is not trivial, as the answer to a query often requires sequential searches within the data itself. A query of an anatomical ontology might be: detail each muscle that is part of the forelimb, and doing this requires knowing that the forelimb has distinct parts, each of which has muscles and, hence performing a sequence of searches within a single table to answer the query. This process is known as recursive searching and is not only technically complex, but can be hard to stop. Second, it is difficult to represent complex knowledge relationships within database tables, particularly as information associated with particular rules can be carried either up or down the hierarchies (see later). The alternative approach is to represent knowledge as sets of structured links; these are known as ontologies, an area of informatics that has received considerable interest in philosophical, linguistic and informatics contexts.
There are several definitions of an ontology, but all have at their core that it is an area of knowledge expressed in a way that is formal and unambiguous, and hence can be used computationally. In practice, it is adequate to see an ontology as a set of connected facts, where a fact is defined as a triad of two terms (concepts, nodes and leaves are equivalent words) linked by a relationship (or edge in mathematical formalism). Examples are ‘the cardiovascular system is a member of the class of organ systems’ (the is-a relationship, this should not be confused with instance of– see below) or ‘the atrium is part of the heart’, although, as we will see, the part of relationship is far more complicated than it appears at first sight. Such sets of facts can of course be set out pictorially and the result is a hierarchical tree (in the simplest case) or a graph in general (this usage of graph is of course different from that for a plot of one variable against another). It should also be noted that this triadic data structure is the standard formalism used by the semantic web* for handling knowledge (Hendler et al. 2002).
The most interesting aspect of these graphs is their relationships and there are two associated properties that need to be mentioned, directivity and propagation. A directed relationship only goes one way and denies its converse: an obvious example is part of: the fact that the tibia is part of the leg implies that the leg isnot part of the tibia. Such relationships often have an inverse (e.g. has part). In contrast, undirected relationships such as continuous with imply a reciprocated relationship: the left atrium is continuous with the left ventricle implies that left ventricle is also continuous with the left atrium (relationships such as continuous with that are their own inverse are known as reflexive). It is also worth noting that relationships may allow multiple parents (the metanephros develops from the ureteric bud and the metanephric mesenchyme) as well as multiple children.
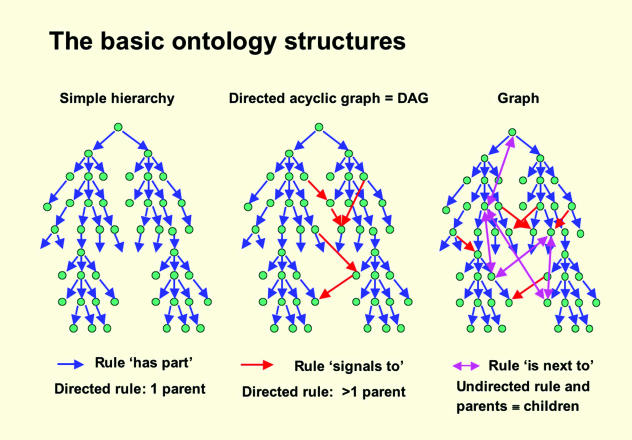
Graphically, there is a key difference between directed and undirected relationships: the former in general forbid circular paths in the graph while the latter allow them (see Fig. 1). In practice, loops are difficult to handle computationally (one has to have a means of knowing when to stop circulating) and best avoided. Directed relationships with a single parent (e.g. evolves from) generate simple hierarchies, while directed relationships with more than one parent generate what are known as directed acyclic graphs or DAGs (see Figs 1 and 2).
Fig. 1.
Pictorial representations of the various sorts of graphs that represent ontologies. (Reprinted from Bard, J.B.L. ‘Ontologies formalizing biological knowledge for bioinformatics’, BioEssays, 25, 501–506, with permission.)
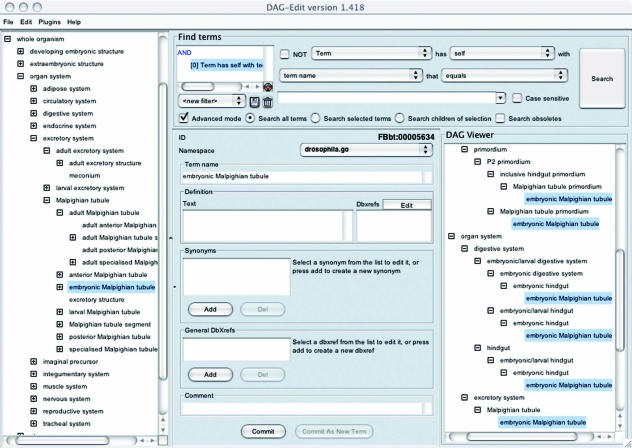
Fig. 2.
Screenshot of the Dagedit program showing the full Drosophila ontology (from Flybase*). In the left panel, the anatomy hierarchy is shown with the embryonic Malpighian tubules highlighted. The upper part of the central panel shows the ID associated with this tissue. The right panel shows some of the subhierarchies within the ontology that include the embryonic Malpighian tubules.
The importance of these relationships is that they allow inferences to be made from the ontology, by a user or computationally. In the latter case, a formal logic system is required and there are a range of possibilities that include first-order predicate logic and description logic; there are also more specialized logics such as that incorporated within Protégé* and GRAIL*, which is used as part of GALEN*, a system for handling clinical information that includes an anatomy ontology (see below). This is a complicated area and will not be discussed further in any detail. The interested reader is referred to Baader et al. (2004) for general comments and to Burger et al. (2003) for a demonstration of how such logics are created for simple anatomies.
Propagation concerns the information associated with a term (e.g. the genes expressed in a tissue) and there are three possibilities: the information can go with the direction of the relationship, against the direction of the relationship, or not be carried by the relationship (no propagation). An anatomical example of a relationship where the information flows with the relationship is part of (if the tibia expresses a gene, so does the hindlimb), and an example where the information flows against is is-a (the tibia is-a bone, and has the general properties of a bone, together with those that are tibia-specific, such as its morphology). An example of a relationship that does not carry inheritance is develops from: the genes expressed in early endoderm may or may not be expressed in gut epithelium.
There is a further aspect of bio-ontologies that needs to be mentioned at this stage and that is the unique identifier (ID) associated with each term. Although the name of a concept or term in an ontology is conventionally unique and helps convey meaning, bio-ontologies have adopted the convention that it is the ID that is unique, and that the name need not be. This ID has two components, an abbreviation for the ontology (e.g. FBbt for the Drosophila ontology) and a number that represents that term within the ontology (thus FBbt:00001896 is the unique ID for Drosophila larval Malpighian tubule). Such an approach makes it possible to give the same name (but different pathways) to similar tissues without any risk of ambiguity (e.g. the epithelium of the ileum and of the jejunum in the mouse).
One importance of the ontology ID is that it can be included within, for example, the tables of a gene-expression database to link data to an anatomy ontology. A request for all the genes expressed within the developing mouse forelimb at E12.5 thus reduces to first searching the E12.5 anatomy ontology for all the IDs of the tissues that are part of the forelimb and then searching the database for all the genes associated with these IDs and collating them. The other importance of this ID is that, as it provides a key that can be used to annotate any database, it can be used by that database to interrogate another, the process known as interoperability. An example of this use in an anatomical context will be given later, but it is worth noting that it is becoming possible to identify what a gene might do in a tissue that expresses it as that gene might already have been associated with a particular process that can be identified on the basis of its Gene Ontology ID (see next section). This is particularly important in microarray work and the analysis software usually includes GO IDs.
Examples of bio-ontologies
A simple and small-scale example of an ontology is ‘sexual phenotype’, which has three members: male, female and hermaphrodite – such an ontology is usually represented as a controlled vocabulary. An obvious large-scale biological example of an ontology is Linnaean taxonomy, in which a typical term represents a specialized set of organisms that share common properties but also defines a new, lower set in the hierarchy that includes organisms that share these common properties but also have other, additional properties (note that properties associated with terms here are propagated down the is-a hierarchy). It is worth noting that a cladistic taxonomy is slightly different: the relationship here is evolves from, but there are no propagation implications because the evolved species is different and may gain new properties and/or lose some from its forebear.
Perhaps the most used bio-ontology at the moment, however, is the Gene Ontology* (or the GO): this encapsulates our knowledge about gene products and thus complements the standard molecular databases, which tend to focus on the data associated with molecules (sequence, references, names, etc.). The GO actually has three ontologies: the first describes the location within or around the cell where the gene product is located (a part of and is-a ontology); the second focuses on the molecular function of the gene product (a hierarchy based on the is-a relationship and includes, for example, a four-level hierarchical description of transcription regulator activity); the third (which is also based on is-a relationships) describes the molecular processes in which gene products are involved (e.g. signal transduction). In all, the go has about 17 000 terms and the ontology is currently linked to a database of about 150 000 genes; the items in the associated database of gene products that are instances of a given concept in the ontology are said to instantiate that term. The go is a formidable and much used resource (many gene resources contain go links) and it, together with its associated database of gene products, can be easily accessed by, for example, the AMIGO* browser.
All the key bio-ontologies (GO, anatomies, cell types, etc.) are now publicly accessible at OBO*, the Open Bio-Ontologies site, and the availability of an ontology there implies a degree of acceptance by the biological community associated with that domain of knowledge.
Anatomical ontologies
Making anatomical ontologies
The general procedure for making an anatomy or indeed any other ontology is, in principle at least, simple. First, one decides on the domain of knowledge to be included (e.g. an adult or a developmental anatomy) and the relationships that are to be used (part of, is a, develops from, etc.). Second, one assembles all the terms to be included: the tissues and classes for the adult or for each developmental stage, and this involves access to textbooks, the literature, experts, one's memory, etc. Finally, one builds the ontology using a computer program that allows the compiler to start with a root term (adult mouse anatomy, say) and to continue adding subordinate terms via the connecting relationships until the ontology appears to be complete. The program stores the terms, adds the IDs and keeps track of relationships in a format that is computer- but not human-readable.
There are three standard tools for building and editing bio-ontologies: Dag-edit* (Fig. 2), COBrA* (Fig. 3) and Protégé* (Fig. 4), which each have their own advantages, while there are several formats for storing the data. Dag-edit* works with GO and OBOL formats, while COBrA* uses GO, OWL and RDFS formats, and has plans to incorporate OBOL. GO and OBOL are flatfile formats, while RDFS and OWL are XML-based formats that can include ever-increasing types of data and relationships, and this diversity reflects what happens in a new field; it is, however, likely that the field will eventually settle on OWL* (Web Ontology Language, the agreed language for the semantic web*) or a subset of it. Protégé, which has its own format, presents the user with a table for each term in the ontology in which all the relationships etc. for that term are stored. The ontology user does not, however, need to know much about these tools and formats and no further attention will be paid to them – interested readers are advised to visit the appropriate websites for additional information.
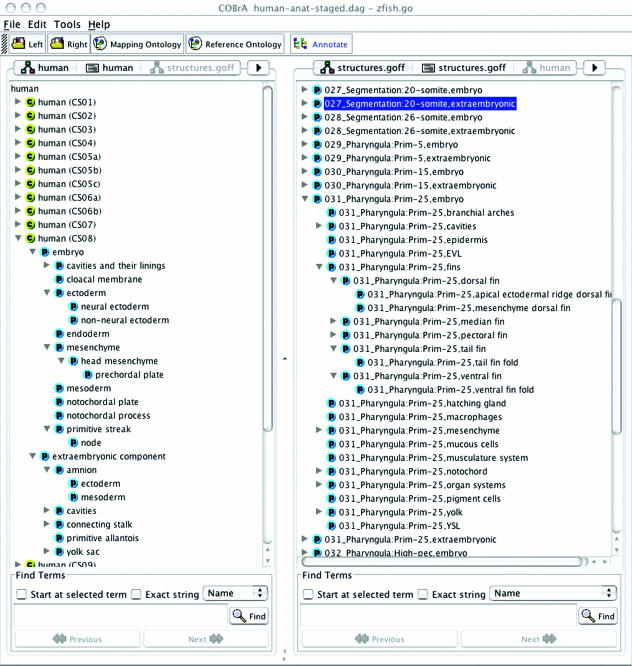
Fig. 3.
Screenshot of the COBrA ontology program. The left panel shows the ontology of human developmental anatomy, which does not include the Carnegie stage in the name for each tissue. The right panel shows the zebrafish anatomy ontology, which does include the stagename for each tissue. Not shown are the alternative panels for these ontologies that give the relationships and IDs for the human (hidden on right) and zebrafish tissues (hidden on left).
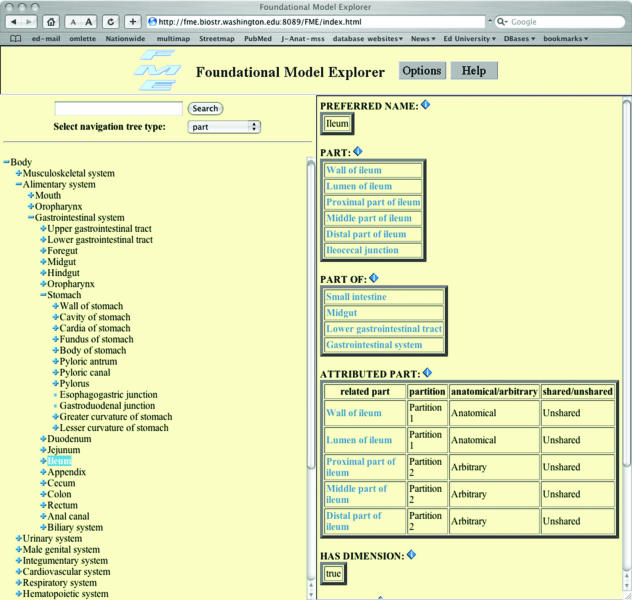
Fig. 4.
Screenshot of the Protégé program displaying the Foundation Model of (Adult Human) Anatomy (or FMA*). The ileum has been highlighted in the left panel, and some of the properties associated with this tissue are displayed in the left panel. Further properties are also accessible from the model.
In practice, there are two general approaches to making an anatomical ontology: the formal and the informal.
Formal ontologies, which aim to provide a full description of their area of knowledge, start by defining each term, and these definitions make explicit the knowledge to be included in the ontology and so eliminate ambiguity. Each term is first given a type identifier (the is-a relationship – the tibia is-a bone), and this defines a taxonomy (it has the semantics of subclass in set-theory). The individual fact triads for each of the other relationships are then added and the ontology is named and saved. If one is making a full ontology for the developmental anatomy of an organism that includes definitions of all tissues so that all terms or concepts are unambiguous, together with their is-a, part of and descends from relationships, it should be obvious that an enormous amount of work is involved. As such an ontology is fully defined; it can be used for making inferences about the terms and their associated properties, without requiring any input from the user.
The two good examples of such formal ontologies are the FMA*, or Foundation Model of (adult human) Anatomy, which aims to capture much of our knowledge of anatomy as a reference resource (Fig. 4; Rosse & Mejino, 2003), and GALEN*, which is a system for handling clinical information and includes a human anatomical ontology (Rogers et al. 2001). The makers of both have made serious efforts to ensure that their ontologies conform to high standards of rigour. It is however, worth noting that only about 70% of the terms in the two ontologies can be viewed as identical (Zhang et al. 2004). A simple example of such a formal ontology is that for cell types*.
Informal ontologies include much less explicit knowledge than formal ontologies. They are typically designed for a single purpose: such an ontology assumes that the user is not naïve but can bring a great deal of implicit knowledge to its use. For such ontologies, the maker may omit the definitions and the IS-A relationships and just include the core relationships needed for its use. Such minimal ontologies are employed, for example, for linking gene expression data to developmental anatomy, and have the advantage that they are extremely easy to use, provided that the user has sufficient knowledge of the anatomy to navigate through the ontology, knows what the terms mean and is prepared to accept the limited degree of automated inference that they can provide. The ontology of human developmental anatomy (Table 1,Fig. 3) is such a minimal ontology as it only includes a simple part of the relationship that is appropriate for handing gene-expression data (see later). The apparently similar lists of tissues present at each Carnegie stage (Hunter et al. 2003; available at Humat*) are not an ontology as they do not include formal linking relationships and IDs; they are intended to provide easy-to-inspect information; they also include links to anatomical notes and the literature.
Although it might seem that making an anatomical ontology merely requires collating and organizing sets of tissues, there are three distinctly anatomical problems that have to be solved before one can begin. The first is about defining what one means by a tissue, the second about considering the spatial resolution required and the third about deciding which explicit relationships should be used. All are more problematic than is initially apparent.
Tissue definitions
The identification of a tissue might seem quite easy for adult anatomies where all the major tissues have well-defined boundaries, but is actually far harder than one might like because it is often hard to recognize a boundary or, even if it is obvious, to decide the tissue to which it is related. The solution when one writes standard text is to brush aside these difficulties and define a tissue by the fact that one can point to its central region in a picture, but this is not really acceptable for an ontology that will be used for making inferences, say, about gene-expression data. Here, the user really does need to link expression with detailed tissue geometry. In principle, this can only be done graphically (see later); in practice, textual ontologies are mainly used as they are far easier to implement.
The definition of a tissue thus needs to include its spatial limits and this is usually straightforward for adult tissues where there are well-defined boundaries. Even here, a difficulty arises when a boundary tissue can be viewed as being part of two neighbouring tissues. For connected bones such as two adjacent phalanges, should one consider the joint as part of the proximal bone, the distal bone or as an independent entity? If one decides on the last, does one associate the hyaline surfaces with the bone or the joint?
In developing tissues, however, it is often far harder than one might like to recognize a boundary on any simple histological basis and this is particularly so as a large tissue regionalizes. Here, we need to distinguish between anatomical and arbitrary parts (Aitken et al. 2004), with the latter being defined by having a boundary that is not clearly defined by an obvious morphological feature. There are two obvious cases of an arbitrary boundary: where the tissue is still developing (e.g. the emergence of the gonadal domain from the mesonephros) and where there is no obvious delimiting morphology between two named tissues – where exactly does the atrial septum become distinguishable from the superior wall of the atrium, even in the adult heart, and can this question really be answered for other than the early stages of septum development when novel growth starts to become apparent in the superior wall of the atrium?
The solution to the problem of boundary assignation in well-defined tissues (e.g. the joint problem) really reduces to one of definition, and, provided that the ontology is explicit about defining the relationships, and that the user understands that definition, it does not really matter what choice is made – no ambiguity results (see Smith, 1996). Things are more complicated when there is a question as to whether a boundary actually exists. Consider the example of the mesonephros: Lhx9 expression evidence in the early mesonephros (Birk et al. 2000) marks the region of early gonad differentiation (E9) well before it becomes a distinct entity (E10). In principle, one could use the expression domain of Lhx9 to define the boundary, but this is unsatisfactory. First, the expression pattern is not particularly sharp and the boundary consequently ill-defined, and, second, it is quite possible that another early marker will appear with a slightly different pattern. As it is unrealisitc for an ontology to contain unstable definitions of when and where a tissue is located, the most practical solution is to use standard, non-molecular-specific stains such as haematoxylin and eosin, and to allow an experienced embryologist to define the time when a new tissue is first apparent on the basis of molecule-neutral histology. As to the boundary here, there is no good solution if the morphology is not clear, and one has to accept a weak definition of the tissue (e.g. expert opinion as to when a part of the mesonephros shows a visible sign of gonad morphogenesis).
These problems derive of course from trying to squeeze spatial anatomy into the confines of a verbal corset, and the best, albeit the most technically complicated solution is to use a three-dimensional (3D) digital map of the embryo and assign tissue names and expression domains to defined volumes (see below).
A further difficulty comes when one has to assign appropriate names to tissues within an ontology: in a textbook, there is no difficulty in doing this without causing ambiguity, but ontological requirements are more stringent. Is, for example, the left ventricle of the E11.5 mouse heart the same tissue as the E12.5 ventricle? Their tissue morphology and detailed patterns of gene expression are clearly different, but is that enough to give them separate names? One could call them the E11.5 and the E12.5 right ventricles, but this solution seems a bit cumbersome as such a distinct name is not really required in a computational context.
Conventionally, an ontology will give unique names to all distinct terms, and any synonyms will be recorded. For Drosophila, where the embryo, the larva and the adult are very different, the ontology explicitly differentiates between the embryonic Malpighian tubules, the larval Malpighian tubules and the adult Malpighian tubules (Fig. 2); the zebrafish ontology takes a similar approach (Fig. 3). The mouse and human developmental anatomies, however, depart from this convention for a number of pragmatic reasons. In the mouse, where many minor tissues in early embryos have the same name at the bottom of the PART OF hierarchy (e.g. epithelium), the solution adopted has been to keep the simple name as a hierarchy term but to ensure that each term in the hierarchy has a unique pathname as well as a unique ID (thus TS11/embryo/ectoderm/surface ectoderm is different from TS12/embryo/ectoderm/surface ectoderm, and TS21/embryo.organs system/visceral system/foregut/oesophagus/epithelium is different from TS21/embryo.organs system/visceral system/foregut/pharynx/epithelium).
Spatial resolution
If the anatomy ontology is to be self-contained and not linked to external datasources, its maker can decide how much information is to be included. If, however, the ontology will be linked to a database of tissue-associated data, a user will naturally expect that the named tissues will be complete in the sense that the whole volume of the organism is included and any expression pattern can be annotated at an appropriate level. This is relatively simple to do for organisms such as the C. elegans worm where every cell is anatomically defined, but impractical for complex organisms such as the mouse where many veins, for example, have no name, while other low-level tissues have the same name (e.g. associated mesenchyme). The situation is worse if one requires the database to hold protein-localization data: here, one also has to allow for the secretion of the proteins into the various cell-free cavities and lumens of the organism.
The only practical solution here is to accept that some parts of the organism are going to be handled at a more detailed level than others. It is also inevitable that, in a vertebrate for example, all the minor vessels and nerve axons will be excluded, and one has to accept that all tissues will include both (an inevitable assumption for all tissue-based microarray work). This acceptance immediately raises the question of how tissue resolution is to be handled. There are no absolute rules here, but, if the ontology is to be easily useable, two key determining organizational principles are that a user should be able to navigate through it intuitively and that the upward propagation of properties should make biological sense.
It is apparent in practice that all anatomy ontologies for animals include functional (or organ or body) systems at a very high level in the ontology (see Fig. 3), and that tissue geometry usually takes a secondary role. In the ontology of mouse developmental anatomy, for example, there are, as yet, no categories for head, neck and thorax (although there are limbs) as it has seemed more useful, in the case of the head for example, to include the cranium as PART OF the skeleton, the brain as PART OF the nervous system, the nose as PART OF the sensory system, etc. This organization is a matter of choice if the ontology is to be represented as a hierarchy where each term has a single parent. If, however, the ontology is to be expressed as a DAG, there is no reason why the cranium cannot be part of both the head and the skeleton. Although it becomes necessary to define where the head ends and the neck begins (e.g. whether the pharynx should be assigned to one or the other), such choices can easily be made and have been incorporated in the DAG ontology of adult mouse anatomy that has been made by the members of the Jackson laboratory (see Table 1).
The advantage of setting out a particular functional system, such as the alimentary system, as a simple hierarchy is that it is straightforward to name each major part from the mouth to the anus as a PART OF subcategory and then to construct a subhierarchy for each of these (Fig. 3) of decreasingly sized parts. This in turn allows the user to move down the hierarchy easily – there is little in the way of a learning curve (a very important practical consideration if one wishes one's ontology to be used by non-experts). The level of tissue size at which the hierarchy stops is decided by several factors that include: the absence of named regions (there are too many small domains of extracellular matrix for each to be recognized, let alone assigned a unique name), the sheer obscurity of some tissues (the salpingopalatine fold of the nasopharynx has attracted no publications accessible in PubMed since 1951), and the practicalities of producing something that can be made available to a field (for both use and criticism) in a reasonable time.
It is not difficult to determine whether an ontology is complete when one deals with low-resolution anatomy but, as one tries to be more and more precise in labelling tissue domains, it becomes harder and harder to ensure that a large tissue is fully partitioned. One clumsy but accurate solution here is to add a term named ‘not-named domain’, but this turns out to be impractical and unhelpful. A better solution is for the ontology makers to do the best they can, and ask users to let them know if a part needs to be added. The importance of community involvement in ontology maintenance will be discussed in more detail in the final section.
The part of relationship
Whereas some relationships are obvious in their meaning (e.g. develops from), the part of relationship, clearly the most important for anatomy, is so complex that its study has its own name, mereology* (Simons, 1987; Winston et al. 1987). A trivial and well-know example demonstrates the sort of problems that can be encountered here: my hand is part of me and I am part of the university, therefore my hand is part of the university! This is obviously untrue in any sense other than as an abusive comment about the demands made by universities on their staff, but the logical error derives from the fact that part of is used in two different senses, to describe first a geometric and then a membership relationship.
Although part of has a wide range of possible meanings in mereology*, the following examples and definitions seem to cover most obvious anatomical relationships.
The left atrium is part of the heart (a component of a single tissue)
The thyroid is part of the glandular system (a component part of a distributed system)
The bone marrow is part of the bone (contained within)
The cardiovascular system is part of the organ systems (ambiguous: could be is-a or part of a distributed system)
Lymphocytes are part of the blood (constituent of a mixture)
The periphery is part of the muscle (usually too ill-defined to be meaningful)
It is quite possible to incorporate any or all of these relationships within an ontology, and the adult human anatomies (FMA and Galen) both include several part of relationships, although their aim is as much accurate and complete description as ease of use.
For an ontology designed to handle tissue-associated data in an intuitive way, however, one really wants to use a single part of relationship. This turns out to be both possible and practical if one accepts a definition of part of that has two related components (Burger et al. 2003). First, it is transitive: this means that, if A is part of B and B is part of C, then A is part of C. Second, it carries upwards propagation so that any property associated with A is also associated with B and C: thus, if the second finger expresses a gene, so do the hand and the arm (albeit at an undefined location). Note that this definition of part of has a corollary that is useful in the context of data storage: if tissue C does not have a particular associated property, then nor do any of its subparts B and A (if a gene is not expressed in the arm, then it is not expressed in either the hand or the second finger).
What this version of part of means in practice, however, is that it should only be applied when upwards propagation is appropriate, and users needs to check the ontology to be sure that an assigned meaning is appropriate for their use. One of the examples given above has part of meaning containment – the bone marrow is part of the bone; and it may or may not be appropriate to equate this definition with one carrying implications of upwards inheritance. Whether one wishes to accept this depends one whether one wishes to consider, say, a blood stem cell as a natural constituent of bone.
Uses of anatomical ontologies
For all the work involved in making anatomic ontologies, they tend not to be particularly glamorous in any visual sense and are just seen on the interface as expandable trees of tissues (Figs 2–4). Their main current use is to provide a user with tissue-associated data (e.g. gene expression and pathological images – see Pathbase*). Although such systems are easy to use, they do not distinguish whether, if a tissue search identifies a particular gene, it is the whole or only part of the tissue that expresses that gene. The solution to this problem that has been adopted by GXD* is to enable users to see the pictures of gene expression that have been published and so decide for themselves whether the data are appropriate and also allow for author annotation. The other solution is to map the gene-expression data onto a 3D model of the organism.
There currently seems to be only one accessible database based on these graphical principles, the Edinburgh Mouse Atlas Project* (Davidson & Baldock, 2001). The basis of this database is a set of voxel models (a voxel is a 3D pixel) of staged mouse embryos in which the domains of all the major tissues are digitally identified (and carry standard IDs). The database allows a user to map gene-expression data to any voxel in a way that is tissue-independent and, of course, search for expression data spatially. One great advantage of the database is that it includes a set of voxel models of tissue-annotated embryos (as yet incomplete – making them is a major effort) that can be visualized in a range of ways, and these can be used, for example, to identify tissue domains in digital sections merely by moving the cursor to that domain (Fig. 5). It is worth noting that such a graphical ontology (Baldock & Burger, 2004) and database is very different from 3D image systems such as the Visible Human Project* as the latter cannot, as it stands, be used for mapping data to volume space.
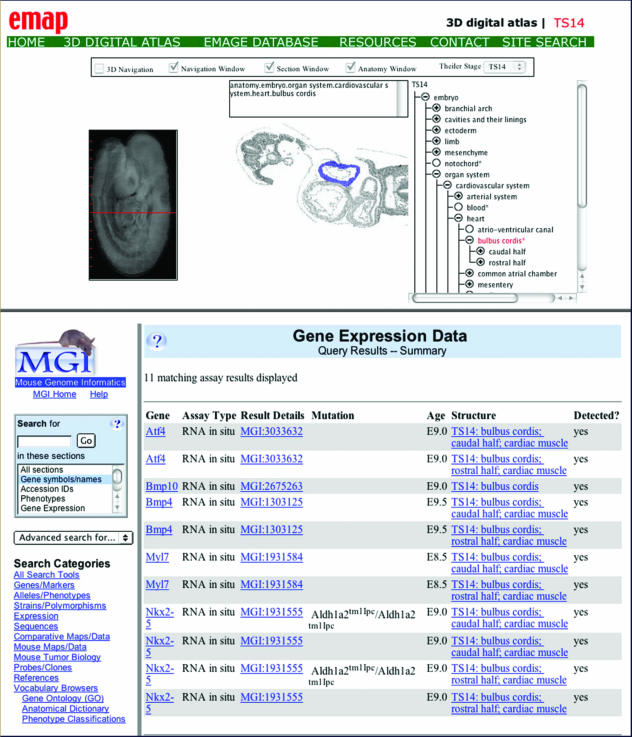
Fig. 5.
Composite picture of two screenshots. The upper screenshot shows the EMAP* site displaying the TS14 (E9) mouse embryo. The red line indicates the plane of the section shown in the central section; here, the cursor overlies the bulbus cordis (blue). The name of the tissue is given in the upper panel and its place in the ontology shown in the right panel. The lower screenshot shows the gene-expression data stored at GXD* for the TS14 bulbus cordis. This example of interoperability was initiated by double clicking on the bulbus cordis image in EMAP*.
This identification has an additional property that illustrates a technology that is becoming of increasing importance and that is known as interoperability, the ability of one database to query another. Because EMAP* and GXD*, the two mouse gene-expression databases, use the same anatomy ontology with the same IDs, there is a linking tool in EMAP*: once a tissue has been identified by the mouse cursor, a right click of the mouse button automatically sends the tissue ID to GXD as part of a search query and GXD returns data on all the genes expressed in that tissue (Fig. 5).
Thus far, it seems that the only data types directly associated with anatomical ontologies are for gene expression (several organisms) and pathological images (mouse). However, with the availability of the new cell-type ontology*, which details all major cell types on the basis of function, lineage, histology, etc., it is now possible to link tissues with their constituent cell types. This is now being done as part of the XSPAN* project, which seeks to make accessible such data, together with links between equivalent tissues across the model organism anatomies (e.g. excretion is handled by the metanephros in the mouse, by the Malpighian tubules in Drosophila, by the mesonephros in the zebrafish and by the excretory canal in C. elegans).
Discussion
There was a time, not so long ago, when Drosophila, C. elegans and mouse anatomists would have had little to say to one another. The molecular revolution has changed that irreversibly as it is now clear that the early molecular networks shaped the tissues of Cambrian organisms in ways that have ramifications for all modern organisms. This, in turn, means that we need tools and concepts that allow us to handle this richness of relationships and associated data. Anatomical ontologies are the key tool that allow us to link anatomical tissues to one another and to their underlying cellular and molecular information, and are now a standard part of the bioinformatics infrastructure.
It should, however, be clear that, even for a simple organism, there is no all-defining anatomical ontology, but that several can be made with different relationships and with more or fewer tissues. Indeed, ontologies are always made for specific purposes and their individual structures of relationships and terms reflect that purpose; they may also reflect the idiosyncrasies of their authors. Once an ontology is in place, however, it is easy to assign to it more authority that it warrants: ontologies are built on relationships, and, unless the meaning and use of these relationships are precise, it is possible to make errors of inference. It should also be clear that, if the anatomical information stored within an ontology is wrong or, more likely, inadequate, then further errors may result from their use. Obvious errors are sins of omission that may be inevitable due to knowledge limitations or accidental as, in a complex ontology, things may just become misplaced or omitted. It cannot be emphasized too strongly that ontology users should be aware of what they are using and place only appropriate trust in their use –caveat emptor!
Although authors of ontologies act in good faith, it is important to realize that, once these ontologies are in the public domain (e.g. posted on the OBO* site or used with a database), they become a public resource and ownership moves to the user community, with the role of author being demoted to that of curator, or servant of both the ontology and the community. With ownership goes responsibility and curators need the user public to provide feedback on the limitations of the ontology. The public might be quite surprised at the number of minor errors of both omission and commission that are present in complex anatomy ontologies: one might suppose that, just because the anatome is quite well defined, it is possible to construct a well-defined ontology. This is not so and makers are more aware than most just what approximations are required. Users of all ontologies are therefore requested to report back to curators any queries or complaints that they have.
A further problem confronting ontology compilers is how to incorporate additional knowledge with their ontologies. An obvious anatomical example is the addition of cell-type data to anatomy ontologies. One approach is to expand the anatomical ontology to include cell types using a new relationship (the triceps HAS CELL TYPE skeletal muscle); this approach is used by the C. elegans ontology, which was made before the cell-type ontology was produced. The alternative is to keep the anatomy and cell-type ontologies separate and provide a mapping between them that links tissue and cell-type IDs; this is the approach adopted for the XSPAN* project. Both are possible, but the latter seems the more appropriate as it uses standard IDs for cell types and so facilitates future interoperability.
A similar problem is facing the Gene Ontology, which is expanding its process ontology to include tissue-associated processes (e.g. eye development, somitogenesis). Thus far, it has included anatomical terms within the ontology, but the sheer number of tissues is requiring the curators to adopt a combinatorial approach: for example, instead of ‘metanephros growth for the mouse’ being introduced as a single term in the GO* process ontology, it is becoming more practical to search databases using the ‘mouse kidney’ ID from the mouse anatomy ontology and the ‘cell proliferation’ ID from the GO* (Hill et al. 2002). In this context, it is worth noting that there is now so much gene expression material in GXD* (e.g. 460 expressed genes for the neural tube) that it is becoming a practical necessity to use GO* terms to restrict the search to specific classes of genes (e.g. restrict the search to membrane receptors).
Perhaps the greatest challenge to the field, however, is providing an ontology rich enough to archive mutant organisms on the basis of their abnormal anatomy and other aspects of their phenotype. One approach, adopted by the Mouse Genome Database for mouse mutants, is to make a single, integrated phenotype ontology*; another is to describe the mutant phenotype using a set of standard ontologies covering the anatomy, the type of trait, etc. (see Bard & Rhee, 2004), that can be associated with the genotype. The practical assay on which all of these choices will be made is ease of access for users and it will be interesting to see how the field handles this problem.
A key subtext to this article has been that, in order to formalize anatomy so that it is computationally useful, one must articulate anatomical concepts and relationships in a way that is far more structured than has been necessary in the past and bears little relationship to the traditional classifications such as that in Terminologica Anatomica. This approach has the benefit of making one look at old material in new ways, and it turns out that, although text-based and computer anatomies describe the same material, they do this so differently that it raises the question of whether the use of the term anatomy is appropriate for the latter.
Here, it is worth noting that the general study of genomes is known as genomics, of the proteome or set of proteins in each organism as proteomics, and of their metabolites as metabolomics. ‘-omes’ and ‘-omics’ neologisms even extend to topics where there is no obvious set of items such as the physiome*. As each organism is composed of a set of tissues (whose number will of course reflect the spatial resolution at which the organism is examined), it seems appropriate to use the terms introduced at the beginning of this review, and consider the sets of tissues that make up organisms as their anatome, and the their formalization and use, at least in a computational context, as anatomics.
Some, like Lederberg & McCray (2001) who have already coined the term vacuomics to express their feelings about jargon, may feel that there are already too many -omics and another one is unnecessary. It does, however, seem entirely appropriate to use the term anatomics for the recasting of the traditional subject of anatomy in a way that meets the computational needs of both anatomists and non-anatomists. One argument for its use is that it meshes with other terms for computer-based topics in the biosciences; another is that it affirms that, for all of its age-old traditions, anatomy is still a key aspect of contemporary biological research.
Acknowledgments
I am grateful to Stuart Aitken for introducing me to formal ontology theory, and I thank him and Richard Baldock and for commenting on the manuscript.
References
- Aitken JS, Webber BL, Bard JBL. Part-of relations in anatomy ontologies: a proposal for RDFS and OWL formalisations. Pacific Symp. Bioinformatics. 2004;9:166–177. doi: 10.1142/9789812704856_0017. [DOI] [PubMed] [Google Scholar]
- Baader F, Horrocks I, Sattler U. Description logics. In: Staab S, Studer R, editors. Handbook on Ontologies. Berlin: Springer; 2004. pp. 3–28. [Google Scholar]
- Baldock R, Burger A. Anatomical ontologies: names and places in biology. Genome Biol. 2005 doi: 10.1186/gb-2005-6-4-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JBL, Rhee SY. Ontologies in biology: design, applications, and future challenges. Nature Rev. Gen. 2004;5:213–222. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- Birk OS, Casiano DE, Wassif CA, et al. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature. 2000;403:909–913. doi: 10.1038/35002622. [DOI] [PubMed] [Google Scholar]
- Burger A, Davidson D, Baldock R. Formalization of mouse embryo anatomy. Bioinformatics. 2003;19:1–9. doi: 10.1093/bioinformatics/btg400. [DOI] [PubMed] [Google Scholar]
- Davidson D, Baldock R. Bioinformatics beyond sequence: mapping gene function in the embryo. Nat. Rev. Gen. 2001;2:409–418. doi: 10.1038/35076500. [DOI] [PubMed] [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Proceedings of the Pacific Symposium on Biocomputing. World Scientific Publishing Co; 2004. Building mouse phenotype ontologies; pp. 178–189. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A. Nucleic Acids Res. Database issue. Vol. 32. 2004. The Gene Ontology (GO) database and informatics resource; pp. D258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendler J, Berners-Lee T, Miller E. Integrating applications of the semantic web. J. Inst. Elec. Eng. Jap. 2002;122:676–680. [Google Scholar]
- Hill DP, Blake JA, Richardson JE, Ringwald M. Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies. Genome Res. 2002;12:1982–1991. doi: 10.1101/gr.580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Kaufman MH, McKay A, Baldock R, Simmen MW, Bard JB. An ontology of human developmental anatomy. J. Anat. 2003;203:347–355. doi: 10.1046/j.1469-7580.2003.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J, McCray AT. ‘Ome Sweet ‘Omics – a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- Parkinson H, et al. The SOFG Anatomy Entry List (SAEL): an annotation tool for functional genomics data. Comp. Func. Genom. 2005 doi: 10.1002/cfg.434. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Eppig JT, Begley DA, et al. Mouse Gene Expression Database (GXD) Nucleic Acids Res. 2001;29:98–101. doi: 10.1093/nar/29.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JE, Roberts A, Solomon WD, et al. GALEN Ten years on: tasks and supporting tools. In: Patel V, et al., editors. Proceedings of the MEDINFO2001. IOS Press; 2001. pp. 256–260. [PubMed] [Google Scholar]
- Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Simons P. Parts: a Study in Ontology. Oxford: Oxford University Press; 1987. [Google Scholar]
- Smith B. Mereotopology: a theory of parts and boundaries. Data Knowledge Engineering. 1996;20:287–303. [Google Scholar]
- Winston ME, Chaffin R, Herrmann D. A taxonomy of part-whole relations. Cognitive Sci. 1987;11:417–444. [Google Scholar]
- Zhang S, Mork P, Bodenreider O. Proceedings of the Pacific Symposium on Biocomputing. World Scientific Publishing Co; 2004. Lessons learned from aligning two representations of anatomy; pp. 250–261. [Google Scholar]