Abstract
A zebrafish M2 muscarinic acetylcholine receptor (mAChR) gene was cloned. It encodes 495 amino acids in a single exon. The derived amino acid sequence is 73.5% identical to its human homologue.
Competitive binding studies of the zebrafish M2 receptor and [3H]-NMS gave negative log dissociation constants (pKi) for each antagonist as follows: atropine (9.16)>himbacine (8.05)⩾4-DAMP (7.83)>AF-DX 116 (7.26)⩾pirenzepine (7.18)⩾tropicamide (6.97)⩾methoctramine (6.82)⩾p-F-HHSiD (6.67)>carbachol (5.20). The antagonist affinity profile correlated with the profile of the human M2 receptor, except for pirenzepine.
Reverse transcription polymerase chain reaction and Southern blotting analysis demonstrated that the M2 mAChR mRNA levels increased during the segmentation period (12 h post-fertilization; h.p.f.) in zebrafish. By whole-mount in situ hybridization, the M2 mAChR was first detectable in the heart, vagus motor ganglion, and vagus sensory ganglion at 30, 48 and 60 h.p.f., respectively.
The muscarinic receptor that mediates carbachol (CCh)-induced bradycardia was functionally mature at 72 h.p.f. The effect of CCh-induced bradycardia was antagonized by several muscarinic receptor antagonists with the order of potency (pIC50 values): atropine (6.76)>methoctramine (6.47)>himbacine (6.10)>4-DAMP (5.72)>AF-DX 116 (4.77), however, not by pirenzepine, p-F-HHSiD, or tropicamide (<10 μM).
The effect of CCh-induced bradycardia was abolished completely before 56 h.p.f. by M2 RNA interference, and the bradycardia effect gradually recovered after 72 h.p.f. The basal heart rate was increased in embryos injected with M2 mAChR morpholino antisense oligonucleotide (M2 MO) and the effect of CCh-induced bradycardia was abolished by M2 MO in a dose-dependent manner. In conclusion, the results suggest that the M2 mAChR inhibit basal heart rate in zebrafish embryo and the M2 mAChR mediates the CCh-induced bradycardia.
Keywords: M2 muscarinic acetylcholine receptor, zebrafish, embryo, bradycardia, RNA interference, antisense, morpholino, knockdown
Introduction
Muscarinic acetylcholine receptors (mAChRs) are members of the superfamily of G-protein-coupled receptors with seven putative transmembrane domains. On the basis of molecular biological characterization, five subtypes (M1–M5) of mAChR genes have been identified (Bonner et al., 1988; Liao et al., 1989), with each of these receptor proteins encoded by an intronless gene exhibiting a unique pharmacological profile (Bonner et al., 1988; Peralta et al., 1987a). The mAChRs can be divided into two groups according to their biological function. M1, M3 and M5 preferentially stimulate phospholipase C activity, whereas M2 and M4 preferentially inhibit adenylyl cyclase activity (Felder, 1995; Wess, 1996).
The mAChRs have been found in cardiac and smooth muscles, central and peripheral neurons, and various exocrine glands (Ashkenazi & Peralta, 1994). In the heart, mAChRs regulate the rate and force of contraction (Caulfied, 1993). In the CNS, mAChRs have been implicated in such diverse processes as the control of movement, learning and memory, circadian rhythms, drinking, nociception, formation of ocular dominance columns, and seizure activity (Caulfield, 1993; Nathanson, 1987; Wess, 1996). The M2 subtype is commonly believed to be the only functional mAChR found in the mammalian heart (Peralta et al., 1987b), and it contributes to decreases in both the heart rate and the force of contraction. However, recent studies have found that other subtypes of mAChR are also expressed in cardiac tissue. A significant amount of M3, M4, and M5 can also be found in the chicken heart (Creason et al., 2000; McKinnon & Nathanson, 1995), and all five subtypes of the mAChRs are expressed in the human heart (Wang et al., 2001).
The zebrafish is a new model system for studies of vertebrate development and genetics. External development and optical clarity during embryogenesis provide advantage with respect to visual analysis of early developmental processes. The zebrafish is particularly amenable to the study of cardiac function such as changes in heart rate, which can be directly observed under a stereomicroscope. Besides, it is easy to inject the circulatory system of zebrafish embryos with trace compounds. All these advantages make the zebrafish a good in vivo model for the analysis of the effects of agonists and antagonists on the heart rate regulation. The zebrafish's high fecundity and short generation time further facilitates genetic analysis. Hundreds of mutant zebrafish phenotypes have been identified (Nusslein-Volhard, 1994) and many of these resemble human clinical disorders. Some studies indicate that double-stranded RNA interference (RNAi) can inhibit gene expression in the zebrafish (Wargelius et al., 1999; Li et al., 2000) and that morpholino-modified antisense oligonucleotides (MOs) are effective and specific translational inhibitors in zebrafish embryos (Nasevicius & Ekker, 2000). The creation of critical genetic reagents and methods, coupled with the rapid progress of the zebrafish genome project, is bringing this model system to its full potential for the study of vertebrate physiology, pharmacology, and human diseases (Dooley & Zon, 2000).
In this paper, we described the cloning and pharmacological characterization of the zebrafish M2 mAChR. We studied the expression patterns and functional maturation of the M2 mAChR during embryogenesis. We established an in vivo assay system to test drugs that act on mAChRs. We also critically investigated the role of M2 mAChR in the regulation of heart rate in zebrafish by RNAi and MO gene knockdown methods. These approaches can be applied to all receptors that play roles in heart rate regulation.
Methods
Animals
Zebrafish (Danio rerio) were obtained from Dr D. Barnes's laboratory (Oregon State University Corvallis, OR, U.S.A.) in 1996 and thereafter bred and reared in the Zebrafish Room, Institute of Zoology at 25–30°C under 14 h : 10 h light–dark conditions. Embryos were raised in a 28.5°C incubator and stated as described by Kimmel et al. (1995). Fertilized eggs were obtained from 3–9-month-old parents. Embryos more than 24 h old were treated with 0.003% phenylthiourea in 10% Hank's solution (Westerfield, 1994) to prevent melanin formation.
Cloning of the zebrafish mAChR genes
To clone the zebrafish mAChR genes, genomic DNA were prepared from adult male zebrafish, and amplified by PCR with the degenerated forward primer 5′-TGGYTIGCIYTIGAYTAYBTIGC-3′ and the reverse primer 5′-RATRTTRTAIGGIGTCCAIGT-3′. The PCR reaction was performed under the following conditions: 35 cycles at 94°C for 30 s, at 60°C for 30 s and at 72°C for 1 min; a final extension at 72°C for 10 min and a final holding at 4°C. Two DNA fragments were amplified and cloned into the pGEM-T vector for DNA sequencing. After DNA sequencing, one 980-bp fragment was identified as the putative zebrafish M2 mAChR, and the other 920-bp fragment was identified as the putative M5 mAChR. The 980-bp fragment was used as a template to synthesize a 32P-labelled probe for screening of a zebrafish genomic DNA library (Provided by Dr C.Y. Chang, Institute of Zoology). The genomic DNA library was consutructed in the FIX lambda vector (Stratagene, La Jolla, CA, U.S.A.). The recombinant phages were amplified with E. coli XL1 Blue MRA (P2) (Stratagene), and plated and screened exactly as described previously (Liao et al., 1989). A MarathonTM cDNA amplification kit was used for rapid amplification of cDNA ends (RACE) of the M2 mAChR. The manufacturer's protocol was followed. The 5′-RACE sequence was used for the design of the double-stranded RNA (dsRNA) and MOs.
DNA sequencing and analysis
After purification of the hybridizing phage DNA, DNA sequencing was carried out with an ABI prism 377 automated DNA sequencer (Applied Biosystems, Foster City, CA, U.S.A.). The DNA sequence and the deduced amino acid sequence were analysed for similarity to known sequences throught the NCBI Blast Network Service. The MAP, PILEUP, FASTA, and BESTFIT programs from the Wisconsin sequence analysis package of the Genetics Computer Group (GCG) were also used.
Transfection of Ltk− cells with zebrafish mAChR expression plasmid DNA
The recombinant phage DNA encoding the complete open reading frame of the zebrafish M2 mAChR was amplified by PCR (forward primer: CGGAATTCGCGAGCTCACCATGGATACAAT; backward primer: CGAGATCTGTGTGTGAGTGTGTGTAAGCGG) and subcloned into an expression vector pcDNA3.1/Neo (Invitrogen, Carlsbad, CA, U.S.A.) as pcDNA–zM2. Murine Ltk-cells was grown in Medium Essential Medium Alpha Medium containing 10% foetal calf serum, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and maintained in a 5% CO2 environment at 37°C. Cells seeded at 1×105 cells per well in 6-well multidishes were transfected with pcDNA-zM2 plasmid DNA by the lipofectamine method 48 h after plating following the manufacturer's procedure. The medium was replaced 24 h after transfection, and 400 μg ml−1 G418 was added 24 h later. Stable cell clones were transferred to 24-well plates, then were expanded to 6-well plates and confirmed by a receptor binding assay using [3H]NMS as the radiolabelled ligand following the procedure described previously (Liao et al., 1989). The cell clone LzM2-42 giving the highest [3H]NMS binding site assays was then used for further experiments.
Saturation binding and competition-binding assays
A saturation-binding assay was performed using nine concentrations of [3H]NMS ranging from 62.5 pM to 5 nM added to the LzM2 cells grown to confluence in 6-well plates as described previously (Liao et al., 1989). Nonspecific binding of [3H]NMS was defined as the radioactivity bound to intact cells in the presence of atropine (1 μM). Linear regression was performed, and the correlation coefficient (r) was calculated using Sigma-Plot. The results were analysed on a Scatchard plot that allows the calculation of the BMax and the KD. The values represent the average of duplicate measurements performed on four individual preparations.
Competition-binding assays with various antagonists were carried out as described previously (Liao et al., 1989). LzM2 cells was incubated with 1 nM [3H]NMS medium containing various concentrations of AF-DX 116, atropine, CCh, 4-DAMP, p-F-HHSiD, himbacine, pirenzepine, or methoctramine, respectively. The follow-on reaction was continued as described above from the saturation-binding assays. Competition experiments were analysed using Sigma-Plot. Antagonist inhibition experiments were analysed by nonlinear curve fitting using the least-squares method, with Ki being calculated by the method of Cheng & Prusoff (1973) using the formula Ki=IC50 (1+[ligand] KD−1)−1. To ensure the validity and accuracy of the competitive binding assays, linear regression was performed on the per cent bound versus the ratio of bound over free ligand, and only data with regression coefficients of >0.9 were accepted for analysis.
Heart rate measurements
Zebrafish embryos of different development stages were collected, and the heart rate was measured. This was done by visually counting the heartbeats on a colour monitor, which transmitted images from an SZH dissecting microscope (Olympus, NY, U.S.A.) by CCD (JVC, Tokyo, Japan). Microelectrodes with 2- to 5-μm tips were made by pulling glass capillaries (1 mm OD) with a glass microelectrode puller (model PA91, Narishige Scientific Instrument, Tokyo, Japan) and a microgrinder (model EG-400, Narishige Scientific Instrument, Tokyo, Japan). The zebrafish embryos were anaesthetized with 0.64 mM tricaine (Sigma Chemical Co., St. Louis, MO, U.S.A.) in embryo medium as described by Westerfield (1994) for 3 min and held for injection using a holding pipette applied to the side of the yolk. About 0.7 nl of CCh (1 mM) with or without different concentrations of antagonists was injected into the right common cardinal vein near the pectoral fin bud by the microelectrode connected to the pressure injection apparatus (model PLI-188, Nikon, Garden City, NY, U.S.A.). In order to control the injection volume, phenol red (final 0.2%) was used to dilute the drugs. Phenol red alone did not influence the embryonic heart rate (data not shown). After injection and for 3-min circulation, the heart rate reached a plateau and was regular. And then the heartbeats were counted for another 1 min with the aid of a counter and a timer. The embryos were then returned to the embryonic medium without tricaine for recovery. The zebrafish heart rate gradually returned to control levels within 30 min. It means that the drugs were cleared or inactivated in a given zebrafish. All experiments were carried out at a room temperature of 25°C. The cardiac output of zebrafish (72 h.p.f.) is 50 nl min−1 (Fritsche et al., 2000) and the heart rate is 142 beats min−1. The injection volume (0.7 nl) is almost equal to two stroke volumes. We observed that it takes about 20 heartbeats for the injected dye to accomplish one circulation. Therefore, the blood concentration of drugs was estimated to be 10 fold diluted than the injection concentration.
Reverse transcription polymerase chain reaction (RT–PCR) and Southern blotting
When different development stage embryos were collected, they were rinsed in ice-cold phosphate buffer saline (mM: NaH2PO4 20, NaCl 150, pH 7.4) and immediately frozen in liquid nitrogen. Total RNA was isolated from these samples using TRIZOL reagent, a phenol/chloroform extraction, and ethanol precipitation. Reverse transcription was performed with oligo-dT primer with 1 μg of total cellular RNA used to synthesize first-strand cDNA using a first-strandTM cDNA synthesis kit. Two microlitres of the cDNA reaction was used in each PCR reaction. PCR was performed with the zebrafish M2-specific primers (sense: 5′-CGAAGATGGCAGGAATGATGAT-3′, antisense: 5′-CATTGGCTGCTGCTGACGAGGG-3′). PCR was run as 35 cycles at 94°C for 30 s, at 65°C for 30 s, and at 72°C for 1 min, with a final extension at 72°C for 10 min and holding at 4°C. RT–PCR for α-actin was conducted the same as described above except that one pair of α-actin-specific primers was used. They are 5′-TCACACCTTCTACAACGAGCTGCG-3′ and 5′-GAAGCTGTAGCCTCTCTCGGTCAG-3′. The PCR products were analysed by electrophoresis on a 1.2% agarose gel and subsequently visualized with ethidium bromide using a gel documentation system. The total RNA concentration was determined spectrophotometrically (Hitachi U-2000).
Southern blot analysis of the RT–PCR products was performed as described earlier (Hwang et al., 1997). Membranes were prehybridized for 2 h in the prehybridization buffer (50% formamide. 5×SSC, 0.5% SDS, 1% blocking solution, and 0.5% sodium lauroyl sarcosine) at 42°C. Hybridization was conducted in prehybridization buffer containing 25 ng ml−1 of 1.5 kb digoxigenin (DIG)-labelled DNA probes at 42°C overnight. CSPD chemiluminescence detection was conducted following the manufacturer's protocol (Roche Applied Science, Mannheim, Germany).
Whole-mount in situ hybridization
DIG-labelled riboprobes were prepared from a pcDNA3.1/Hygro plasmid harbouring the sense or antisense M2 receptor coding region following the manufacturer's protocols (Roche Applied Science). T7 RNA polymerase was used for in vitro transcription of all probes. Whole mount in situ hybridization was based on the method described by Chen & Fishman (1996). Embryos were fixed in 4% paraformaldehyde (PFA) in phsophate-buffered saline (PBS) at 4°C overnight, rinsed in PBS and manually dechorionated. Embryos were dehydrated in 100% methanol and stored at −20°C. In the beginning, embryos were rehydrated in a graded series of 75%, 50%, 25% methanol/PBS containing 0.1% Tween-20 (PBST), and washed in PBST. The embryos were treated with proteinase K (5, 10, 15, 25 and 50 μg ml−1, 25 min for 30, 36, 48, 60 and 72 h.p.f. embryos, respectively) to facilitate the probe penetration during in situ hybridization. Embryos were refixed in 4% PFA for 20 min, washed with PBS, and incubated in 1 ml of 0.1 M triethanolamine containing 2.5 μl acetic anhydride for 1 h. After washing, the embryos were incubated in hyb-buffer (50% formamide, 5×SSC, 0.1% Tween 20, pH 5.0) at 68° for 5 min, and prehybridized with hyb+-buffer (hyb-buffer plus 500 μg ml−1 yeast tRNA and 50 μg ml−1 heparin) at 68°C for 2 h. The hybridization was performed with sense or antisense M2 DIG-labelled probes at 68°C for 60 h. Then the embryos were washed in a series of 75, 50, 25% hyb-buffer/2×SSC, 2×SSC and 0.2×SSC at 68°C followed by washing in a series of 75, 50, 25% 0.2×SSC/PBST at room temperature. Embryos were immersed in blocking solution (PBST containing 2 mg ml−1 bovine serum albumin and 5% sheep serum) for 2 h and incubated with pre-absorbed anti-DIG-alkaline phosphatase (1 : 2000 in blocking solution) for 2 h. After washing with PBST, the embryos were incubated with freshly made detection buffer (0.1 M Tris-HCl, pH 9.5, 50 mM MgCl2, 0.1 M NaCl, 0.1% Tween-20) and colour development was carried out using detection buffer containing NBT/BCIP. The reaction proceeded for 3–4 h. After colour development, embryos were fixed in 4% PFA, mounted in 50% glycerol and photographed under a stereomicroscope.
Synthesis and microinjection of dsRNA
To prepare dsRNA for injection, a 174-bp M2 5′UTR sequence was amplified by PCR with the sense primer 5′-GAGAAGAGCTCCGACTGACCGTC-3′ and reverse primer 5′-TGGTGAGCTCGCTCAGGTACAG-3′. This fragment was then subcloned into the pGEM-T Easy vector. A 742-bp EGFP fragment from the pEGFP-1 vector was subcloned into the NotI and BamHI restriction sites of the pcDNA3.1/Hygro vector. Synthesis and annealing of the dsRNA were performed essentially as described by Li et al. (2000). The l-cell-stage embryos were microinjected with 100 fg (M2) or 430 fg (EGFP) dsRNA (approx 5.25×106 molecules) under a dissecting microscope with pulled microcapillary pipettes to deliver the dsRNA solution. In order to control the injection volume, phenol red (final 0.2%) was used to dilute the dsRNA products. The injected embryos were subsequently incubated in embryo medium at 28.5°C. Finally, the heart rate was measured as described above.
Microinjection of MOs
The MOs were obtained from Gene Tools, LLC (Philomath, OR, U.S.A.). The selected sequences were based on design parameters according to the manufacturer's recommendations, namely 25-mer MOs of ∼50% G+C content with no predicted internal hairpins. Sequences were as follows: M2 MO, 5′-CACTCAGATCGCTATTGGCAGGACG-3′; oligonucleotide with four mismatch base (M2 4 mp MO) was used as control for M2 MO, 5′-CACTCAGATGGCTATTCGCACGTCG-3′. MOs were solubilized in water at the concentration of 5 mM (∼40 μg μl−1). The stock solution was diluted to a working concentration with water and phenol red before injection as described above. The l-cell-stage embryos were injected with one of four solutions at different dose concentrations (10, 100, 300, 1000 μM) and the injected volume was about 1 nl. This was equivalent to 0.08, 0.8, 2.4 and 8 ng per embryo. The heart rate was then measured as described above. Higher doses (8 ng per embryo) of the M2 MO resulted in phenotypic changes (data not shown) in about 50% of fishes, which were not measured for heart rate.
Materials
Atropine sulphate, CCh, 4-DAMP, p-F-HHSiD, (+)-himbacine, methoctramine, and pirenzepine were obtained from Research Biochemicals International (Natick, MA, U.S.A.). AF-DX 116 and tropicamide were obtained from Tocris Cookson (Bristol, U.K.). The oligonucleotide primers were synthesized by DNAFax (Taipei, Taiwan). 1-[[N-methyl-3H]scopolamine methyl chloride (78 Ci/mmol) was purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Cell culture medium, G418, Lipofectamine and TRIZOL reagent were purchased from Life Technologies (Carlsbad, CA, U.S.A.). The pGEM-T vector was obtained from Promega (Madison, WI, U.S.A.). The MarathonTM cDNA amplification kit, first-strandTM cDNA synthesis kit and pEGFP-1 vector were obtained from Clontech (Palo Alto, CA, U.S.A.).
Results
Cloning of the zebrafish M2 mAChR gene and amino acids alingment
To clone the muscarinic acetylcholine receptor, PCR was performed on zebrafish genomic DNA using degenerate primers based upon conserved residues of transmembrane domain IV and transmembrane domain VII. Agarose gel electrophoresis demonstrated the presence of two bands. The amplified fragments were cloned into the pGEM-T Vector for sequencing. Sequence analysis demonstrated that these fragments show a higher sequence identity to M2 and M5 (data not shown) mAChR. The fragment of M2 mAChR was used as a probe to screen a genomic zebrafish library in order to isolate a full-length clone encoding the zebrafish M2 mAChR. Several positive phage clones were obtained from genomic library and a number of the clones were sequenced. The predicted amino acid sequence encoded by the putative M2 proteins was 495 amino acids. The entire coding region sequence corresponding to the M2-subtype mAChR is shown in Figure 1a. The zebrafish M2 mAChR shared high (73.5%) amino acid identity with human M2 while it shared only 45.4, 44.0, 58.8, and 43.6% amino acid identity with the human M1, M3, M4, and M5, respectively. Comparisons of the deduced amino acid sequence with other vertebrate mAChRs demonstrated that the zebrafish M2 mAChR shares a high degree of identity with other vertebrate M2 mAChR (Figure 1b). The predicted zebrafish M2 mAChR has 73.5, 73.3, 73.3 and 74.8% amino acid identity to human, pig, rat and chicken M2, respectively.
Figure 1.
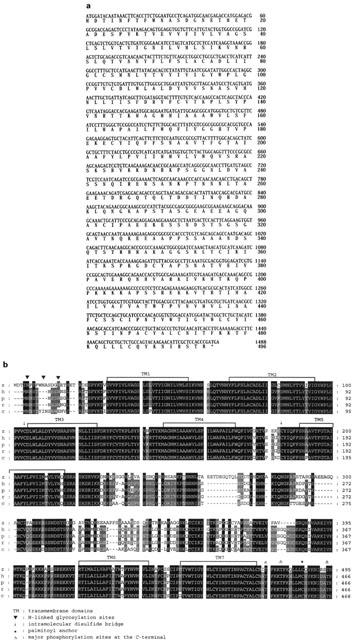
(a) Nucleotide and deduced amino acid sequences of the zebrafish M2 mAChR gene. Nucleotide residues are numbered in the 5′ to 3′ direction, beginning with the 1st nucleotide encoding the 1st in-frame methionine. The predicted amino acid sequence is shown below the nucleotide sequence. (b) Alignment of M2 mAChR peptide sequences from zebrafish (z), human (h), pig (p), rat (r) and chicken (c). Common features of mAChRs are indicated above the zebrafish sequence. GenBank accession numbers of zebrafish, human, pig, rat and chicken M2 mAChR are AY039653, P08172, P06199, P10980, and A40972, respectively.
Pharmacological characterization of M2 mAChR in transfected cell line
To confirm that the genomic clone encodes a functional mAChR, the expression construct pcDNA-zM2 containing the entire coding region was transfected into murine Ltk− cells. The stable LzM2-42 subclone exhibited highest levels of specific binding and was selected for subsequent ligand binding and functional analysis. Figure 2a depicts the results of a typical saturation analysis. The results shown in Figure 2a are specific binding (total binding subtracted non-specific binding). Linear regression was obtained on the replotted equilibrium binding data according to the Scatchard plot. This radioligand identified a single class of high-affinity binding sites associated with LzM2-42 cells with an apparent dissociation constant (KD) of 0.18±0.03 nM (n=3) and Bmax of 12,000±1100 sites/cell for [3H]NMS (Figure 2a).
Figure 2.
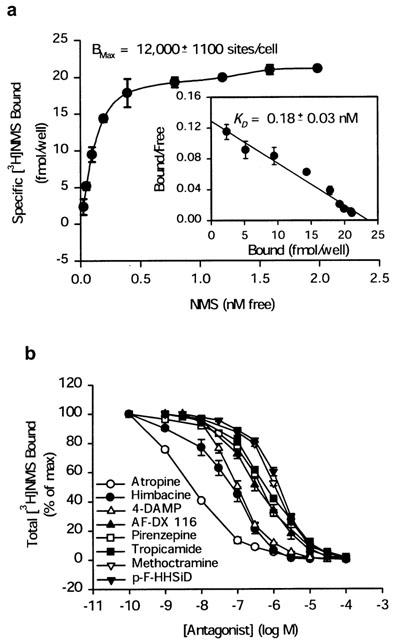
(a) Specific binding of [3H]NMS to LzM2 cells. LzM2 cells were grown confluently in 6-well plate and receptor binding assay was performed as described in Methods. Specific binding was determined by total binding subtracted the nonspecific binding in the presence of 1 μM atropine. A Scatchard plot of the specific binding data is shown in the inner panel. Bound and free are the concentrations of specifically bound and free ligands, respectively, at the time at which equilibrium was reached. The correlation coefficients (r=0.98) and KD (0.18±0.03 nM) were calculated from linear regression analysis. Each point represents the mean and s.e.mean for four independent triplicate experiments. (b) Competitive receptor binding assay of LzM2 cells. LzM2 cells were grown confluently in 6-well plate, and incubated with 1 nM [3H]NMS in the presence or absence of unlabelled muscarinic antagonists as described in Methods. Effects of atropine, pirenzepine, AF-DX 116, himbacine, methoctramine, 4-DAMP, p-F-HHSiD, and tropicamide are shown. Each point represents the mean with the s.e.mean from independent three–six duplicate experiments.
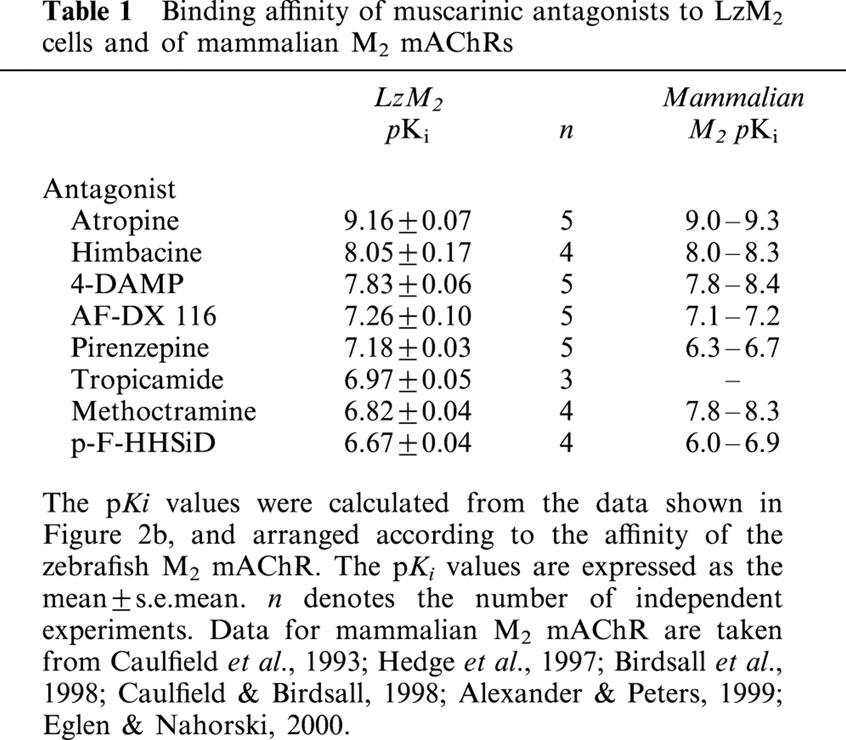
The affinity of the zebrafish mACHR for several mAChR antagonists was determined by a series of competitive receptor binding assays. These equilibrium displacement studies were carried out with the mAChR antagonists as shown in Figure 2b. The rank order of the negative log dissociation constants (pKi) for each antagonist was: atropine (9.16)>himbacine (8.05)⩾4-DAMP (7.83)>AF-DX 116 (7.26)⩾pirenzepine (7.18)⩾tropicamide (6.97)⩾methoctramine (6.82)⩾p-F-HHSiD (6.67) (Table 1).
Table 1.
Binding affinity of muscarinic antagonists to LzM2 cells and of mammalian M2 mAChRs
Expression patterns of the M2 mAChR
The temporal expression of the zebrafish M2 mAChR transcripts was studied throughout embryogenesis from the early cleavage stages to 30 day (d.p.f.) using RT–PCR and Southern blotting (Figure 3). The M2 mAChR was found at only trace levels before the segmentation period (12 h.p.f.). The M2 mRNA was present at low levels at 12 h.p.f. and was obviously expressed by 24 h.p.f. In addition, the M2 mRNA reached a maximum level between 3–3.5 d.p.f. and slightly decreased at 30 d.p.f. By whole-mount in situ hybridization, the DIG-labelled antisense RNA hybridized with the endogenous mRNA. It showed that the M2 mAChR was not expressed in the embryonic heart at 22 h.p.f. (Figure 4a). At 30 h.p.f., the M2 mAChR transcripts began to be visible in the heart (Figure 4b). The M2 mAChR was detected bilaterally along the heart to the sinus venosus at 36 h.p.f. (Figure 4c) and began to be expressed in the vagus motor ganglion at 48 h.p.f. (Figure 4d). After 60 h.p.f., it was observed in the vagus sensory ganglion and mid-hindbrain (Figure 4e) with further increase of expression at 72 h.p.f. (Figure 4f).
Figure 3.
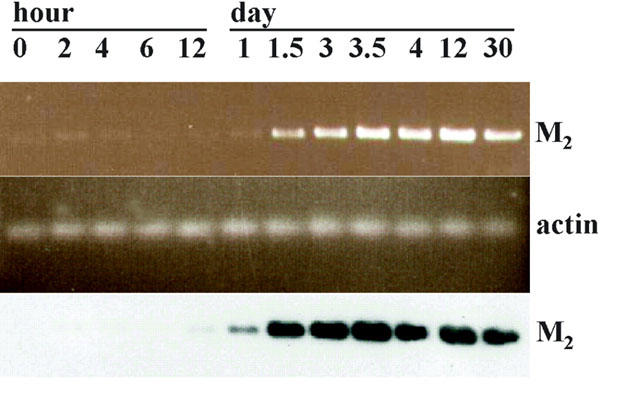
Expression of M2 mAChR mRNA during zebrafish embryonic development analysed by RT–PCR and Southern blot. DNase I treated total RNAs from the indicated developmental stages were used as a template for the RT–PCR reaction. Equal aliquots of cDNA were amplified with oligonucleotides specific for M2 mAChR (582 b.p. product) or α-actin (341 b.p. product). Five μl of the RT–PCR reaction products were loaded on a 1.2% agarose gel and stained with ethidium bromide as shown in first panel (M2) and second panel (α-actin). Southern blot analysis using a DIG-labelled M2 mAChR DNA probe was carried out to confirm the identity of the amplified DNA as shown in the third panel. Hour postfertilization (h.p.f.) or days postfertilization (d.p.f.) of the embryo stage are shown on the upper edge.
Figure 4.
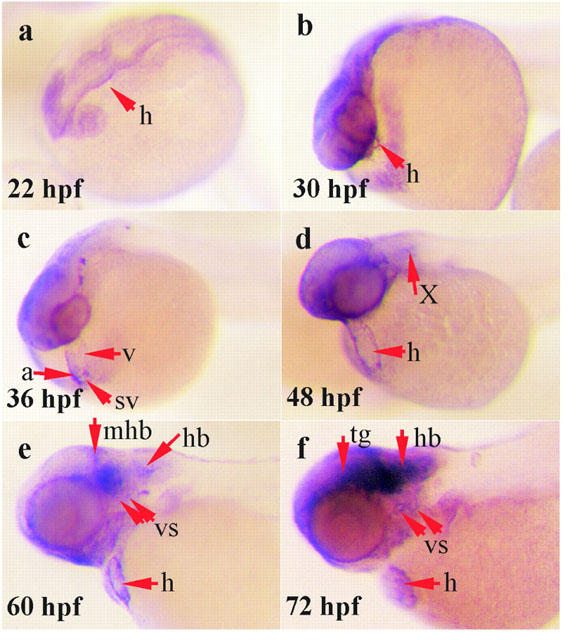
M2 mAChR mRNA was detected by whole mount in situ hybridization during the zebrafish development. Lateral view of whole zebrafish embryos was shown with dorsal side on the top. Embryos, more than 24 h.p.f. old, were treated with 0.003% phenylthiorea in 10% Hank's solution to prevent melanin formation (Westerfield, 1994). (a) Twenty-two h.p.f. stage. M2 mAChR expression has not appeared in the heart (h). (b) Thirty h.p.f. stage. M2 mAChR is beginning to express in the heart. (c) Thirty-six h.p.f. stage. M2 mAChR is strongly expressed in the heart. (d) Forty-eight h.p.f. stage. M2 mAChR expression is observed in the heart and it is beginning to express in the vagus motor ganglion (X). (e–f) Sixty and 72 h.p.f. stage. M2 mAChR is strongly expressed in the heart, detected in vagus sensory ganglion (vs), mid-hindbrain (mhb) and obviously extends out from the ventral site of the tegmentum (tg) and hindbrain (hb). The figures shown are representative of four independent experiments.
Developmental regulation of zebrafish embryo heart rate by M2 mAChR
In order to set up the zebrafish embryo as a model for cardiac mAChR research, it was necessary to first address the question of when the embryo heart is under mAChR regulation. Therefore the embryo heart rate changes during development were examined. The heart rates of the unrestrained embryos were measured directly under a stereomicroscope starting from the embryonic stage at 24–168 h.p.f. From the pharyngula period (24 h.p.f.) to the hatching period (48–72 h.p.f.), heart rate increased dramatically from 75.0±3.0 beats min−1 to 146.0±3.4 beats min−1 and was maintained at 155.7±4.2 beats min−1 thereafter (Figure 5). When hatched embryos were anaesthetized with tricaine, the heart rate decreased significantly at an early developmental stage but varied only slightly after 72 h.p.f. (Figure 5). When CCh was injected into the embryos at different developmental stages under tricaine anaesthesia, the bradycardia effect of CCh (100 μM) varied dramatically from partial inhibition (14.8±0.3% at 48 h.p.f., 58.7±3.2% at 56 h.p.f.) to maximal inhibition (79.7±2.2% at 72–168 h.p.f.) (Figure 5, insertion panel). The antagonism of CCh-induced bradycardia was studied by co-injection with mAChR antagonists at 72 h.p.f. The pIC50 values of antagonists, measured by antagonism of CCh-induced bradycardia in the zebrafish heart, were atropine (6.76±0.11)>methoctramine (6.47±0.05)>himbacine (6.10±0.04)>4-DAMP (5.72±0.11)>AF-DX 116 (4.77±0.13). The bradycardia was not antagonized by pirenzepine, p-F-HHSiD, or tropicamide (<10 μM) (Figure 6a,b).
Figure 5.
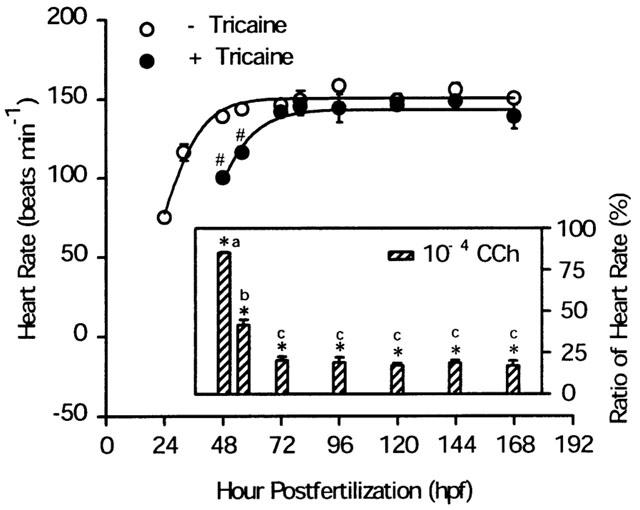
Heart rate changes and CCh-induced bradycardia during zebrafish embryogenesis. The heart rate of zebrafish embryos at different developmental stages were measured before and after being anaesthetized with tricaine. Fitted curved lines were plotted by a sigmoid 3 parameter model (Sigma-Plot). CCh-induced bradycardia (inner panel) was performed in vivo. Each embryo was anaesthetized with tricaine and microinjected with CCh. Heart rate was measured before and after CCh stimulation as described in Methods. The ratio of the heart rate (y-axis) represents the percentage of heart rate after CCh related to before CCh stimulation. All experiments were carried out at a room temperature of 25°C. Each value represents the mean and s.e.mean from three independent experiments, each carried out on 8–10 individuals. Absence of error bars indicates that the magnitude of the error is less than the symbol size. The differences between before and after treatment were tested using the Student's t-test and P<0.01 (*,#) was considered significant. Different lower-case letters (a, b, c) indicate a significant difference (P<0.01) among groups using the ANOVA and multiple range t-test.
Figure 6.
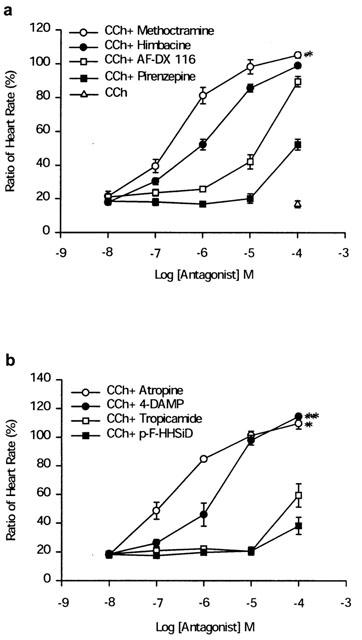
Antagonism of CCh-induced bradycardia by mAChR antagonists in zebrafish embryos. Each 72 h.p.f. embryos was anaesthetized with tricaine, the heart rate was counted first and then CCh alone or CCh with antagonists was injected into the posterior cardinal vein as described in Methods. (a) Effects of M1-selective antagonist (pirenzepine) and M2-selective antagonists (himbacine, AF-DX 116, and methoctramine) on CCh-induced bradycardia. (b) Effects of non-selective antagonist (atropine), M3-selective antagonists (p-F-HHSiD and 4-DAMP), and M4-selective antagonist (tropicamide) on CCh-induced bradycardia. The ratio of the heart rate (y-axis) represents the percentage of heart rate after drug related to before drug injection. All results are expressed as the mean±s.e.mean derived from three independent experiments, each carried out on 3–6 individuals. The differences were tested using the Student's t-test and P<0.05 (*) or P<0.01 (**) was considered significant.
Effect of RNAi and MOs of M2 mAChR on CCh-induced bradycardia
Because the embryo may contain more than one-subtype of mAChR, the pharmacological effects based on in vivo experiments may result from a mixture of mAChRs. In order to test this possibility, an RNAi technique was used to knockdown endogenous mAChR synthesis to test which is the major mAChR associated with the bradycardia effect in zebrafish embryos. The effect of the control EGFP RNAi on CCh-induced bradycardia (Figure 7) was similar to those for the wild-type at three different developmental stages. However, M2 RNAi blocks CCh-induced bradycardia by almost 100% at the 56 h.p.f. developmental stage. From the 72-h.p.f. stage, the RNAi effect is gradually lost; this is possibly due to degradation of the dsRNA. At the 96-h.p.f. stage, the RNAi effect of M2 on CCh-induced bradycardia is still significantly different from the wild-type and EGFP.
Figure 7.
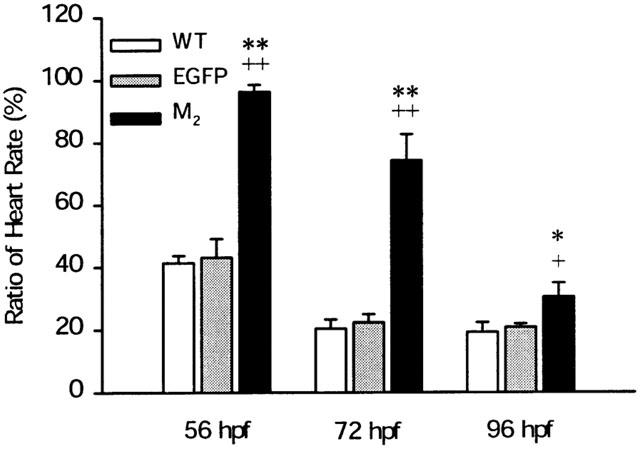
Effects of M2 and EGFP RNAi on CCh-induced bradycardia in zebrafish embryos. Each one-cell stage embryos was injected with M2 or EGFP dsRNA, and then incubated at 28.5°C until the heart rate was measured at room temperature. The same embryo was assayed for CCh-induced bradycardia at 56, 72 and 96 h.p.f. CCh-induced bradycardia was performed as described in Methods. The ratio of the heart rate is defined as in Figure 6 legend. The data represent the mean and the s.e.mean for three independent experiments, each with 8–10 individuals. A difference between the M2 dsRNA and the wild-type, or EGFP was considered significant with P<0.05 (*,+) or P<0.01 (**,++) using Student's t-test.
To confirm the results, we used MO to knockdown the endogenous M2 mAChR translation. Microinjection of M2 MO increased the zebrafish embryo basal heart rate in a dose-dependent manner but not the M2 4 mp MO (Figure 8a). The M2 MO also attenuates the CCh-induced bradycardia (Figure 8b) in a dose-dependent manner. It can block CCh-induced bradycardia by 90.5±4.6% at a concentration of 8 ng per embryo. However, the M2 4 mp MO had no influence on the CCh-induced bradycardia.
Figure 8.
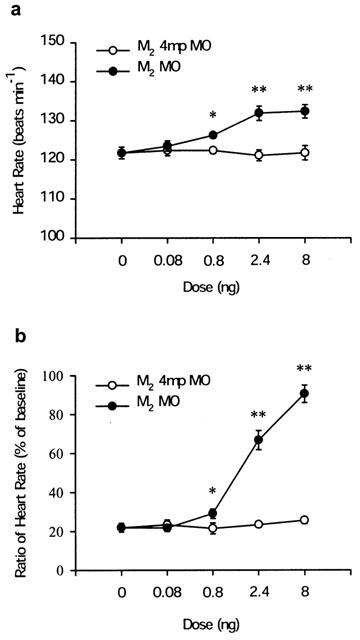
Dose-response effects of M2 MOs on zebrafish embryo basal and CCh-induced heart rate changes. Each one-cell stage embryo was injected with different doses (0.08, 0.8, 2.4 and 8 ng per embryo) of M2 MO or four mismatch base M2 MO (M2 4 mp MO), and then the embryos were incubated at 28.5°C until 72 h.p.f. for the experiments. (a) The basal heart rate of each 72 h.p.f. embryo was measured at room temperature. (b) The 72 h.p.f. embryo was injected with 10−4 M CCh at room temperature. The ratio of heart rate of CCh-induced bradycardia is defined as in Figure 6 legend. The data represent the mean and s.e.mean from three independent experiments, each with 8–10 individuals. A differences in the M2 MO compared to the M2 4 mp MO control was tested with the Student's t-test and considered significant with P<0.05 (*) or P<0.01 (**).
Discussion
In the present study, we first cloned a zebrafish M2 mAChR gene and expressed the M2 mAChR in cell line for mAChR pharmacological characterization. The temporal and spatial gene expression pattern of the M2 mAChR suggested it is involved in the regulation of heart rate. We then developed a functional assay to demonstrate that the machinery for CCh-induced bradycardia matures in the 72 h.p.f. zebrafish. By using the 72 h.p.f. zebrafish heart as an in vivo bioassay system, we compared the mAChR antagonists' potency to block the CCh-induced bradycardia. Finally, by RNAi and MO knockdown methods, we demonstrated that the CCh-induced bradycardia is mediated through the M2 mAChR.
There are some notable features of the zebrafish M2 mAChR. Firstly, the human, pig, rat and chicken M2 mAChR gene sequences all predict a polypeptide containing 466 amino acids (GenBank accession nos. P08172, P06199, P10980, and A40972, respectively), while the zebrafish M2 mAChR gene encodes a 495-amino-acid protein (Figure 1b). Secondly, zebrafish M2 mAChR has the same three putative N-glycosylation sites as do the rat, pig, and chicken receptors, compared to four N-glycosylation sites in the human receptor. All these 5 M2 mAChR genes share a common feature with several other members of the G-protein coupled receptor gene superfamily; their open reading frames are contained within a single exon. Rapid amplification of cDNA ends (RACE) and sequence analysis of zebrafish M2 mAChR show that the coding sequence is contained in a single exon, as has been observed for mammalian mAChR (Caulfield, 1993).
To confirm that the genomic clone encoded a functional mAChR, the entire coding region of the M2 mAChR was expressed in stably transfected Ltk− cells for pharmacological characterization. The apparent dissociation constant for the binding of [3H]NMS was determined by Scatchard analysis (Figure 2a) and competitive receptor binding with various muscarinic antagonists demonstrated the order of potency in the inhibition of [3H]NMS binding (Figure 2b). The antagonist affinity profile of the zebrafish M2 mAChR significantly correlates with that of mammalian M2 receptors (Table 1). However, the zebrafish M2 receptor binding affinity for pirenzepine is higher than the mammalian M2 receptor binding affinity (Caulfield & Birdsall, 1998) (Table 1). It may be due to the presence of a threonine at the ligand binding site. In the human M1/M2 chimeric receptors, each chimeric receptor that possesses a threonine (i.e., M1 sequence) at the position corresponding to M2401 had a higher affinity for pirenzepine than did the wild-type M2 receptor (Kubo et al., 1988; Lai et al., 1992). The M2 receptor mutant (A401T) also expressed enhanced affinity for pirenzepine (Ellis & Seidenberg, 2000). Furthermore, previous studies have shown that the chicken M2 receptor, which also has a threonine at this position, has a significantly higher affinity for pirenzepine than does mammalian M2 mAChR (Tietje & Nathanson, 1991).
In close association with the neurotransmitter acetylcholine (ACh) are two enzymes, choline acetyltransferase and acetylcholinesterase (AChE), which are involved in its synthesis and hydrolysis, respectively. When ACh binds to the mAChR, the mAChR couples with G protein, then the βγ subunit can bind to and directly opens a cardiac K+ channel. All of these components are related to the mediation of the mAChR signal transduction to a biological response of heart contraction. There is an appearance of AChE in the ventrolateral hindbrain region and reiculospinal neurons (Hanneman et al., 1988; Ross et al., 1992) and γ3 mRNA in the ventral hindbrain (Kelly et al., 2001) of zebrafish embryos at late somitogenesis (18–19 h.p.f.). The zebrafish K+ channel is functioning in a day 3 embryo (Baker et al., 1997). From whole-mount in situ hybridization, M2 mAChR transcripts were abundantly expressed in the sinus venosus and atrium (Figure 4c) at 36 h.p.f. and it was visible in the vagus motor and sensory ganglion before 60 h.p.f. (Figure 4). A recent study has indicated that, in the teleost heart, branches of the vagus nerves innervate the sinus venosus and atrium, causing bradycardia via muscarinic receptors (Preston & Courtice, 1995). We have shown that with tricaine treatment, the heart rate was significantly reduced at the 48–56 h.p.f. embryos but not at 72 h.p.f. embryos and thereafter (Figure 5). Tricaine can prevent transient increases in sodium ion permeability, thereby decrease excitability and block impulse conduction of nervous tissue (Letcher, 1992). Besides, they also act as noncompetitive cholinergic antagonists (Nogrady & Keshmirian, 1986). Thus tricaine may inhibit the zebrafish heart rate by blocking sodium ion permeability in the early embryos before mAChR appearance. The inhibitory effect of tricaine on heart rate is reversed after 72 h.p.f. when the mAChR regulation of heart rate become mature (Figure 5) and tricaine can function as a cholinergic antagonist. In CCh-induced bradycardia, we found that bradycardia occurred at 48 h.p.f. and reached a maximum at 72 h.p.f. The effect was blocked by atropine (Figure 6b). From the results of both RT–PCR and whole-mount in situ hybridization, the temporal and spatial expression of M2 mAChR suggests its involvement in the regulation of heart rate. We propose that the machinery and the muscarinic receptor subtypes, most likely the M2 mAChR, which mediate CCh-induced bradycardia are functionally mature in the 72-h.p.f. embryonic heart.
Since the embryo heart may contain more than one-subtype of mAChR, the in vivo effect of CCh may result from the activation of multiple mAChRs. Although the M2 subtype is commonly believed to be the only functional mAChR found in the mammalian heart (Peralta et al., 1987b), M1 receptor mRNAs have been identified in guinea-pig and rat ventricular cardiomyocytes (Colecraft et al., 1998; Gallo et al., 1993) and M3 and M4 receptors were reported to probably have a functional role in the control of K+ channels in atrial tissue of the dog heart (Shi et al., 1999). In order to analyse which subtype(s) of mAChR mediate the CCh-induced bradycardia, we utilize the pharmacological approach with various muscarinic antagonists to study their effect on CCh-induced bradycardia in zebrafish embryos (Figure 6). The results demonstrated that M2-specific antagonists had stronger blocking effect. However, since the subtype-selective antagonists are not highly specific, we further explore the muscarinic regulation of heart rate with molecular approaches.
RNAi is a method whereby an injection of dsRNA into cells results in the degradation of the existing cognate mRNA and repression of further synthesis. This technique is receiving increased attention (Gura, 2000). The success of RNAi in the invertebrates, Caenorhabditis elegans and Drosophila, is well known and now it has been extended to targeting gene expression in the vertebrate mouse (Wianny & Zernicka-Goetz, 2000; Zernicka-Goetz, 2000). Some studies indicate that RNAi can specifically knockdown gene expression in the zebrafish (Wargelius et al., 1999; Li et al., 2000), although it has nonspecific effects on early zebrafish development such as causing abnormal phenotypes (Oates et al., 2000). When injected at a low concentration (5.25×106 molecules), the nonspecific effects can be minimized. In the RNAi experiments, we found that M2 dsRNA could block CCh-induced bradycardia (Figure 7) and that the M2 dsRNA is stable in zebrafish embryo for about 3 days.
Additionally, MOs have produced well-accepted phenotypes in the analysis of zebrafish embryonic development (Nasevicius & Ekker, 2000). MOs function through an RNase–H-independent mechanism by blocking translational initiation (Summerton, 1999). We found that the antisense M2 MO could both increase the basal heart rate and blocked the CCh-induced bradycardia (Figure 8). In addition, the CCh-induced bradycardia was reversed by atropine, methoctramine, or 4-DAMP (100 μM) (Figure 6). The heart rate reflects the balance between sympathetic and parasympathetic nervous system (Swynghedauw et al., 1997). The vagal tone may decrease the heart rate and the antagonist atropine can accelerate the heart rate. It has been reported that mouse has no vagal tone, and atropine injection does not increase the heart rate as it does in humans (Swynghedauw et al., 1997). The basal heart rate was not increased on the M2 mAChR knockout mice (Stengel et al., 2000; Birdsall et al., 2001). We found, however, that M2 MO can significantly increase the basal heart rate in zebrafish. This novel finding in zebrafish further suggests species-dependent M2 mAChR mediated regulation of the basal heart rate. Besides, after fluorescein tagged MO injection (8 ng per embryo), the fluoresence in embryo can be seen for one month (Mark Chen, personal communication). Thus, the MO is much more stable than dsRNA in zebrafish embryo. It is also easy to perform double knockdowns with MOs. This will help to solve the arduous problem of gene redundancy and non-specific agonists or antagonists.
We have also cloned the zebrafish M5 mAChR gene. However, the M5 mAChR transcripts were not detectable in the zebrafish heart by the whole mount in situ hybridization. The in vivo pharmacological experiments, RNAi or MO knockdown studies showed M5 mAChR had no effect on CCh-induced bradycardia and basal heart rate at 72 h.p.f. embryos (data not shown). M5 mAChR lacked functional significance in the regulation of heart rate.
Our study represents the first paradigm of application of zebrafish in mAChR pharmacology research. Our approaches can be applied to study other receptors and drugs involved in the regulation of heart rate.
Accession number GenBank accession number of zebrafish M2 mAChR: AY039653.
Acknowledgments
We are grateful to Dr Chi-Yao Chang for the zebrafish genomic DNA library and Mr Jyh-I Wang for animal care and technical assistance. The authors wish to thank Ms Ching-Chun Lin and Dia-Nan Lin for their help with DNA sequencing and Drs Ching-Feng Weng, Eileen Jea Chien and Shue-Fen Luo for reviewing the manuscript and helpful discussion. This work was supported by a grant from the National Science Council of Taiwan (NSC-87-2311-B-001-036-B24, NSC-88-2611-B-001-005-B24 and NSC-89-2311-B-001-096-B24).
Abbreviations
- AF-DX 116
[[2-[diethylamino)methyl]-1-piperidinyl]acetyl]-5,11-dihydro-6H-pyrido[2,3b][1,4]-benzodiazepine-6-one
- 4-DAMP
4-diphenylacetoxy-N-methylpiperidine methiodide
- dsRNA
double-stranded RNA
- p-F-HHSiD
p-fluoro hexahydro-sila-difenidol hydrochloride
- mAChR
muscarinic acetylcholine receptor
- MO
morpholino-modified antisense oligonucleotide
- [3H]NMS
[3H]N-methylscopolamine; RACE, rapid amplification of cDNA ends
- RNAi
RNA interference
- tropicamide
N-ethyl-3-hydroxy-2-phenyl-N-(pyridinylmethyl)
References
- ASHKENAZI A., PERALTA E.G.Muscarinic acetylcholine receptors Handbook of receptors and channels 19941Florida: Boca Raton; 1–27.ed. Peroutka, S.J. pp [Google Scholar]
- ALEXANDER S.P.H., PETERS J.A. Trends in Pharmacological Sciences Receptor and Ion Channel Nomenclature Supplement 1999Cambridge: Elsevier Trends Journals; 10th edn [Google Scholar]
- BAKER K., WARREN K.S., YELLEN G., FISHMAN M.C. Defective ‘pacemaker' current (Ih) in a zebrafish mutant with a slow heart rate. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4554–4559. doi: 10.1073/pnas.94.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIRDSALL N.J.M., BUCKLEY N.J., CAUFIELD M.P., HAMMER R., KILBINGER H.J., LAMBRECHT G., MUTSCHLER E., NATHANSON N.M., SCHWARZ R.D.Muscarinic acetylcholine receptors The IUPHAR Compendium of Receptor Characterization and Classification 1998London: IUPHAR Media; 36–45.ed. Girdlestone, D. pp [Google Scholar]
- BIRDSALL N.J.M., NATHANSON N.M., SCHWARZ R.D. Muscarinic receptors: it's a knockout. Trends Pharmacol. Sci. 2001;22:215–219. [Google Scholar]
- BONNER T.I., YOUNG A.C., BRANN M.R., BUCKLEY N.J. Cloning and expression of the human and rat M5 muscarinic acetylcholine receptor genes. Neuron. 1988;1:403–410. doi: 10.1016/0896-6273(88)90190-0. [DOI] [PubMed] [Google Scholar]
- CAULFIELD M.P. Muscarinic receptors – characterization, coupling and function. Pharmacol. Ther. 1993;58:319–379. doi: 10.1016/0163-7258(93)90027-b. [DOI] [PubMed] [Google Scholar]
- CAULFIELD M.P., BIRDSALL N.J. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 1998;50:279–290. [PubMed] [Google Scholar]
- CHEN J.N., FISHMAN M.C. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- CHENG Y., PRUSOFF W.H. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- COLECRAFT H.M., EGAMINO J.P., SHARMA V.K., SHEU S.S. Signaling mechanisms underlying muscarinic receptor-mediated increase in contraction rate in cultured heart cells. J. Biol. Chem. 1998;273:32158–32166. doi: 10.1074/jbc.273.48.32158. [DOI] [PubMed] [Google Scholar]
- CREASON S., TIETJE K.M., NATHANSON N.M. Isolation and functional characterization of the chick M5 muscarinic acetylcholine receptor gene. J. Neurochem. 2000;74:882–885. doi: 10.1046/j.1471-4159.2000.740882.x. [DOI] [PubMed] [Google Scholar]
- DOOLEY K., ZON L.I. Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- EGLEN R.M., NAHORSKI S.R. The muscarinic M5 receptor: a silent or emerging subtype. Br. J. Pharmacol. 2000;130:13–21. doi: 10.1038/sj.bjp.0703276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLIS J., SEIDENBERG M. Site-directed mutagenesis implicates a threonine residue in TM6 in the subtype selectivities of UH-AH 37 and pirenzepine at muscarinic receptors. Pharmacology. 2000;61:62–69. doi: 10.1159/000028382. [DOI] [PubMed] [Google Scholar]
- FELDER C.C. Muscarinic acetylcholine receptors: signal transduction through multiple effectors. FASEB J. 1995;9:619–625. [PubMed] [Google Scholar]
- FRITSCHE R., SCHWERTE T., PELSTER B. Nitric oxide and vascular reactivity in developing zebrafish, Danio rerio. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000;279:R2200–R2207. doi: 10.1152/ajpregu.2000.279.6.R2200. [DOI] [PubMed] [Google Scholar]
- GALLO M.P., ALLOATTI G., EVA C., OBERTO A., LEVI R.C. M1 muscarinic receptors increase calcium current and phosphoinositide turnover in guinea-pig ventricular cardiocytes. J. Physiol. 1993;471:41–60. doi: 10.1113/jphysiol.1993.sp019890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GURA T. A silence that speaks volumes. Nature. 2000;404:804–808. doi: 10.1038/35009245. [DOI] [PubMed] [Google Scholar]
- HANNEMAN E., TREVARROW B., METCALFE W.K., KIMMEL C.B., WESTERFIELD M. Segmental pattern of development of the hindbrain and spinal cord of the zebrafish embryo. Development. 1988;103:49–58. doi: 10.1242/dev.103.1.49. [DOI] [PubMed] [Google Scholar]
- HEDGE S.S., CHOPPIN A., BONHAUS D., BRIAUD S., LOEB M., MOY T.M., LOURY D., EGLEN R.M. Functional role of M2 and M3 muscarinic receptors in the urinary bladder of rats in vitro and in vivo. Br. J. Pharmacol. 1997;120:1409–1418. doi: 10.1038/sj.bjp.0701048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HWANG S.P., TSOU M.F., LIN Y.C., LIU C.H. The zebrafish BMP4 gene. sequence analysis and expression pattern during embryonic development. DNA Cell Biol. 1997;16:1003–1011. doi: 10.1089/dna.1997.16.1003. [DOI] [PubMed] [Google Scholar]
- KELLY G.M., VANDERBELD B., KRAWETZ R., MANGOS S. Differential distribution of the G protein gamma3 subunit in the developing zebrafish nervous system. Int. J. Dev. Neurosci. 2001;19:455–467. doi: 10.1016/s0736-5748(01)00002-8. [DOI] [PubMed] [Google Scholar]
- KIMMEL C.B., BALLARD W.W., KIMMEL S.R., ULLMANN B., SCHILLING T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- KUBO T., BUJO H., AKIBA I., NAKAI J., MISHINA M., NUMA S. Location of a region of the muscarinic acetylcholine receptor involved in selective effector coupling. FEBS Lett. 1988;241:119–125. doi: 10.1016/0014-5793(88)81043-3. [DOI] [PubMed] [Google Scholar]
- LAI J., NUNAN L., WAITE S.L., MA S.W., BLOOM J.W., ROESKE W.R., YAMAMURA H.I. Chimeric M1/M2 muscarinic receptors: correlation of ligand selectivity and functional coupling with structural modifications. J. Pharmacol. Exp. Ther. 1992;262:173–180. [PubMed] [Google Scholar]
- LETCHER J. Intracelomic use of tricaine methanesulfonate for anesthesia of bullfrogs (Rana catesbeniana) and leopard frogs (Rana pipiens) Zoo Biol. 1992;11:243–251. [Google Scholar]
- LI Y.X., FARRELL M.J., LIU R., MOHANTY N., KIRKBY M.L. Double-stranded RNA injection produces null phenotypes in zebrafish. Dev. Biol. 2000;217:394–405. doi: 10.1006/dbio.1999.9540. [DOI] [PubMed] [Google Scholar]
- LIAO C.F., THEMMEN A.P., JOHO R., BARBERIS C., BIRNBAUMER M., BIRNBAUMER L. Molecular cloning and expression of a fifth muscarinic acetylcholine receptor. J. Biol. Chem. 1989;264:7328–7337. [PubMed] [Google Scholar]
- MCKINNON L.A., NATHANSON N.M. Tissue-specific regulation of muscarinic acetylcholine receptor expression during embryonic development. J. Biol. Chem. 1995;270:20636–20642. doi: 10.1074/jbc.270.35.20636. [DOI] [PubMed] [Google Scholar]
- NASEVICIUS A., EKKER S.C. Effective targeted gene ‘knockdown' in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- NATHANSON N.M. Molecular properties of the muscarinic acetylcholine receptor. Annu. Rev. Neurosci. 1987;10:195–236. doi: 10.1146/annurev.ne.10.030187.001211. [DOI] [PubMed] [Google Scholar]
- NOGRADY T., KESHMIRIAN J. Rotifer neuropharmacology–II. Synergistic effect of acetylcholine on local anesthetic activity in Brachionus calyciflorus (Rotifera, Aschelminthes) Comp. Biochem. Physiol. C. 1986;83:339–344. doi: 10.1016/0742-8413(86)90133-7. [DOI] [PubMed] [Google Scholar]
- NUSSLEIN-VOLHARD C. Of flies and fishes. Science. 1994;266:572–574. doi: 10.1126/science.7939708. [DOI] [PubMed] [Google Scholar]
- OATES A.C., BRUCE A.E., HO R.K. Too much interference: injection of double-stranded RNA has nonspecific effects in the zebrafish embryo. Dev. Biol. 2000;224:20–28. doi: 10.1006/dbio.2000.9761. [DOI] [PubMed] [Google Scholar]
- PERALTA E.G., ASHKENAZI A., WINDLOW J.W., SMITH D.H., RAMACHANDRAN J., CAPON D.J. Distinct primary structures, ligand-binding properties and tissue-specific expression of four human muscarinic acetylcholine receptors. EMBO J. 1987a;6:3923–3929. doi: 10.1002/j.1460-2075.1987.tb02733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERALTA E.G., WINSLOW J.W., PETERSON G.L., SMITH D.H., ASHKENAZI A., RAMACHANDRAN J., SCHIMERLIK M.I., CAPON D.J. Primary structure and biochemical properties of an M2 muscarinic receptor. Science. 1987b;236:600–605. doi: 10.1126/science.3107123. [DOI] [PubMed] [Google Scholar]
- PRESTON E., COURTICE G.P. Physiological correlates of vagal nerve innervation in lower vertebrates. Am. J. Physiol. 1995;268:R1249–R1256. doi: 10.1152/ajpregu.1995.268.5.R1249. [DOI] [PubMed] [Google Scholar]
- ROSS L.S., PARRETT T., EASTER S.S., JR Axonogenesis and morphogenesis in the embryonic zebrafish brain. J. Neurosci. 1992;12:467–482. doi: 10.1523/JNEUROSCI.12-02-00467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHI H., WANG H., WANG Z. Identification and characterization of multiple subtypes of muscarinic acetylcholine receptors and their physiological functions in canine hearts. Mol. Pharmacol. 1999;55:497–507. [PubMed] [Google Scholar]
- STENGEL P.W., GOMEZA J., WESS J., COHEN M.L. M2 and M4 receptor knockout mice: muscarinic receptor function in cardiac and smooth muscle in vitro. J. Pharmacol. Exp. Ther. 2000;292:877–885. [PubMed] [Google Scholar]
- SUMMERTON J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- SWYNGHEDAUW B., JASSON S., CLAIRAMBAULT J., CHEVALIER B., HEYMES C., MEDIGUE C., CARRE F., MANSIER P. Myocardial determinants in regulation of the heart rate. J. Mol. Med. 1997;75:860–866. doi: 10.1007/s001090050177. [DOI] [PubMed] [Google Scholar]
- TIETJE K.M., NATHANSON N.M. Embryonic chick heart expresses multiple muscarinic acetylcholine receptor subtypes. Isolation and characterization of a gene encoding a novel M2 muscarinic acetylcholine receptor with high affinity for pirenzepine. J. Biol. Chem. 1991;266:17382–17387. [PubMed] [Google Scholar]
- WANG H., HAN H., ZHANG L., SHI H., SCHRAM G., NATTEL S., WANG Z. Expression of multiple subtypes of muscarinic receptors and cellular distribution in the human heart. Mol. Pharmacol. 2001;59:1029–1036. doi: 10.1124/mol.59.5.1029. [DOI] [PubMed] [Google Scholar]
- WARGELIUS A., ELLINGSEN S., FJOSE A. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 1999;263:156–161. doi: 10.1006/bbrc.1999.1343. [DOI] [PubMed] [Google Scholar]
- WESS J. Molecular biology of muscarinic acetylcholine receptors. Crit. Rev. Neurobiol. 1996;10:69–99. doi: 10.1615/critrevneurobiol.v10.i1.40. [DOI] [PubMed] [Google Scholar]
- WESTERFIELD M.Recipes The Zebrafish Book 1994University of Oregon: Eugene, U.S.A; pp 10.16. [Google Scholar]
- WIANNY F., ZERNICKA-GOETZ M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell. Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- ZERNICKA-GOETZ M. Jumping the gun on mouse gene expression. Nature. 2000;405:733. doi: 10.1038/35015787. [DOI] [PubMed] [Google Scholar]


