Abstract
The midbrain dopaminergic (mDA) neurons play a key role in the function of a variety of brain systems, including motor control and reward pathways. This has led to much interest in these neurons as targets for intervention in human disorders such as Parkinson's disease and schizophrenia. A major area of interest is to direct embryonic stem (ES) cells to differentiate into mDA neurons in vitro, which can then be used for cell therapy or drug screening. At present, our understanding of mDA development in vivo is limited. However, recent studies have identified a number of regulatory factors that influence the development of mDA neurons in vivo. Such studies will not only increase our understanding of mDA development in vivo, they may also promote new paradigms for regulating mDA production from ES cells in vitro. Here we review the current knowledge on mDA development in vivo and mDA differentiation.
Keywords: differentiation, dopaminergic neuron, embryonic stem cells, Pitx3, transcription factor
Introduction
Dopamine-containing neurons are present in various areas of the central nervous system, including the retina, olfactory bulb and periventricular hypothalamus (Bear et al. 2001). However, the largest assembly of dopaminergic (DA) neurons in the brain are located in the midbrain. Midbrain dopaminergic (mDA) neurons are separated into functionally distinct subgroups called the substantia nigra (SN) and the ventral tegmental area (VTA) based on their position within the midbrain and the target structures which they innervate (Bjorklund & Lindvall, 1984). Dopaminergic neurons of the SN regulate motor function via nigro-striatal projections to the dorsolateral striatum. In humans, the degeneration of SN neurons results in Parkinson's disease (PD). PD affects 1% of the population aged over 50 years, and is characterized by a slowness of movement (bradykinesia) and a difficulty in initiating movement (akinesia). Dopaminergic neurons of the VTA project to the ventromedial striatum, cortical areas and the limbic system, forming the mesolimbocortical system. Defects of the mesolimbocortical system are implicated in psychiatric disorders such as schizophrenia (Wallen & Perlmann, 2003).
Midbrain DA development in vivo
Diverse neural cell types are generated in the nervous system from multipotent neural progenitor cells. Accordingly, the specification of cell fate is complex and is governed by the interplay of extrinsic and intrinsic signalling molecules, combined with cell-type- and temporal-specific factors. A grid-like set of positional identities is given to neural progenitors via gradients of signalling molecules secreted throughout the dorso-ventral and rostro-caudal axes of the neural tube. In addition, secondary organizers act as local signalling centres to further refine cell fates at the rostrocaudal level. Once neuronal fate has been restricted by these extrinsic cues, intrinsic signals – often transcription factors – direct further differentiation into a mature, post-mitotic neuron (Edlund & Jessell, 1999).
Specification of mDA neuron progenitors
The correct specification and development of mDA neurons depends upon a number of signalling/transcription factors and the proper development of the ventral midbrain. Early in embryogenesis, mDA neurons are specified at the ventral mesencephalon just rostral to the isthmic organizer (IsO). The IsO is a signalling centre located at the border between the midbrain and the hindbrain and its position determines the location and size of the mDA neuron population (Brodski et al. 2003). Accordingly, the correct specification of the IsO is necessary for the development of midbrain structures. Many molecules involved in establishing and patterning the midbrain are expressed in a complicated network of regulation in the isthmic region, which has not yet been fully uncovered. At the end of gastrulation, embryonic day (E) 7.5 in mouse, the future position of the IsO is identified where the expression domains of the transcription factors Otx2 and Gbx2 meet. Otx2 is expressed on the rostral side and Gbx2 is expressed on the caudal side of the IsO, and they are required to suppress hindbrain and midbrain development, respectively (Li & Joyner, 2001). Also expressed from this stage is the homeobox transcription factor Lmx1b, which is initially expressed throughout the ventral mesencephalon and diencephalon and is later restricted to post-mitotic mDA neurons (Smidt et al. 2000). At E8, the transcription factor Pax2 is expressed across the Otx2/Gbx2 border and expression of the secreted molecule Wnt1 is predominantly limited to the Otx2 domain (Wurst & Bally Cuif, 2001). Following this, the transcription factors En1, En2 and Pax5 are expressed across the Otx2/Gbx2 border, and fibroblast growth factor 8 (FGF8) is expressed on the Gbx2-positive side (Wurst & Bally Cuif, 2001). By E9.5 the expression of Wnt1 and FGF8 domains have sharpened into rings on the rostral and caudal sides of the IsO, respectively (Wurst & Bally Cuif, 2001). Otx2, Lmx1b and Wnt1 are expressed on the midbrain side of the IsO whilst Gbx2 and FGF8 are expressed on the hindbrain side. The intersection of the floor plate-derived signal Sonic hedgehog (Shh) with the isthmus-derived FGF8 creates an inductive centre for mDA neurons just rostral to the IsO. It has been shown that Shh is necessary and sufficient for the induction of mDA neurons along the dorso-ventral axis, whereas FGF8 is responsible for the position of mDA neurons along the anterior–posterior axis of the neural tube (Ye et al. 1998). FGF8 also functions to induce and maintain the expression of early regulatory genes, which pattern the mid/hindbrain region and establish the isthmus.
Development of post-mitotic mDA neurons
The first post-mitotic DA neurons are detected at around E10.5 in the mouse, and studies in the developing rat brain have demonstrated that most post-mitotic cells of the mDA system emerge between E11 and E15, with a peak of SN neurons emerging at E13–14 and VTA neurons at E14–15 (Hanaway et al. 1971; Lauder & Bloom, 1974; Altman & Bayer, 1981). At E11.5 of mouse development, the first and rate-limiting enzyme of dopamine biosynthesis, tyrosine hydroxylase (TH), is expressed. The mechanisms involved in the regulation of TH expression in mDA neurons remain unclear. However, it appears that TH is not required for mDA development, as TH-deficient mice have an apparently intact mDA system with normal projections to target tissues in the telencephalon (Zhou & Palmiter, 1995). Here we discuss regulatory factors that are implicated in mDA neuron development and their potential in directing mDA fate from stem cells.
Pitx3
Pitx3 is a bicoid-related homeobox protein, which within the CNS is expressed exclusively in mDA neurons starting from E11.5 in mice (Smidt et al. 1997). The expression specificity and the fact that many homeobox genes are key mediators during development suggests that Pitx3 may play an important role in mDA neuron development.
Aphakia is a spontaneously occurring recessive phenotype that is associated with a double genomic deletion in the Pitx3 gene. Aphakia mice have a 652-bp deletion 2.5 kb upstream from the putative Pitx3 transcription start site, and a 1423-bp deletion that removes non-coding exon 1 and the putative Pitx3 promoter region in intron 1 (Semina et al. 2000; Rieger et al. 2001). Recent studies on adult aphakia mice demonstrated a loss of TH-expressing cells in the SN, whereas the VTA is affected to a lesser degree. Immunostaining in the striatum revealed a loss of TH immunoreactivity in the dorsal striatum but not in the nucleus accumbens, which are the target structures for projections from the SN and VTA, respectively (Hwang et al. 2003; Nunes et al. 2003; Van Den Munckhof et al. 2003; Smidt et al. 2004). Examination of earlier stages in mDA development of aphakia mice has led to conflicting results. One study reported an altered distribution of TH-positive cells in E12.5 aphakia mice, although the total number of TH-positive cells does not appear to be reduced (Smidt et al. 2004). Another study, however, revealed no changes in TH immunoreactivity in E12.5 aphakia midbrain (Van Den Munckhof et al. 2003). We have generated Pitx3 knockout mice, which also carry an eGFP reporter under the control of the endogenous Pitx3 promoter, via homologous recombination (Fig. 1) and have found a striking reduction in the number of mDA neurons in homozygous Pitx3 mutant mice at E12.5 (unpublished data). This apparent more severe mDA defect in the Pitx3 knockout is probably due to the creation of a true null mutation by gene targeting, as the major part of the Pitx3 coding region is deleted in the targeted Pitx3 locus (Zhao et al. 2004a,b). By contrast, the entire coding sequence remains intact in aphakia mice, which have 1 ± 5% of the wild-type level of Pitx3 transcript from E11 to newborn (Rieger et al. 2001).
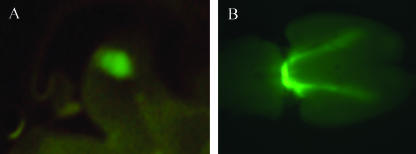
Fig. 1.
Direct visualization of the GFP reporter, which is under the control of the endogenous Pitx3 promoter (Pitx3–GFP), in developing mouse embryos that were heterozygous for Pitx3 locus. (A) Sagittal section of E13.5 embryo showing GFP expression in the ventral mesencephalon. (B) Ventral view of an E14.5 brain showing GFP expression in the midbrain and axonal projection to the forebrain.
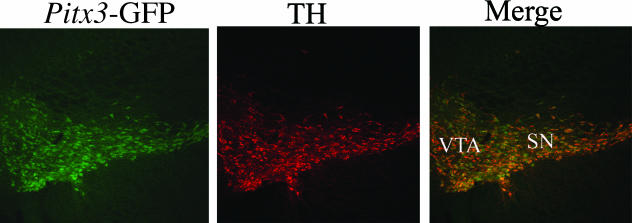
Van Den Munckhof et al. (2003) suggest that the SNc-specific loss of DA neurons in later fetal development of aphakia mice is due to differential expression of Pitx3 in ventral part of the SNc. However, this observation was neither supported by our studies nor by those of Smidt et al., which show that Pitx3 and TH expression overlap completely throughout the SN and VTA (Fig. 2) (Smidt et al. 2004; (Zhao et al. 2004a,b). As Pitx3 is expressed in both the SN and the VTA it is intriguing that the defects in aphakia and Pitx3 knockout mice are SN specific. This suggests that there may be differential dependence on Pitx3 by different mDA neurons. A better understanding of the transcriptional profile of mDA neurons may reveal SN-specific factors, which either function cooperatively with or downstream of Pitx3.
Fig. 2.
A coronal section of the ventral midbrain from a Pitx3–GFP heterozygous adult mouse. Double antibody staining against GFP and TH was performed. Panels from left to right show GFP staining, TH staining and a merge of GFP and TH images. The vast majority of the GFP-positive cells also express TH.
A potential role for Pitx3 in the induction of TH expression has been suggested by the demonstration that Pitx3 can bind to response elements and activate the TH promoter in a cell-type-dependent manner (Cazorla et al. 2000; Lebel et al. 2001). However, overexpression of Pitx3 in adult hippocampus-derived progenitor cells had no effect on the expression of DA neuron markers, including the dopamine synthesizing enzymes TH and l -aromatic amino acid decarboxylase (AADC), the glial cell line-derived neurotrophic factor (GDNF) signal transducing receptors c-ret and GFRalpha-1 and the D2 dopamine receptor (D2R) (Sakurada et al. 1999). This implies that, unlike Nurr1, which can induce a DA phenotype in both neuronal and non-neuronal cells (Sonntag et al. 2004b), Pitx3 requires cell-type-specific factor(s) to regulate TH expression. Cazorla et al. (2000) suggest that Pitx3 and Nurr1 cooperate to regulate TH expression, and that Pitx3 may be involved in the maintenance of TH expression.
Nurr1
Nurr1 is a member of the orphan nuclear receptor family of transcription factors and is expressed in the ventral mesencephalic flexure from mouse E10.5 onwards (Zetterstrom et al. 1997). In addition, Nurr1 is also expressed in other brain regions, including the neocortex, hypothalamus, hippocampus and cerebellum (Zetterstrom et al. 1996). Nurr1-deficient mice lack TH-expression in the ventral midbrain from E11.5; other DA neuronal markers including the retinoic acid-converting enzyme aldehyde dehydrogenase 2 (AHD2), c-ret and the D2R are also absent from the adult SNc and VTA (Zetterstrom et al. 1997). A later study revealed that Nurr1-deficient neuroepithelial cells undergo normal ventralization and differentiate into Pitx3-expressing neurons (Saucedo-Cardenas et al. 1998). Further studies have demonstrated the expression of En1, En2, AHD2 and AADC in midbrain DA cells in Nurr1 mutant midbrains, although the expression of these markers is reduced or absent by E15.5 (Wallen et al. 1999; Smits et al. 2003). These Nurr1-deficient midbrain neurons do not express DA neuron markers such as TH, vesicular monoamine transporter 2 (VMAT2) and dopamine transporter (DAT) (Smits et al. 2003) and subsequently die by apoptosis, resulting in a severe loss of SNc and VTA cells by neonate stage. Thus, Nurr1 is required for specific features of the dopaminergic phenotype and is necessary for the survival of late dopaminergic neurons (Saucedo-Cardenas et al. 1998). In addition, the mDA markers that were expressed in Nurr1 mutant mice by E15.5 are medially located. A further investigation into the distribution of the mutant Nurr1 mRNA expression in Nurr1 null brains revealed that Nurr1–/– mDA neurons fail to migrate laterally to their normal positions during development and are unable to innervate their target tissue, the striatum (Wallen et al. 1999).
Engrailed
Engrailed is a homeodomain transcription factor that has two vertebrate homologues, En1 and En2. Both En1 and En2 are expressed in the developing mouse brain from E8 on either side of the IsO in the midbrain and hindbrain (Davis & Joyner, 1988). In adulthood En1 and En2 have a more limited expression pattern. In the midbrain, En1 is expressed throughout the SN and VTA at high levels, whereas En2 is expressed at lower levels and is restricted to a subset of the En1-expressing cells (Simon et al. 2001). In addition, En1 and En2 are expressed in motor nuclei of the pons and En2 is expressed in the cerebellum (Davis & Joyner, 1988). En1 null animals have major brain defects during embryonic development, including deficits in the midbrain, hindbrain, colliculi, cerebellum and the third and fourth cranial nerves (Wurst et al. 1994). Yet, mDA neurons are present in En1 null mice at P0, and En2 is expressed at higher levels in these cells (Simon et al. 2001). The En2 knockout phenotype is milder with no gross mid-hindbrain abnormalities (Joyner et al. 1991). By contrast, the compound mutation of En1 and En2 results in a complete absence of both SN and VTA DA neurons by E14. At E12, however, TH-positive neurons are present in the ventral midbrain of En1/2 double mutants although there are fewer cells than in the wild-type controls. At E13 the number of TH-positive cells is more reduced in the En1/2 double mutants and some of the remaining TH-positive cells display signs of apoptosis (Alberi et al. 2004). However, there is a substantial truncation of the midbrain and anterior hindbrain in the engrailed double mutants resulting in a loss of the isthmus and reduced levels of Shh and FGF8 (Simon et al. 2001). Accordingly, it is difficult to make assumptions about the function of En1 and En2 in mDA neuron development in a situation where key midbrain structures and developmental signalling factors are aberrant. Recently this has been addressed by in vitro studies on mDA neurons, which demonstrated that En1 and En2 are cell-autonomously required for mDA neuron survival (Alberi et al. 2004).
Lmx1b
Lmx1b is a LIM class homeobox (LIM-HD) gene that is expressed in the CNS and in the periphery where it is necessary for dorsoventral patterning of limbs (Riddle et al. 1995). During development Lmx1b is expressed in many areas of the CNS, including those that give rise to SN, VTA, Raphe nuclei, subthalamic nucleus (STN), posterior hypothalamus and the spinal cord (Asbreuk et al. 2002). Recently it has been reported that Lmx1b is required for serotonergic neuron development in the hindbrain (Ding et al. 2003). Lmx1b is expressed throughout the mesencephalon and diencephalon from E7.5 in mice and expression in the SNc and VTA has been reported from E12 onwards (Smidt et al. 2000; Nunes et al. 2003). Studies on Lmx1b null mice have revealed that a loss of Lmx1b results in the failure of proper mDA neuron development. At E12.5 there is a reduction in the number of TH-expressing cells and an absence of Pitx3 expression in the ventral midbrain of Lmx1b null mice. However, Nurr1 expression in the ventral midbrain is intact in Lmx1b null mice at E12.5. From E16 onwards, TH expression is not detected in the mutant ventral midbrain (Smidt et al. 2000). As Lmx1b is expressed early (E7.5 in mouse) during CNS patterning events and has a widespread expression pattern in the brain, it is difficult to draw conclusions about a direct role for Lmx1b in mDA neurons from this study. Further experiments are needed to address the cell-autonomous functions of Lmx1b in mDA neurons. However, the knockout study suggests that Nurr1 is not regulated by Lmx1b and that Pitx3 might be a target of Lmx1b.
Ldb1 is a co-factor that is able to bridge between LIM-HD proteins and Pitx proteins (Bach et al. 1997). The expression of Lmx1b not only overlaps with Pitx3 in mDA neurons, but also with Pitx2 in the STN, posterior hypothalamus and some mammillary nuclei where Ldb1 is also expressed (Asbreuk et al. 2002). Therefore, Ldb1 may mediate cooperation between Lmx1b and Pitx factors in these regions.
Wnt
The Wnt family of glycoproteins are signalling molecules with key roles in regulation of patterning, cell proliferation and cell determination in the embryo. Furthermore, within the nervous system Wnt proteins are involved in virtually all significant patterning events (Patapoutian & Reichardt, 2000). Wnt1 is expressed around the IsO, predominantly on the midbrain side. A key role for Wnt1 in midbrain development was demonstrated by loss of function studies that revealed a failure by Wnt1-deficient embryos to develop any midbrain or anterior hindbrain structures (McMahon & Bradley, 1990; McMahon et al. 1992). Later, complementary misexpression studies in the chick demonstrated a role for Wnt1 in the formation and maintenance of the IsO. In that study, Wnt1 was shown to be able to induce Fgf8 expression, which in turn induces the expression of Lmx1b (Matsunaga et al. 2002).
As well as Wnt1, Wnt5a is also expressed at a high level in the ventral midbrain, with a peak at E11.5 in rat (Castelo-Branco et al. 2003). Addition of Wnt5a to rat E14.5 ventral midbrain primary cultures resulted in an increase in the number of TH-expressing neurons. Furthermore, there was an increase in the overall expression levels of Pitx3 and c-ret in cultures treated with exogenous Wnt5a. It was concluded that Wnt5a increases the total number of mDA neurons by promoting the acquisition of mDA phenotype, whilst Wnt1 acts as a general midbrain mitogen (Castelo-Branco et al. 2003).
Generation of DA neurons in vitro from ES cells
Neural differentiation from ES cells
Embryonic stem (ES) cells are pluripotent, clonal cell lines derived from the inner cell mass of the pre-implantation embryonic blastula (Evans & Kaufman, 1981). When injected into the blastocyst of a host mouse embryo, ES cells can give rise to all cell lineages, including germ cells. In vitro ES cells are capable of unlimited self-renewal and upon removal of leukemia inhibitory factor (LIF) can differentiate into a myriad of cell types, including neurons, cardiomyocytes, adipocytes and pancreatic β-cells (O'Shea, 1999). Therefore, ES cells provide an important cellular system for developmental studies allowing the processes of fate determination and differentiation to be dissected in detail, which otherwise may be difficult to study in experimental animal systems. Because ES cells are easily accessible for genetic modification without compromising pluripotency, they can be used to test certain transgene expression during differentiation (Aubert et al. 2002; Kim et al. 2003). In addition, ES cells can potentially be used for drug discovery and to provide an unlimited source of material for cell-based therapy for treating diseases, such as PD (Kawasaki et al. 2000; Lee et al. 2000; Barberi et al. 2003).
Methods for general neural differentiation from ES cells include formation of embryoid bodies (EBs) with retinoic acid (RA) treatment (Bain et al. 1995), co-culture with stromal cells (Kawasaki et al. 2000b) and monolayer differentiation in serum-free conditions (Ying et al. 2003). Neural lineages are evaluated by the presence of neural precursor markers such as the intermediate filament protein nestin and SRY-related transcription factors Sox1 and Sox2 which are expressed specifically in developing neuroepithelium (Lendahl et al. 1990; Pevny et al. 1998) or general neuronal marker such as βtubulin3, MAPs and neurofilament proteins.
Because the exact requirements for differentiation of ES cells into specific cell lineages remain elusive, current differentiation protocols yield heterogeneous cell types. In the absence of a system for directed differentiation from ES cells to neuroectoderm, we have developed a genetically based neuroepithelial lineage selection strategy that allows the purification of a highly enriched population of neural progenitors from heterogeneous ES cell differentiated progeny (Li et al. 1998).
The strategy is based on targeted insertion of a βgeo marker/reporter gene into the Sox2 gene via homologous recombination in ES cells. These ES cells were induced to differentiate into neural cells in EBs (Bain et al. 1995; Li et al. 1998), which usually results in around 50% of the culture expressing Sox2. The Sox2-positive cells were then enriched for by applying G418 selection, which resulted in over 90% of the cells expressing Sox1 or Sox2. Neural precursor markers including Pax3, Pax6, Mash1, Math4A, Delta1 and Islet1 were found in neural precursor cultures indicating that a range of distinct neural subpopulations were present. In differentiated cultures neuronal fates were identified, which included GABA and glutamate neurotransmitter phenotypes (Li et al. 1998).
More recently, a Sox1 lineage marking technique has been used to identify neural precursors in differentiating ES cells. Sox1 has a more restricted expression pattern than Sox2 during embryo development. Importantly, unlike Sox2, Sox1 is not expressed in undifferentiated ES cells and therefore provides a better reporter to track neural fate determination without the ambiguous effects of other cell types in the culture (Ying et al. 2003). So far, this genetic selection/marking approach has not been used to enrich for or track a specific neuronal subtype derived from ES cells. The Pitx3–GFP ES cells that have been generated in our laboratory could potentially be used to purify mDA neurons and precursors (see below, Zhao et al. 2004).
Dopaminergic differentiation in vitro
Although neural progenitors and neurons in general can be derived from ES cells with high frequency, the production of DA neurons is less efficient. Two protocols have been developed to differentiate ES cells towards a DA fate, one involving the application of Shh and FGF8 and the other via co-culture of ES cells with PA6 stromal cells (Kawasaki et al. 2000; Lee et al. 2000). General DA neuronal markers including TH, VMAT, DAT and AADC were found to be expressed in these ES cell-derived cultures. Furthermore, some cells in these cultures also express the mDA-specific marker genes En1 and Pitx3. However, the extent to which those ES derived DA neurons express midbrain phenotype and the relative proportion of cells that are of midbrain character, remains to be established.
Co-culture with stromal cells
Kawasaki and co-workers have identified a stromal cell-derived inducing activity (SDIA) that efficiently induces neural differentiation from ES cells, with a high proportion of TH-positive neurons (Kawasaki et al. 2000). This method involves co-culture with bone marrow-derived PA6 stromal cells and does not require EB formation or RA treatment. The nature of SDIA is unclear as PA6 neural inducing activity remains when the cells are fixed and thus unable to secrete soluble factors, or when the PA6 cells are separated from the ES cells by a filter. The authors suggest that SDIA may be a secreted factor that is secondarily tethered to the cell surface, as treatment with heparin removes the neural inducing activity. TH-positive neurons appeared between days 6 and 8 of the induction period and constituted 30% of the total neurons in the culture.
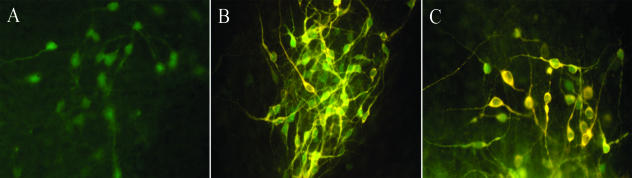
We have analysed DA differentiation of Pitx3–GFP ES cells with the PA6 co-culture method. We found that the vast majority of Pitx3–GFP-positive neurons express TH (Fig. 3). However, only around 10–15% of all TH- or DAT-expressing cells co-express Pitx3–GFP, suggesting that not all of the DA neurons generated by this method display a mesencephalic character (Zhao et al. 2004). Indeed, many in vitro studies assess mDA neuronal phenotype by determining TH expression in cultured cells. However, this has limitations as TH is expressed in all catecholaminergic neuronal types, including adrenergic and noradrenergic neurons. Furthermore, other DA markers such as DAT, AADC and dopamine receptors are expressed in DA cells in other CNS regions, including the olfactory bulb and the diencephalic DA cell groups located in the arcuate nucleus, periventricular nucleus, caudal thalamus and hypothalamic regions (Bjorklund & Lindvall, 1984). Accordingly, a system with an mDA-specific reporter such as Pitx3–GFP is a more relevant tool with which to study mDA neuronal differentiation.
Fig. 3.
Pitx3–GFP expression in DA neurons derived from Pitx3–GFP ES cells, which are heterozygous for Pitx3 locus, using the PA6 co-culture method after 14 days in culture. (A) Direct visualization of GFP in ES-derived neurons. (B,C) Double immunostaining for GFP and TH.
Inductive factors and cytokines
Another approach for DA fate induction from ES cells involves the addition of Shh and FGF8 (Lee et al. 2000). This protocol starts with a short period of EB differentiation followed by medium-based selection for nestin-positive neural precursor cells. The neural precursors are then expanded using FGF2 before being induced to a DA fate by addition of FGF8, Shh and ascorbic acid. The authors demonstrated that it was possible to obtain TH expression in over 30% of the neurons.
Using a similar approach, Rolletschek et al. (2001) use an array of survival-promoting factors in the production of DA neurons from ES cells. They found that a cocktail containing interlukin-1β (IL-1β), glial cell line-derived neurotrophic factor (GDNF), neurturin (NTN), transforming growth factor-β3 (TGF-β3) and dibutyryl-cAMP (db-cAMP) could enhance RNA expression levels of En-1, Mash-1, dopamine receptor 2 (D2R), TH and Nurr1 and increase the proportion of TH-positive and DAT-positive neurons from around 20% to around 40% (Rolletschek et al. 2001).
Nurr1 overexpression
Wagner et al. (1999) were the first to exploit Nurr1 in promoting DA differentiation by engineering an immortalized neural stem cell line to overexpress Nurr1 stably. In this system, Nurr1 overexpression alone did not result in the production of DA neurons. However, 80% of the Nurr1-overexpressing cells initiate TH expression when they were co-cultured with ventral midbrain type 1 astrocytes. Later studies demonstrated that Nurr1 is able to induce TH expression in CNS precursors derived from cortex, midbrain and lateral ganglionic eminence and from ES cells (Chung et al. 2002; Kim et al. 2002; Hwang et al. 2003). Enhanced production of TH-positive neurons was associated with an increase in the expression level of AADC, DAT, VMAT and Pitx3 in Nurr1-expressing cultures as determined by RT-PCR. Recently, Nurr1 has been shown to induce a DA-like cell phenotype in non-neuronal cells, specifically inducing expression of TH, AADC, AHD2 and calbindin (Sonntag et al. 2004). This suggests that Nurr1 may be involved in controlling the biochemical neurotransmitter phenotype of DA neurons independent of their neuronal cell fate specification.
Pitx3–GFP as reporter to track midbrain-specific DA differentiation from stem cells
We have generated ES cells and transgenic mice with a green fluorescent protein (GFP) reporter gene inserted into the Pitx3 locus (Zhao et al. 2004). DA differentiation of these ES cells was investigated by co-culturing with PA6 stromal cells. Pitx3–GFP-positive cells were detected after13–14 days in culture (Fig. 3) and the vast majority of them express the pan neuronal marker Tublin beta 3 and the DA marker TH. ES cell-derived Pitx3-expressing neurons respond to the trophic factors GDNF, NT3 and IGF1 and are sensitive to the DA neuronal neurotoxin MPTP. Therefore, ES cell-derived Pitx3-expressing DA neurons have similar biological properties to primary mDA cells. However, we have shown that Pitx3–GFP is co-expressed only in a proportion of ES cell-derived TH-positive, DAT-positive neurons and that many TH-positive neurons are GFP-negative. These results suggest that ES cell-derived dopaminergic neurons are heterogeneous and do not all exhibit mesencephalic identity. This is a significant finding in the field because functional repair of PD appears to be specifically associated with mesencephalic grafts, as demonstrated by transplantation experiments (Hudson et al. 1994). Our study has highlighted the need for using mDA-specific markers in studies relevant to PD instead of general DA neuronal markers.
We have previously established protocols for efficient generation of neuroepithelial stem cells from ES cells by exploiting Sox-B expression (Li et al. 1998; Ying et al. 2003). Such a system has been employed in testing and screening for key molecules regulating neural differentiation by microarray and cDNA subtraction experiments, as well as being used as a tool for functional testing of gene candidates derived from the screen (Aubert et al. 2002, 2003; Zhao et al. 2004). Similarly, the Pitx3–GFP ES cells could be used to track the fate of mDA neuron differentiation from ES cells and/or neural stem cells derived from embryonic or adult Pitx3–GFP mouse brain. In combination with transgene expression or knock-down approaches in ES cells, the Pitx3–GFP system may be used to identify key determinants governing mDA fate and to investigate mechanisms of regulatory genes in mDA neuron differentiation. Furthermore, the Pitx3–GFP ES cells could be used for pharmacological assays to analyse the activity of potential drugs, and as a source of mDA neurons for transplantation studies in animal models of PD.
In summary, recent studies have identified some of the pathways responsible for early patterning events within the midbrain and number of regulatory factors that are important for transcriptional control of mDA neuron development. However, many questions remain; for example, do the known mDA transcription factors interact to regulate an mDA neuronal fate, and what are their downstream genetic targets and upstream activators? Furthermore, neurons of the SN and VTA are functionally distinct yet no known genetic markers have been identified to distinguish between them. Therefore, there is a need to gain a better understanding of the transcriptional profile of mDA neurons. These studies would facilitate the identification of potential mDA regulators and the knowledge gained would enable directed differentiation in vitro for biomedical applications.
References
- Alberi L, Sgado P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131:3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. V. Thymidine-radiographic study of the time of origin of neurons in the midbrain tegmentum. J Comp Neurol. 1981;198:677–716. doi: 10.1002/cne.901980409. [DOI] [PubMed] [Google Scholar]
- Asbreuk CH, Vogelaar CF, Hellemons A, Smidt MP, Burbach JP. CNS expression pattern of Lmx1b and coexpression with ptx genes suggest functional cooperativity in the development of forebrain motor control systems. Mol Cell Neurosci. 2002;21:410–420. doi: 10.1006/mcne.2002.1182. [DOI] [PubMed] [Google Scholar]
- Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- Aubert J, Stavridis MP, Tweedie S, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci USA. 2003;100:11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- Barberi T, Klivenyi P, Calingasan NY, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol 21, 1200–7 gene promoter. J Neurochem. 2003;74:1829–1837. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- Bear MF, Connors BW, Paradiso MA. Neuroscience: Exploring the Brain. USA: Lippincot, Williams & Wilkins; 2001. [Google Scholar]
- Bjorklund A, Lindvall O. Dopamine-containing systems in the CNS. In: Bjorklund A, Hokfelt T, editors. Classical Transmitters in the CNSPart 1. Vol. 2. Amsterdam: Elsevier; 1984. pp. 55–111. [Google Scholar]
- Brodski C, Weisenhorn DM, Signore M, et al. Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J Neurosci. 2003;23:4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G, Wagner J, Rodriguez FJ, et al. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla P, Smidt MP, O'Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–1837. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- Chung S, Sonntag KC, Andersson T, et al. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Joyner AL. Expression patterns of the homeo box-containing genes En-1 and En-2 and the proto-oncogene int-1 diverge during mouse development. Genes Dev. 1988;2:1736–1744. doi: 10.1101/gad.2.12b.1736. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, et al. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Hanaway J, McConnell JA, Netsky MG. Histogenesis of the substantia nigra, ventral tegmental area of Tsai and interpeduncular nucleus: an autoradiographic study of the mesencephalon in the rat. J Comp Neurol. 1971;142:59–73. doi: 10.1002/cne.901420105. [DOI] [PubMed] [Google Scholar]
- Hudson JL, Bickford P, Johansson M, Hoffer BJ, Stromberg I. Target and neurotransmitter specificity of fetal central nervous system transplants: importance for functional reinnervation. J Neurosci. 1994;14:283–290. doi: 10.1523/JNEUROSCI.14-01-00283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res. 2003;114:123–131. doi: 10.1016/s0169-328x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Herrup K, Auerbach BA, Davis CA, Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science. 1991;251:1239–1243. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kim JH, Auerbach JM, Rodriguez-Gomez JA, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- Kim JY, Koh HC, Lee JY, et al. Dopaminergic neuronal differentiation from rat embryonic neural precursors by Nurr1 overexpression. J Neurochem. 2003;85:1443–1454. doi: 10.1046/j.1471-4159.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- Lauder JM, Bloom FE. Ontogeny of monoamine neurons in the locus coeruleus, Raphe nuclei and substantia nigra of the rat. I. Cell differentiation. J Comp Neurol. 1974;155:469–481. doi: 10.1002/cne.901550407. [DOI] [PubMed] [Google Scholar]
- Lebel M, Gauthier Y, Moreau A, Drouin J. Pitx3 activates mouse tyrosine hydroxylase promoter via a high-affinity binding site. J Neurochem. 2001;77:558–567. doi: 10.1046/j.1471-4159.2001.00257.x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, McKay RDG. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Li M, Pevny L, Lovell-Badge R, Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129:5269–5277. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, McMahon J, Bradley A. The midbrain-hindbrain phenotype of Wnt-1/Wnt-1 mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci USA. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea KS. Embryonic stem cell models of development. Anat Rec. 1999;257:32–41. doi: 10.1002/(SICI)1097-0185(19990215)257:1<32::AID-AR6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Reichardt LF. Roles of Wnt proteins in neural development and maintenance. Curr Opin Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, Tabin C. Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell. 1995;83:631–640. doi: 10.1016/0092-8674(95)90103-5. [DOI] [PubMed] [Google Scholar]
- Rieger DK, Reichenberger E, McLean W, Sidow A, Olsen BR. A double-deletion mutation in the Pitx3 gene causes arrested lens development in aphakia mice. Genomics. 2001;72:61–72. doi: 10.1006/geno.2000.6464. [DOI] [PubMed] [Google Scholar]
- Rolletschek A, Chang H, Guan K, Czyz J, Meyer M, Wobus AM. Differentiation of embryonic stem cell-derived dopaminergic neurons is enhanced by survival-promoting factors. Mech Dev. 2001;105:93–104. doi: 10.1016/s0925-4773(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Murray JC, Reiter R, Hrstka RF, Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9:1575–1585. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- Simon HH, Saueressig H, Wurst W, Goulding MD, O'Leary DD. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci. 2001;21:3126–3134. doi: 10.1523/JNEUROSCI.21-09-03126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, van Schaick HS, Lanctot C, et al. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Smits SM, Bouwmeester H, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- Smits SM, Ponnio T, Conneely OM, Burbach JP, Smidt MP. Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci. 2003;18:1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x. [DOI] [PubMed] [Google Scholar]
- Sonntag KC, Simantov R, Kim KS, Isacson O. Temporally induced Nurr1 can induce a non-neuronal dopaminergic cell type in embryonic stem cell differentiation. Eur J Neurosci. 2004;19:1141–1152. doi: 10.1111/j.1460-9568.2004.03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Munckhof P, Luk KC, Ste-Marie L, et al. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- Wagner J, Akerud P, Castro DS, et al. Induction of a midbrain dopaminergic phenotype in Nurr1-overexpressing neural stem cells by type 1 astrocytes. Nat Biotechnol. 1999;17:653–659. doi: 10.1038/10862. [DOI] [PubMed] [Google Scholar]
- Wallen A, Zetterstrom RH, Solomin L, Arvidsson M, Olson L, Perlmann T. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999;253:737–746. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- Wallen A, Perlmann T. Transcriptional control of dopamine neuron development. Ann NY Acad Sci. 2003;991:48–60. doi: 10.1111/j.1749-6632.2003.tb07462.x. [DOI] [PubMed] [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Williams R, Perlmann T, Olson L. Cellular expression of the immediate early transcription factors Nurr1 and NGFI-B suggests a gene regulatory role in several brain regions including the nigrostriatal dopamine system. Brain Res Mol Brain Res. 1996;41:111–120. doi: 10.1016/0169-328x(96)00074-5. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- Zhao S, Maxwell S, Jimenez-Beristain A, et al. Generation of embryonic stem cells and transgenic mice expressing green fluorescence protein in midbrain dopaminergic neurons. Eur J Neurosci. 2004;19:1133–1140. doi: 10.1111/j.1460-9568.2004.03206.x. [DOI] [PubMed] [Google Scholar]
- Zhou QY, Palmiter RD. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209. doi: 10.1016/0092-8674(95)90145-0. [DOI] [PubMed] [Google Scholar]