Abstract
Transcriptional control of the human β2 adrenergic receptor gene (ADRB2) predominantly resides within a 549 base pair region immediately 5′ to the start of translation. Within this region, four naturally occurring polymorphisms, −468 C→G, −367 T→C, −47 T→C, and −20 T→C, have been identified.
To determine the individual site and haplotype effects of these polymorphisms, we generated 16 luciferase-based mutant constructs which were transiently transfected into HEK293 cells, and measured ADRB2 promoter-driven luciferase activity.
Two of the 16 mutant constructs, GCCT (−468G, −367C, −47C, −20T) and CTCT, showed a highly significant 3 fold decrease in luciferase induction relative to the reference CTTT. These haplotype effects could not be accounted for by the separate and additive effects of each site.
These findings indicate that promoter polymorphisms interact to significantly alter β2 adrenergic receptor expression, and should be examined further for their association with disease-related phenotypes.
Keywords: ADRB2; polymorphisms; promoter; human β2 adrenergic receptor; beta2-adrenergic receptor; transfection; luciferase; HEK293 cells, haplotype
Introduction
Adrenergic receptors are members of the superfamily of seven transmembrane-spanning domain receptors that carry out signal transduction. They are an integral part of the sympathetic nervous system and have been shown to play important roles in cardiovascular, respiratory and metabolic functions (Kobilka, 1991). Adrenergic receptors were first identified in pharmacological studies and have been subdivided into two major types, α and β, on the basis of agonist-mediated responses, with subsequent classification into subtypes based on differential tissue localization. To date, nine different subtypes have been cloned and pharmacologically characterized, including three β subtypes (Guimarães & Moura, 2001).
The human β2 adrenergic receptor gene (ADRB2) is encoded by a single intronless gene consisting of 1239 nucleotides, located on the distal portion of the long arm of chromosome 5 (5q32–q34). This chromosomal region has been shown in linkage studies to be significantly related to blood pressure control (Krushkal et al., 1999). Positional candidate gene studies have identified the ADRB2 gene as a susceptibility locus for human hypertension (Bray et al., 2000). In addition, genetic association studies have also implicated ADRB2 polymorphisms in other disease phenotypes (Turki et al., 1995; Yamada et al., 1999). Studies of deletion fragments of a 1470 bp sequence 5′ to the start of translation indicate that the majority of promoter activity resides within a 549 bp fragment immediately upstream of the ATG start codon (Scott et al., 1999). This fragment not only contains important known and predicted regulatory elements (Kobilka et al., 1987), but also multiple genetic polymorphisms: −468 C→G, −367 T→C, −47 T→C, and −20 T→C. This study was undertaken to delineate the functional significance of these promoter polymorphisms individually and as haplotypes, using luciferase-based expression studies. We report here highly significant alterations in luciferase expression among the 16 haplotypes, indicating the potential for variations in ADRB2 expression depending on the promoter haplotypes.
Methods
Recombinant DNA constructs
A 570 bp fragment of the 5′ regulatory region of the human ADRB2 was obtained by PCR using human genomic DNA as the template. Primers were designed, based on GenBank accession number M15169, to include in the product the four polymorphic sites at −20, −47, −367 and −468 (relative to the ATG start codon), as well as the restriction sites XhoI and MluI for direct subcloning into the promoterless, enhancerless firefly luciferase expression vector pGL3-Basic (Promega).
Construction of mutant reporter constructs
In vitro site-directed mutagenesis was used to generate the 24 (=16) possible combinations of the four polymorphic sites from the recombinant ADRB2 promoter fragment cloned into pGL3-Basic. Two oligonucleotide primers, a mutagenic primer, designed to mutate the desired base pair, and a selection primer, which mutates a unique restriction site in the plasmid backbone, were simultaneously annealed to denatured template DNA. The mutant DNA strand was synthesized using T4 DNA polymerase and re-ligated using T4 DNA ligase. Mutants were initially transformed into the DNA mismatch repair-deficient strain of E. coli BMH71-18 mutS. Mutant plasmid DNA was isolated and restriction digested at the site mutated by the selection primer to linearize parental and non-mutant plasmid DNA. A second transformation into XL1-Blue Supercompetent E. coli cells (Stratagene) allowed amplification of the mutant plasmids (Deng & Nickoloff, 1992). Confirmation of all sequences was obtained by dye terminator chemistry (Big-Dye™, Applied Biosystems) and an Applied Biosystems 3700 sequencer.
Transfection of HEK-293 cells
HEK293 cells cultured to passage number ⩽45 were seeded into 24-well plates at a density of 50,000 cells per well in DMEM, supplemented with 10% foetal calf serum, and grown to approximately 75% confluency. Cells were transfected, using liposomal mediated DNA transfer (LipofectAMINE™, Gibco BRL), with 500 ng of ADRB2 mutant plasmid DNA and co-transfected with 10 ng per well of pRL-CMV plasmid DNA, a Renilla based luciferase construct (Promega). The medium was aspirated after 48 h and the cells were washed with PBS once, and lysed with passive lysis buffer (Dual Luciferase® Reporter Assay System, Promega). Firefly and Renilla luciferase activity of cell lysates were measured using the Monolight™ 3010 luminometer (Pharmingen). Firefly luciferase activity was adjusted to Renilla luciferase activity to control for variations in cell viability and transfection efficiency.
Statistical analysis
Because standardized luciferase activity was not assumed to be normally distributed, non-parametric statistical methods were used throughout. The mean fold-induction of luciferase activity among the 16 haplotypes were compared using the Kruskal-Wallis test. Pair-wise comparisons between individual haplotypes were carried out using the Wilcoxon rank sum test. To help control for post-hoc multiple comparisons when analysing the 16 haplotypes, a P-value of 0.0065 was used to assess statistical significance. Luciferase activity between alleles at each polymorphic site were compared using the Wilcoxon rank sum test.
Results
Plasmid constructs were named according to the nucleotide at each of the four sites, beginning with −468 and proceeding in a 3′ direction to the −20 site. The original fragment obtained from PCR of human genomic DNA, and cloned into pGL3-basic yielded two haplotypes, pCTTT (−468C, −367T, −47T, −20T) and pGCCC (−468G, −367C, −47C, −20C). Both plasmids were used to generate the remaining 14 mutant constructs used in the luciferase assays (see Methods). Analysis of the contribution of the individual loci to luciferase induction indicated a significant difference between the −47C allele compared to −47T allele (see Table 1). Figure 1 shows the relative activity of the 16 promoter constructs normalized to the pCTTT construct. The non-parametric analysis of variance of luciferase induction among the 16 haplotypes revealed significant variation in luciferase expression (P=0.0001). The constructs pGCCT and pCTCT showed an approximate 3 fold decrease in luciferase expression relative to pCTTT (P<0.004). Two other constructs, pCTTC and pCCTC showed a statistically significant increase in luciferase expression of approximately 30% (P<0.004) (see Figure 1).
Table 1.
Contribution of separate ADRB2 5′ polymorphisms to mean fold-induction of luciferase activity
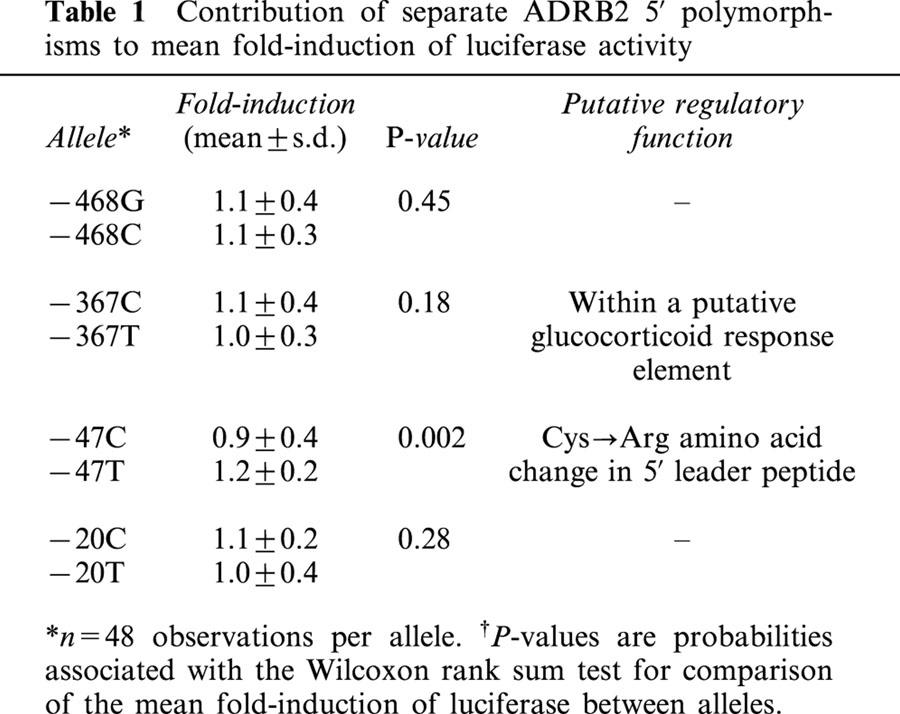
Figure 1.
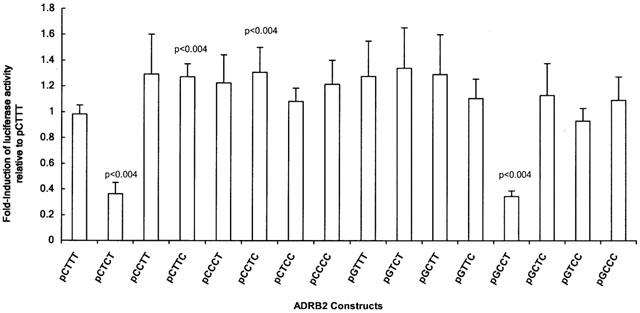
Relative promoter activity of the human ADRB2 gene in transiently transfected HEK293 cells. Constructs are designated ‘p' for ‘plasmid', and named according to the nucleotide starting at position −468 and proceeding in a 3′ direction to −20. For each transfection, firefly luciferase activity was adjusted to Renilla luciferase activity to control for transfection efficiency. Fold-induction of each transfection is the adjusted luciferase activity standardized to the median value of the pCTTT construct for each experiment. Each bar represents the mean±standard deviation of n=6 observations derived from triplicate transfections of two independent experiments. P-values are probabilities associated with the Wilcoxon Rank-Sum Test for comparisons with pCTTT.
Discussion
The data presented here show significant effects of ADRB2 haplotype variation on promoter activity that cannot be attributed to the individual and additive effects of each polymorphic site. Post-hoc analyses identified four haplotypes of ADRB2 promoter polymorphisms which significantly impact ADRB2 expression. Two of these, GCCT and CTCT exhibited a highly significant three-fold decrease in luciferase expression relative to CTTT. Published GenBank sequence data (Accession number M15169) shows the nucleotides G, C, C, and T at −468, −367, −47 and −20 respectively, suggesting that this haplotype exists in human populations. In on-going population studies, both the CTCT and GCCT haplotypes were found to exist. Of 296 non-Hispanic white individuals that have been genotyped to date, two individuals carried the GCCT haplotype and one carried the CTCT haplotype. For purposes of this study, the CTTT haplotype was chosen as the reference sequence since the original PCR product derived from human genomic DNA yielded two distinct haplotypes, CTTT and GCCC. In addition, population studies of ADRB2 promoter haplotypes indicate that these two haplotypes are the most common in human populations (Drysdale et al., 2000). Standardization to the GCCC haplotype did not alter the main finding that the GCCT and CTCT haplotypes resulted in a highly significant 3 fold decrease in luciferase expression.
Exploratory analysis of the genotypic contribution to luciferase induction revealed a highly significant decrease in luciferase induction due to the presence of the C variant at the −47 site compared to the T (P=0.002). The −47 site is within a short open reading frame extending from nucleotide −102 to −42, which encodes a 19 amino acid polypeptide thought to modulate ADRB2 mRNA translation (Parola & Kobilka, 1994). The C variant at nucleotide −47 introduces a Cys→Arg amino acid change which has been shown to downregulate receptor expression in both 125Iodine radio-ligand binding experiments and luciferase assays (Mcgraw et al., 1998). Of speculative importance was the −367 site which lies within a putative glucocorticoid response element (GRE) located at positions −379 to −365 in the 5′ flanking region of the rat ADRB2 gene (Cornett et al., 1998). GREs are known to increase receptor density by increasing the rate of ADRB2 transcription in a variety of tissues and cell types. However analysis of indivdual contribution by this site did not attain statistical significance with respect to luciferase induction.
Two other haplotypes, CTTC and CCTC, showed a statistically significant increase in luciferase induction of approximately 30% relative to CTTT (P<0.004). Further studies would be necessary to determine the physiological impact of these haplotypes on ADRB2 receptor expression and their potential to influence disease phenotypes. Based on the observed effect of the haplotype on luciferase induction, these 16 haplotypes may be subdivided into three groups with regard to ADRB2 expression, downregulators (CTCT and GCCT), upregulators (CTTC and CCTC), and no significant change (the remaining 12 haplotypes). Post-hoc analysis of variance of the mean fold induction of these three groups using the Kruskal–Wallis test for significance indicate a highly significant difference among these groups (P=0.0001).
These findings demonstrate that polymorphisms in the 5′ regulatory region of ADRB2 significantly alter ADRB2 expression, the degree and direction of the alteration being haplotype dependent with a significant impact attributable to the −47C variant. Assessment of the physiological consequences of these haplotypes requires further study in 125Iodine radio-ligand binding assays to determine alterations in receptor density, as well as genetically modified animal models to determine phenotypic changes. Targeted disruption of ADRB2 in mouse models have been shown to produce a hypertensive phenotype associated with exercise-induced stress, as well as alterations in both vascular tone and energy metabolism (Chruscinski et al., 1999). While experimental alterations in ADRB2 expression in animal models may provide insight into physiological consequences, the interpretation of such studies in relation to human disease phenotypes may be limited and should be interpreted with caution. Association studies, particularly involving high risk population or patient groups, would also be necessary to assess the physiological impact of ADRB2 promoter haplotypes and its potential effect on disease progression.
The precise mechanism by which mutations in this region may alter receptor expression is speculative at this point. Regardless of the choice of standard, it is clear from the data that both CTCT and GCCT significantly downregulate ADRB2 expression and as such it is conceivable that both haplotypes may contribute to interindividual blood pressure variation and hypertension (Bray et al., 2000), nocturnal asthma (Turki et al., 1995), and obesity and higher triglyceride levels (Yamada et al., 1999). The hypothesis that genetic polymorphisms might predispose an individual to disease, or alter the clinical progression of disease, is important not only from a prognostic standpoint but also in the context of individual responses to treatment. This study provides insight into the mechanism or pathway involved in disease progression and hence a mechanism-based rationale for predicting efficacy and toxicity.
Acknowledgments
This work was supported by grants from the National Heart Lung and Blood Institute.
Abbreviations
- ADRB2
beta2-adrenergic receptor
- bp
base pairs
- HEK
human embryonic kidney
- PCR
polymerase chain reaction
References
- BRAY M.S., KRUSHKAL J., LI L., FERREL R., KARDIA S., SING C.F., TURNER S.T., BOERWINKLE E. Positional genomic analysis identifies the β2-adrenergic receptor gene as a susceptibility locus for human hypertension. Circulation. 2000;101:2877–2882. doi: 10.1161/01.cir.101.25.2877. [DOI] [PubMed] [Google Scholar]
- CHRUSCINSKI A.J., ROHRER D.K., SCHAUBLE E., DESAI K.H., BERNSTEIN D., KOBILKA B.K. Targeted disruption of the β2 adrenergic receptor gene. J. Biol. Chem. 1999;274:16694–16700. doi: 10.1074/jbc.274.24.16694. [DOI] [PubMed] [Google Scholar]
- CORNETT L.E., HILLER F.C., JACOBI S.E., CAO W., MCGRAW D.W. Identification of a glucocorticoid response element in the rat β2-adrenergic receptor gene. Mol. Pharmacol. 1998;54:1016–1023. doi: 10.1124/mol.54.6.1016. [DOI] [PubMed] [Google Scholar]
- DENG W.P., NICKOLOFF J.A. Site-directed mutagenesis of virtually any plasmid by elimination of unique sites. Anal. Biochem. 1992;200:81. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- DRYSDALE C.M., MCGRAW D.W., STACK C.B., STEPHENS J.C., JUDSON R.S., NANDALBAN K., ARNOLD K., RUANO G., LIGGETT S.B. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUIMARÃES S., MOURA D. Vascular adrenoceptors: an update. Pharmacol. Rev. 2001;53:319–356. [PubMed] [Google Scholar]
- KOBILKA B. Molecular and cellular biology of adrenergic receptors. Trends Cardiovasc. Med. 1991;1:189–194. doi: 10.1016/1050-1738(91)90036-E. [DOI] [PubMed] [Google Scholar]
- KOBILKA B.K., FRIELLE T., DOHLMAN H.G., BOLANOWSKI M.A., DIXON R.A.F., KELLER P., CARON M.G., LEFKOWITZ R.J. Delineation of the intronless nature of the genes for the human and hamster β2-adrenergic receptor and their putative promoter regions. J. Biol. Chem. 1987;262:7321–7327. [PubMed] [Google Scholar]
- KRUSHKAL J., FERREL R., MOCKRIN S., TURNER S., SING C., BOERWINKLE E. Genome-wide linkage analyses of systolic blood pressure using highly discordant siblings. Circulation. 1999;99:1407–1410. doi: 10.1161/01.cir.99.11.1407. [DOI] [PubMed] [Google Scholar]
- MCGRAW D.W., FORBES S.L., KRAMER L.A., LIGGETT S.B. Polymorphisms of the 5′ leader cistron of the human β2-adrenergic receptor regulate receptor expression. J. Clin. Invest. 1998;102:1927–1932. doi: 10.1172/JCI4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAROLA A.L., KOBILKA B.K. The peptide product of a 5′ leader cistron in the beta2-adrenergic receptor mRNA inhibits receptor synthesis. J. Biol. Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- SCOTT M.G.H., SWAN C., WHEATLEY A.P., HALL I.P. Identification of novel polymorphisms within the promoter region of the human β2 adrenergic receptor gene. Br. J. Pharmacol. 1999;126:841–844. doi: 10.1038/sj.bjp.0702385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURKI J., PAK J., GREEN S.A., MARTIN R.J., LIGGETT S.B. Genetic polymorphisms of the β2-adrenergic receptor in nocturnal and nonnocturnal asthma: evidence that Gly16 correlates with the nocturnal phenotype. J. Clin. Invest. 1995;95:1635–1641. doi: 10.1172/JCI117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMADA K., ISHIYAMA-SHIGEMOTO S., ICHIKAWA F., YUAN X., KOYANAGI A., KOYAMA W., NONAKA K. Polymorphism in the 5′-leader cistron of the β2-adrenergic receptor gene associated with obesity and Type 2 diabetes. J. Clin. Endocrinol. Metab. 1999;84:1754–1757. doi: 10.1210/jcem.84.5.5822. [DOI] [PubMed] [Google Scholar]


